Abstract
Background:
Recent large scale genomic profiling and clinical correlation studies of chronic lymphocytic leukemia (CLL) identified clinically significant gene mutations with important clinical prognostic and potential therapeutic implications. Integration of individual clonal and subclonal gene mutation information with existing prognostic markers such as somatic hypermutation and chromosomal aberrations provide comprehensive risk stratification for CLL patients. In addition, mutations in BTK and PLCG2 are associated with resistance to targeted therapies using BTK inhibitors (ibrutinib). We implemented routine clinical testing for CLL patients. We describe our initial experience with EndCLL Assay V1, a high yield targeted NGS assay for routine clinical testing of clinically significant mutations in CLL and B-cell neoplasms.
Materials and Methods:
DNA from total of 376 blood or bone marrow samples containing >10% CLL cells were tested during the panel validation and subsequent routine clinical testing. Targeted sequencing was performed on 29 genes reported to be mutated in CLL and other B-cell neoplasms. These include ATM, BIRC3, BTK, CALR, CARD11, CD79A, CD79B, CHD2, CSMD3, CXCR4, DDX3X, EZH2, FAT1, FBXW7, KLHL6, LRP1B, MAPK1, MUC2, MYD88, NOTCH1, PLCG2, PLEKHG5, POT1, SF3B1, SPEN, TGM7, TP53, XPO1 and ZMYM3. Panel design included a combination of hotspots, limited exons and all exon coverage as required to detect known clinically significant mutations in the genes targeted in the panel. A total of 500 ng input DNA was used to prepare sequencing libraries using Agilent Haloplex HS chemistry, which incorporates unique molecular barcodes in each DNA molecule being sequenced. Sequencing was performed on MiSeq sequencers (Illumina) followed by data analysis on Agilent SureCall v3.0. The average per base coverage depth ranged from 1500x to 4000x based on the number of samples per run. The analytical sensitivity of the assay was determined to be at 2% to 5% for reliable and reproducible detection of mutations depending on the sequencing coverage and percentage of mutant reads.
Results:
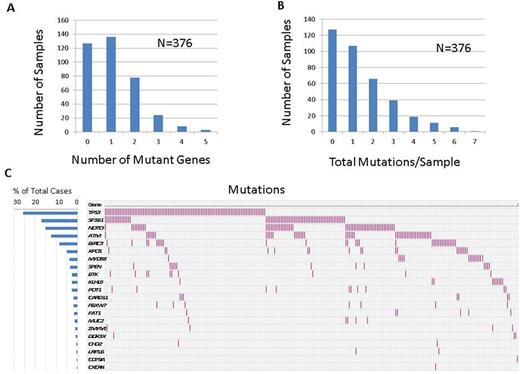
For cases with available information, the average age was 63.3 years (range: 22-87, median: 64). The male:female ratio was 2:1 and the ratio of cases with unmutated:mutated IGHV was 1.2:1. Positive FISH results for -11q, +12, -13q and -17p were detected in 18%, 23%, 59% and 15% cases respectively. Out of 376 samples tested, 249 (66.2%) showed at least one mutation. Average number of genes mutated per sample was 1.1 (range: 0-5, median: 1) and the average number of total mutations per sample was 1.4 (range 0-7, median: 1). Top 10 mutated genes included TP53, SF3B1, NOTCH1, ATM, BIRC3, XPO1, MYD88, SPEN, BTK and KLHL6 (Figure 1). Patterns of co-mutations and mutual exclusivity were observed (Figure 1). Top 10 mutations (hotspots) are listed in the table. Mutations in 3'UTR of NOTCH1 were detected in 5/376 (%) cases. Resistance mutations in BTK codon 481 were detected in 10 patients on ibrutinib treatment, including 5 cases with >1 mutations and 2 cases with subclonal mutations indicating early treatment resistant clone. Clinical findings in all 10 cases were consistent with proven or emerging treatment resistance. No mutations were detected in PLCG2 (codons 646-685). One patient initially on ibrutinib and on idelalisib treatment showed acquisition of 17p deletion and a subclonal TP53 mutation at the time of ibrutinib resistance and progression to an aggressive disease course. Use of molecular barcodes allowed efficient detection of low level subclonal mutations. Correlations with a comprehensive set of clinicopathologic parameters as well as treatment outcomes are in progress.
Conclusion:
The targeted EndCLL Mutation Assay V1 allowed a high yield detection of clinically significant mutations in 66% of the patient samples using a clinically sustainable panel design accommodating up to 24 multiplexed samples in a single sequencing run. Use of molecular barcodes allows removal of PCR duplicates and enables accurate calling of subclonal mutations for prognostication. This was evident in simultaneous detection of subclonal BTK mutations allows early detection of emerging ibrutinib resistant clone while obtaining additional prognostic information. Overall, the EndCLL Assay V1 is a valuable addition for CLL patient care.
Jain:Incyte: Research Funding; BMS: Research Funding; ADC Therapeutics: Consultancy, Honoraria, Research Funding; Celgene: Research Funding; Novimmune: Consultancy, Honoraria; Pfizer: Consultancy, Honoraria, Research Funding; Seattle Genetics: Research Funding; Abbvie: Research Funding; Infinity: Research Funding; Novartis: Consultancy, Honoraria; Genentech: Research Funding; Pharmacyclics: Consultancy, Honoraria, Research Funding; Servier: Consultancy, Honoraria. Wierda:Gilead: Research Funding; Genentech: Research Funding; Acerta: Research Funding; Novartis: Research Funding; Abbvie: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal