Abstract
BACKGROUND
Chronic myeloid leukemia in chronic phase (CML CP) is successfully treated with tyrosine kinase inhibitors (TKIs) but a proportion of patients show primary or secondary resistance to TKI therapy. BCR-ABL1 mutations are the primary cause for secondary resistance to TKI therapy, but the causes for primary resistance are poorly known. Putative variants in genes affecting drug absorption, distribution, metabolism and excretion (ADME) have not been comprehensively analyzed. We set out to screen CML CP patients who either had primary resistance to TKI therapy or suboptimal response with large ADME gene panel to examine if these genes play a role in poor TKI responses.
METHODS
We selected samples from 21 CML CP patients treated in our institute who either had primary resistance (N=6) or responded suboptimallyto TKI treatment (n=15) but did not have BCR-ABL1 mutations. DNA samples from the time of diagnosis were sequenced with massively parallel sequencing using a comprehensive custom ADME gene panel covering exons and introns of 382 genes. Over 200 healthy volunteers who had been sequenced with the same gene panel served as population controls. Loss-of-function and novel variants were validated using capillary sequencing.We tested the differences of variant allele frequencies between CML patients and healthy controls by performing the Cochran-Armitage trend test using additive coding (0,1,2) for genotypes homozygous for the major allele, heterozygous, and homozygous for the minor allele, respectively. CML patients with optimal response (n=24) were used as a control group.
RESULTS
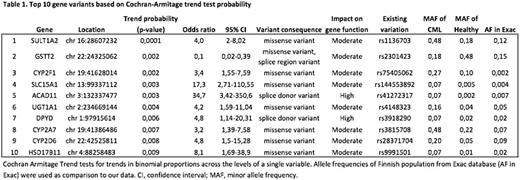
Sequencing of 21 poor responding CML CP patients revealed a total of 658 nonsynonymousgene variants. Majority of the variants (94%) were missense variants whose effects on gene function are uncertain. Eigthvariants were previously unreported. 18 variants were predicted to lead to a complete loss-of-function of the genes affected. Principal component analysis did not reveal clear global differences between the CML patients and healthy controls in the frequencies of ADME gene variants. However, the Cochran-Armitage trend test showed a number of variants whose frequencies in CML samples were significantly different from the healthy controls (table 1). Two of the loss-of-function variants were in the top 10 variants, whose frequencies differed between CML patients and healthy controls. ACAD11 rs41272317 variant was observed in 14.2% of patients whereas its frequency in healthy controls was 0.5% (p=0.003). Similarly DPYD rs3918290 was observed in 14.2% of patients and 3.3% of healthy controls (p=0.006). ACAD11 encodes an acyl-CoA dehydrogenase enzyme with a preference for carbon chain lengths between 20 and 26, and has been shown to be an important metabolic target for p53 pro-survival function. DPYD codesdihydropyrimidine dehydrogenase, which is involved in the metabolism ofpyrimidines and the anticancer agent 5-fluorouracil. Sanger sequencing confirmed both variants in patient samples. From the top10 missense variants, SLC15A1 rs144553892 variant was significantly more often observed in poor responding CML patients than in healthy controls (14.2 and 0.9%, respectively, p=0.003). SLC15A1 is a solute carrier family 15 member 1 which has a known function as a co-transporter foroligopeptides. Recently solute carriers have been identified to have significant impact on cancer therapy. Screening of additional CML patients is ongoing to confirm the frequency of these variants in optimally responding CML patients.
CONCLUSIONS
CML patients with primary resistance or suboptimal response to TKI therapy have mutations in genes affecting drug intake and metabolism, which can be causally associated with poor therapy responses. Validation in larger patient cohorts is warranted.
Porkka:Novartis: Honoraria, Research Funding; Bristol-Myers Squibb: Honoraria, Research Funding; Pfizer: Honoraria, Research Funding. Mustjoki:Bristol-Myers Squibb: Honoraria, Research Funding; Pfizer: Honoraria, Research Funding; Novartis: Honoraria, Research Funding; Ariad: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal