Abstract
Background: The prognostication of cytogenetically normal AML (CN-AML) continues to evolve with the use of NGS-based risk stratification. Emerging data indicate that the presence of additional mutations in good- and intermediate-risk patients (as defined by conventional cytogenetic and molecular analyses) changes the behavior of their disease, suggesting that more personalized treatment approaches are needed.
Methods: We analyzed the mutational profile of newly diagnosed AML patients with DNMT3A mutations (N = 48) seen at our center and described their clinical characteristics and associated mutations. These patients were identified as part of the Advanced Genomics in Leukemia (AGILE) clinical project currently underway at the Princess Margaret Cancer Centre, Toronto. NGS molecular profiling was performed using the TruSight Myeloid Sequencing Panel (TMSP; Illumina) on the MiSeq benchtop genome sequencer (Illumina). This process permitted profiling of 54 genes (39 in the hotspot region; 15 complete coding region coverage) using amplicon-based library preparation and sequencing by synthesis. 160 patients with newly diagnosed AML were evaluated for this project from February 2015 to February 2016. All were analyzed in parallel by the standard cytogenetic and molecular (NPM1, FLT3-ITD/TKD) diagnostic algorithm.
Results: 48 unique patients were identified bearing mutated DNMT3A. Of these, 31 patients had a normal karyotype. Their clinical characteristics are depicted in Table 1.
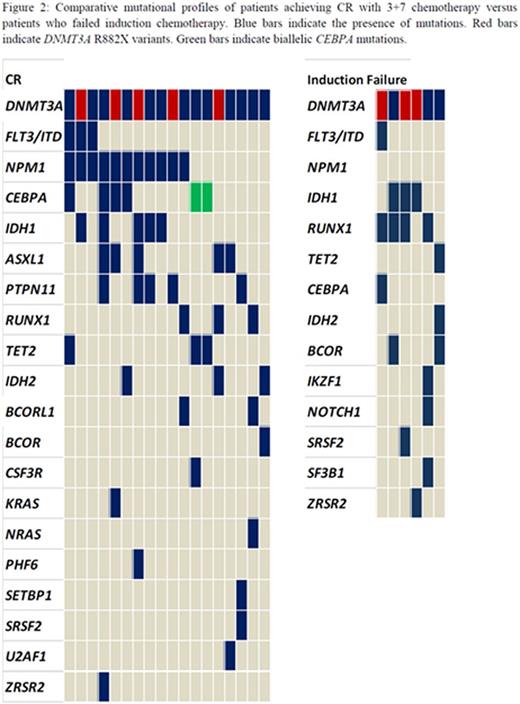
The R882X DNMT3A variant was detected in 12 patients. A total of 100 additional mutations were identified on sequencing (Figure1). The most common associated mutations were in NPM1 (38.7%) followed by IDH1 (29%), RUNX1 (25.8%) and TET2 (22.6%). PTPN11 mutations were identified in 6 patients, 75% of which also had mutated NPM1. Two patients were found to have biallelic CEBPA mutations. Potential gene-gene interactions were also examined and led to the identification of subgroups such as NPM1-FLT3-DNMT3A mutated (n=4), DNMT3A-IDH2R140 (n=3) and DNMT3A-IDH2R172 (n=3) based on recent data by Papaemmanuil, et al (New England Journal of Medicine, 2016) defining these subgroups as being prognostically relevant.( Patients >60 years of age had more frequent mutations in NRAS, BCOR, BCORL1 and TET2. NRAS and BCOR mutations were mutually exclusive with NPM1. RUNX1, IDH2, SRSF2 and U2AF1 mutations were also seen exclusively in the NPM1 negative group.
Eligible patients (n=25) received induction therapy with daunorubicin and cytarabine leading to CR1 in 18 patients (72%). Primary induction failure occurred in 6 cases (24%). All 6 cases were NPM1 negative and had missense mutations in DNMT3A including 3 R882X variants. (Figure 2) Additional mutations were identified in IDH1, RUNX1 and/or TET2 in all 6 patients. During the median follow up of 8 months (range 1 - 15 months), 4 patients relapsed, 2 of which had mutated NPM1 with wild type FLT3-ITD. Sixteen patients are alive at this point and 12 are in CR, six having received allogeneic stem cell transplants. One patient with relapsed disease entered a clinical trial of an IDH1 inhibitor
Conclusion: Genomic analysis is increasingly recognized as a vital adjunct to conventional diagnostic and prognostic approaches. With ongoing advancements in technology leading to increasing cost effectiveness and decreased turnaround times, the use of NGS is likely to become an up-front investigation resulting in a more personalized approach to therapy. Also, unique patient subgroups defined by gene-gene interactions can be identified to further predict clinical behavior and potentially identify druggable targets for therapy.
Gupta:Novartis: Consultancy, Honoraria, Research Funding; Incyte Corporation: Consultancy, Research Funding. Schimmer:Novartis: Honoraria. Yee:Novartis Canada: Membership on an entity's Board of Directors or advisory committees, Research Funding. Kamel-Reid:BMS: Research Funding. Schuh:Amgen: Membership on an entity's Board of Directors or advisory committees.
Author notes
Asterisk with author names denotes non-ASH members.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal