Abstract
Background:DNA methyltransferase 3A(DNMT3A) is a member of the DNA methyltransferase family primarily involved in de novo gene methylation. Mutations in DNMT3A are associated with a wide range of hematological malignancies, most frequently acute myeloid leukemia (AML). DNMT3A mutations are thought to produce a pre-leukemic state, rendering cells vulnerable to secondary oncogenic mutations and malignant transformation. Mutations in DNMT3A often coexist with secondary lesions in leukemia-related genes such as NPM1 and FLT3 (Ley T et al., N Engl J Med, 2010). Furthermore, healthy individuals harboring DNMT3A-driven clonal hematopoiesis are at increased risk of future hematologic malignancies and all-cause mortality (Jaiswal S et al., N Engl J Med, 2014). Despite these important clinical associations, the mechanisms by which DNMT3A mutations contribute to malignant transformation have not been well-defined. Dnmt3a-knockout (KO)mouse hematopoietic stem cells (HSCs) preferentially self-renew rather than undergo differentiation, leading to their accumulation in the bone marrow (Challen GA et al., Nat Genet, 2011). DNMT3A loss has also been shown to drive hypomethylation and subsequent activation of leukemia-related genes (Lu R et al., Cancer Cell, 2016; Yang L et al., Cancer Cell, 2016). However, these findings have not been recapitulated using human tissue. The goals of this study were thus to determine the transcriptional and biological effects of DNMT3A mutations which contribute towards malignant transformation in human cells.
Methods:To elucidate the effects of DNMT3A mutation, we introduced DNMT3A frameshift mutations into K562 cells using the CRISPR/Cas9 gene-editing system. We then performed various functional and genomic assays to better elucidate effects of DNMT3A loss.
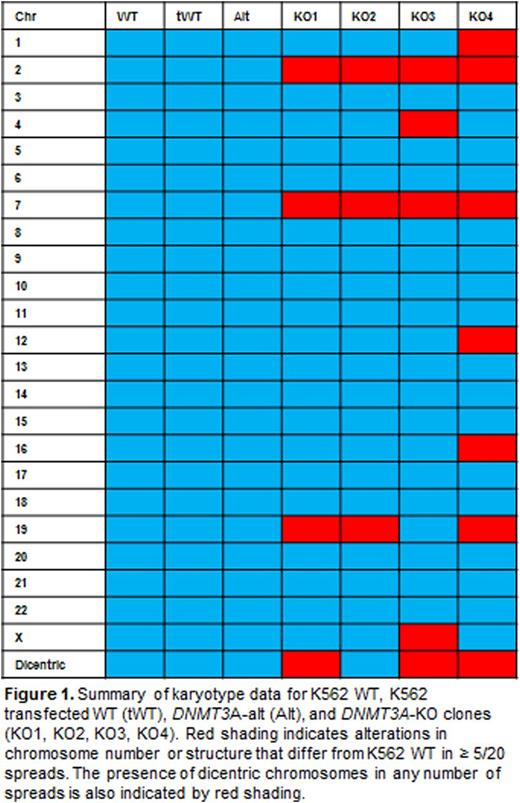
Results and Discussion:We successfully created 4 DNMT3A-KO K562 clones and 1 clone containing a mutation that produces an altered DNMT3A protein with an intact catalytic domain (DNMT3A-alt). We first assessed effects of DNMT3A loss on cell growth and apoptosis. DNMT3A-KO clones exhibited impaired growth compared to wild-type (WT) cells. DNMT3A-KO clones also displayed significantly increased apoptotic activity after exposure to 5-fluorouracil (5-FU). The DNMT3A-alt clone had similar growth and apoptotic activity to WT cells. We examined how DNMT3A loss impacted differentiation using phorbal 12-myristate 13-acetate (PMA), known to induce megakaryocytic differentiation of K562 cells. After overnight exposure to PMA, DNMT3A-KO clones exhibited less CD61 expression, a marker of megakaryocytic differentiation, than did WT cells. Again, the differentiation of the DNMT3A-alt clonewas comparable to WT. Finally, we performed karyotype analysis to elucidate a potential role of DNMT3A in maintaining genomic integrity. Surprisingly, DNMT3A-KO clones exhibited profound cytogenetic variability and genomic instability compared to WT, with most DNMT3A-KO clones containing dicentric chromosomes and ring forms in multiple spreads (Figure 1). The DNMT3A-alt clone had a karyotype identical to WT. CRISPR/Cas9-edited K562 clones without DNMT3A mutation (transfected WT or tWT) also had identical karyotypes to WT K562. TA cloning and mRNA sequencing were employed to elucidate whether loss of DNMT3A would lead to transcriptome instability. DNMT3A-KOand DNMT3A-altclones exhibited distorted splicing patterns, while tWT cell lines were comparable to WT. To further assess the effect of DNMT3A ablation on genomic integrity, we examined DNA-damage responses by measuring DNA double-stranded breaks (DSBs) after treatment with 5-FU. DNMT3A-KO clones were significantly more susceptible to DNA damage than were WT cells, while the DNMT3A-alt clone exhibited more DNA DSBs compared to WT only at high concentrations of 5-FU.
Conclusion:CRISPR/Cas9-mediated DNMT3A-KO K562 cells may be used to model effects of DNMT3A mutations in human cells. Consistent with previous reports, our data suggest that DNMT3A is involved in the differentiation of multipotent progenitors. Novel to this approach, our findings implicate induction of genomic instability as a mechanism by which DNMT3A mutations might predispose to malignancy.
Hosokawa:Aplastic Anemia and MDS International Foundation: Research Funding. Townsley:Novartis: Research Funding. Young:GSK/Novartis: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal