Abstract
DNA cytosine methylation is one of the best-characterized epigenetic modifications that play important roles in diverse cellular and pathological processes. The mechanism underlying the dynamic regulation of the level and distribution of 5-methylcytosine (5mC) as well as the biological consequence of DNA methylation deregulation have been interesting research topics in recent years. TET1, first identified as a fusion partner of the histone H3 Lys4 (H3K4) methyltransferase MLL (mixed-lineage leukemia) in acute myeloid leukemia (AML), is the founding member of the Ten-Eleven-Translocation (TET) family of DNA hydroxylases which are capable of converting 5mC to 5hmC (5-hydroxymethylcytosine) and lead to gene activation. Our group has previously demonstrated that TET1 plays an oncogenic role in MLL-rearranged leukemia (Huang H, et al. PNAS 2013; 110(29):11994-9). The expression of the TET1 protein and the global level of its enzymatic product, 5hmC, are significantly up-regulated in MLL-rearranged leukemia, whereas the opposite has been reported in other cancers where TET1 functions as a tumor suppressor. Therefore, a global understanding of the targets of TET1 in MLL-rearranged leukemia would greatly help to understand the role of TET1 in this specific type of AML.
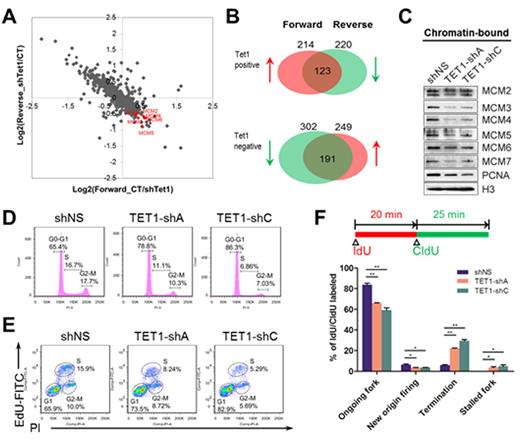
To this end, we performed proteomics study in parallel with RNA-seq to systematically explore the functional targets of TET1 in a genome-wide and unbiased way. Stable isotope labeling by amino acids in cell culture (SILAC)-based proteomic profiling showed that when Tet1 was knocked down in MLL-ENL-estrogen receptor inducible (ERtm) mouse myeloid leukemia cells, a total of 123 proteins were down-regulated whereas 191 were up-regulated with a fold-change cutoff of 1.2 (Fig. 1A and B), representing positively and negatively regulated targets of TET1, respectively. Most of the proteins with altered expression upon Tet1 knock-down showed a corresponding change at the mRNA level as reflected by the RNA-seq data. Interestingly, gene ontology (GO) analysis indicated enrichment on genes associated with DNA replication and cell cycle progression. Among these genes, the minichromosome maintenance complex genes, including MCM2, MCM3, MCM4, MCM5, MCM6, and MCM7, showed significant downregulation when Tet1 expression was depleted. We further conducted chromatin immunoprecipitation (ChIP) assays and demonstrated that TET1 binds directly to the CpG islands in the promoters of these MCM genes, suggesting that the regulation of the MCM genes by TET1 may occur at the transcriptional level.
The six main minichromosome maintenance proteins (MCM2-7) are recruited to DNA replication origins in early G1 phase of the cell cycle and constitute the core of the replicative DNA helicase. We showed that not only the total levels of the MCM2-7 proteins, but also their binding to chromatin (Fig. 1C), were decreased by shRNAs against TET1 in human leukemia cell lines. Examination on cell cycle distribution revealed a significant decrease in the S phase population upon TET1 knockdown (Fig. 1D), which could be phenocopied by silencing of individual MCM genes. Consistently, incorporation of 5-ethynyl-2'-deoxyuridine (EdU) into newly synthesized DNA in the S phase can be inhibited by TET1 shRNAs (Fig. 1E), indicating the inhibition on DNA replication by TET1 silencing. Furthermore, DNA combing assays suggest that TET1 knockdown inhibits new origin firing (Fig. 1F) but does not influence replication fork speed. Collectively, our findings reveal a novel role of TET1 on regulating DNA replication in MLL-rearranged leukemia through targeting of MCM genes and highlight the therapeutic implication of targeting the TET1/MCM signaling.
Role of TET1 in regulate DNA replication by controlling expression of MCM genes
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal