Abstract
INTRODUCTION:
Multiple myeloma is a heterogeneous disease featured by recurrent translocations involving the IgH region. Such cytogenetic events have a driver role in early transformation of a normal plasma cell into a MM cell. Although several studies have reported the presence of limited number of other structural chromosomal events using different approaches, including conventional cytogenetics, high-resolution genome mapping, interphase fluorescence in situ hybridization (FISH) and whole exome sequencing, the full catalogue of genomic rearrangements in MM samples has never been carried out systematically. Here, we have utilized whole-genome sequencing technologies to perform a systematic, genome-wide analysis to uncover the frequency and nature of rearrangements in MM.
MATERIAL AND METHODS:
We performed Whole genome sequencing (WGS) using the Illumina X10 platform in 68 serial samples from 30 patients including 11 patients with smoldering myeloma, 13 newly-diagnosed patients and 44 relapsed patient samples to provide further insight into evolution of rearrangements in MM. Structural variations (translocations, deletions, inversions, internal tandem duplications, fusions) and copy number changes were analyzed using the analysis pipeline at the Wellcome Trust Sanger Institute as recently described (Nik-Zainal Nature 2016).
RESULTS:
We observed a total of 1295 rearrangements for a median of 27 per sample (range 2-138) including a median of 6 (range 1-36) inversions, 5 (range 1-33) internal tandem duplications, 10 (range 1-40) deletions, 7 (range 1-32) translocations and 5 fusions (0-20). While the vast majority of events was non-recurrent, the high prevalence of rearrangements at smoldering stage and in myeloma at diagnosis and further increase at the time of relapse suggest a much more complex genomic landscape than previously thought. Translocations involving the IGH locus were identified including t(11;14) in 6 (20%), t(4;14) in 4 (13%) and t(8;14) in 3 (10%) of 30 unique patients. We also report frequent involvement by light chain loci in the rearrangements. The MYC locus was recurrently affected by non-IGH rearrangements in 11/30 (36%) patients. The other main MYC partners were IGL (4/30) and IGK (2/30), while about one-third of cases were involved by rearrangements not involving immunoglobulins or other obvious partners. MYC is therefore frequently involved by rearrangements through immunoglobulin-independent mechanisms.
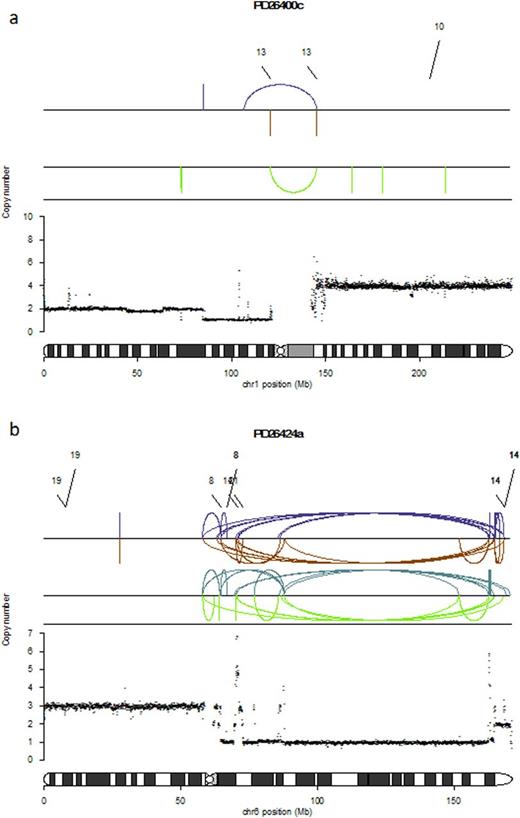
Interestingly, many regions affected by recurrent copy number abnormalities (CNAs) were associated with rearrangements. In particular 7/14 (50%) 1q gains and 6/8 (75%) 1p deletions were involved by translocations and inversions respectively (i.e Figure 1a). Overall 15/22 chromosome 1 CNAs were associated with a specific rearrangements. A similar association between copy number changes and rearrangement breakpoints was observed among other recurrent genomic aberrations such as 6q deletions (6/12, 50%), 8p deletions (4/7, 57%) and 16q deletions (7/13, 53%).
In addition to deletions, inversions, internal tandem duplications (ITDs) and translocations, we observed at least one and often more regions of chromothripsis in 10/30 (33%) patients. Chromothripsis represents a complex event characterized by localized chromosome shattering and repair occurring in a one-off catastrophic event (Korbel J. et al. Cell 2013) (Figure 1b) and known to be associated with worse prognosis in MM. In our series, chromothriptic events were always conserved during every investigated evolution process: suggesting an early onset of this complex event in myelomagenesis.
CONCLUSION:
We report for the first time a comprehensive catalogue of rearrangements in MM based on whole-genome sequencing data. Our data provide evidence that the genomic landscape of rearrangements in MM is very complex and heterogeneous than speculated before and besides IgH involves number of other recurrent chromosomal alteration mechanisms. These diverse aberrations, in many cases acquired early, may deregulate oncogenes as illustrated by the MYC locus.
Moreau:Celgene: Honoraria; Amgen: Honoraria; Takeda: Honoraria; Janssen: Honoraria, Speakers Bureau; Novartis: Honoraria; Bristol-Myers Squibb: Honoraria. Avet-Loiseau:sanofi: Consultancy; celgene: Consultancy; amgen: Consultancy; janssen: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal