Abstract

Intrachromosomal amplification of chromosome 21 (iAMP21) defines a distinct cytogenetic subgroup of 2% childhood acute lymphoblastic leukaemia (ALL). iAMP21-ALL patients have precursor B-cell ALL, are older (median age 9 years), generally present with low white cell counts and have an inferior outcome when treated with standard therapy. Stratification to high risk treatment arms has significantly reduced their relapse risk, thus accurate diagnosis is essential. We have identified iAMP21 as a complex structure of one copy of chromosome 21, comprising multiple regions of gain, amplification, inversion and deletion, initiated through breakage-fusion-bridge cycles and chromothripsis.
Currently, fluorescence in situ hybridisation (FISH), using probes directed to RUNX1, is the most reliable and convenient detection method, in which four or more copies of RUNX1 on a single abnormal chromosome 21 defines iAMP21. From our examination of several hundred cases of iAMP21-ALL, we have noted that, in the absence of metaphase FISH and/or in cases with an unusual cytogenetic presentation, reliance on FISH alone for accurate detection may be problematic. Among a collection of 210 patients with iAMP21-ALL, we performed SNP6.0 and Multiplex Ligation-dependent Probe Amplification (MLPA) using the SALSA MLPA kit P327 iAMP21-ERG (MRC Holland) on 57 and 45 patients, respectively. Although chromosome 21 structure is highly variable between patients, a characteristic copy number profile emerged from the SNP6.0 data (Figure 1). In common, all cases were amplified across a 5.1Mb region, between 32,813,553 and 37,941,425bp that includes RUNX1 and 46 other known protein-coding genes. Asub-telomeric deletion occurred in 88% patients. We propose that this distinctive chromosome 21 copy number profile is used in addition to FISH for the definitive diagnosis of iAMP21-ALL in problematic cases.
Support for this approach is provided by results from 5 patients from the Children's Oncology Group (North America, Australia and New Zealand) and ALL2003 (UK) treatment trials who met the iAMP21 FISH criteria but had atypical karyotypes or an unusual distribution of the additional RUNX1 signals that made confident diagnosis challenging. Clinical and cytogenetic data were collected and SNP6.0 and MLPA were performed to clarify the genomic alterations present in these cases. In patient #1, although five RUNX1 signals were observed per cell, two normal copies of chromosome 21 (each with one RUNX1 signal) were present with only three signals located to the abnormal chromosome 21. In patient #2, the additional signals were distributed between 2 different abnormal copies of chromosome 21. Metaphase FISH of patient #3 indicated that the RUNX1 signals were distributed between one normal copy of chromosome 21 and four small chromosomes, which were identified to originate from chromosome 21 by chromosome painting with a chromosome 21 specific probe (wcp21). In a further 2 cases (patients #4 and #5) the signals were too tightly clustered for the number to be discerned. In these 5 cases the characteristic SNP6.0 profile definitively confirmed the suspected diagnosis of iAMP21-ALL (Figure 2).
Our previous SNP6.0 data have shown that the copy number profile of the iAMP21 chromosome remains stable between diagnosis and relapse, as well as in serial xenografts successfully transplanted with iAMP21-ALL cells over several generations of mice. However, while FISH analysis of patient #6 at diagnosis and relapse showed the same signal pattern at both time points, there was a change in the karyotype, involving translocation of part of the iAMP21 chromosome onto chromosome 11 at relapse. These observations indicate that the iAMP21 chromosome may become fragmented and distributed throughout the genome without changing its genomic profile.
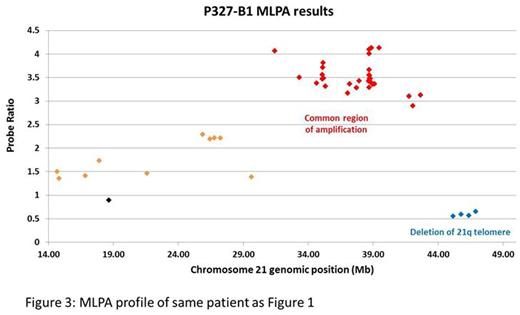
Collectively these observations indicate that the amplified segments resulting from the formation of iAMP21 chromosomes, with typical copy number profiles, can be distributed throughout the genome, either by translocation or fragmentation. Thus FISH together with chromosome 21 copy number profiling provide more accurate diagnosis of iAMP21-ALL than cytogenetics, which may be misleading. For laboratories with no access to SNP6.0 or other copy number arrays, we have shown that MLPA, with a kit specifically designed to detect chromosome 21 copy number changes, provides a reliable alternative (Figure 3).
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This icon denotes a clinically relevant abstract



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal