Key Points
ADAMTS13 contains complex type N-linked glycans, which contain terminal mannose, sialic acids, and fucose residues.
TSP1 repeats are modified by O-fucosylation and C-mannosylation; O-fucosylation was also observed in the disintegrin domain.
Abstract
Patients suffering from acquired thrombotic thrombocytopenic purpura develop autoantibodies directed toward the plasma glycoprotein ADAMTS13. Here, we studied the glycan composition of plasma-derived ADAMTS13. Purified ADAMTS13 was reduced, alkylated, and processed into peptides with either trypsin or chymotrypsin. Glycopeptides were enriched using zwitterionic HILIC zip-tips and analyzed by tandem mass spectrometry employing higher-energy collision dissociation fragmentation. Upon detection of a diagnostic ion of a glycan fragment, electron transfer dissociation fragmentation was performed on the same precursor ion. The majority of N-linked glycans were of the complex type containing terminal sialic acids and fucose residues. A high mannose-containing glycan was attached to Asn614 in the spacer domain. Six O-linked glycans mostly terminating in sialic acid were found dispersed over ADAMTS13. Five O-linked glycans were attached to a Ser and one to Thr. All 6 O-linked glycans contained a terminal sialic acid. O-fucosylation is a common posttranslational modification of thrombospondin type 1 repeats. We identified 7 O-fucosylation sites in the thrombospondin (TSP) type 1 repeats. Unexpectedly, one additional O-fucosylation site was found in the disintegrin domain. This O-fucosylation site did not meet the proposed consensus sequence CSX(S/T)CG. C-mannosylation sites were identified in TSP1, linker TSP4-TSP5, and TSP8. Overall, our findings highlight the complexity of glycan modifications on ADAMTS13, which may have implications for its interaction with immune- or clearance receptors containing carbohydrate recognition domains.
Introduction
ADAMTS13 (a disintegrin and metalloproteinase with a thrombospondin type 1 motif, member 13) is a metalloproteinase responsible for cleaving ultralarge von Willebrand factor multimers in the circulation. Absence or lack of activity of ADAMTS13 results in the microvascular angiopathy thrombotic thrombocytopenic purpura (TTP), which is characterized by systemic aggregation of the platelets in the microvasculature, fragmented erythrocytes, renal, and neurological disturbances.1 Clinical assays measuring ADAMTS13 activity are used to diagnose TTP.2 The reduction in activity of ADAMTS13 is predominantly caused by the formation of autoantibodies against the spacer domain.3-6 Antispacer domain antibodies inhibit the binding of ADAMTS13 to von Willebrand factor but may also accelerate its clearance from the circulation.7,8 In a subset of patients, autoantibodies directed toward the thrombospondin (TSP) 2-8 and CUB1-2 domains have also been identified.6,8,9 Autoantibodies directed against the carboxyterminal domains of ADAMTS13 have been proposed to promote its clearance from the circulation.8,10 Development of high-affinity autoantibodies is crucially dependent on the activation of CD4+ T cells.11 These CD4+ T cells are activated by dendritic cells that present ADAMTS13-derived peptides on major histocompatibility complex class II.12 Previously, we have shown that ADAMTS13 is endocytosed by dendritic cells via the macrophage mannose receptor.13 This observation suggests that high-mannose glycans are present on ADAMTS13. Previous work in our department identified 9 N-linked glycosylation, 6 O-fucosylation, and 2 C-mannosylation sites on plasma-derived ADAMTS13 (pADAMTS13).14 In this study, we determined the glycan composition on pADAMTS13 and discuss the possible role of glycans in the immune recognition and clearance of ADAMTS13.
Experimental procedures
Purification of plasma-derived ADAMTS13
Purification of pADAMTS13 was performed as described previously.15 Briefly, ADAMTS13 from cryosupernatant was purified using the A10 antibody coupled to CNBr sepharose (GE Healthcare, Eindhoven, The Netherlands). Bound ADAMTS13 was washed to remove impurities, subsequently eluted using 40% dimethyl sulfoxide, dialyzed, and stored in 10 mM Tris-HCl pH 7.4 at −30°C until use.
Sample preparation
Samples for analysis by tandem mass spectrometry (MS) were prepared as follows. Five micrograms of purified pADAMTS13 was diluted in 50 mM ammonium bicarbonate buffer pH 8.0 to a final volume of 50 µL. To reduce the cysteine bridges, dithiothreitol (Thermo Scientific, Bremen, Germany) was added to a final concentration of 10 mM and incubated for 30 minutes at 56°C. Iodoacetamide (Thermo Scientific) was added to a final concentration of 55 mM and incubated for 45 minutes at 25°C (in the dark). Trypsin Gold (Promega, Madison, WI) or chymotrypsin (Thermo Scientific) was added to a final concentration of 5 ng/µL and incubated overnight at 37°C. Peptide samples were dried using a speed-vac (Savant, SPP1110; Thermo Scientific) and purified using ZIC-HILIC Protea Tips according to the manufacturer’s protocol (Protea, Morgantown, WV). Next, the samples were eluted in 20 µL ZIC-HILIC Elution Solution and subsequently dried using the speed-vac. Purified and dried samples were reconstituted in 8 µL of 1% formic acid. The flow through of the ZIC-HILIC columns was purified using C18 stage-tips prepared in-house (3M, Neuss, Germany) to further increase the peptide coverage.
MS analysis of plasma-derived ADAMTS13 glycopeptides
Glycopeptide-enriched samples were separated by nanoscale C18 reverse-phase chromatography coupled on line to an Orbitrap Fusion mass spectrometer (Thermo Scientific) via a nanoelectrospray ion source (Nanospray Flex Ion Source; Thermo Scientific). Peptides were loaded on a 20-cm 75 to 360-µm inner- to outer-diameter fused silica emitter (New Objective, Woburn, MA) packed in-house with ReproSil-Pur C18-AQ, 1.9-μm resin (Dr. Maisch, Ammerbuch-Entringen, Germany). The column was installed on a Dionex Ultimate3000 RSLC nanoSystem (Thermo Scientific) using a MicroTee union formatted for 360-μm outer-diameter columns (IDEX, Erlangen, Germany) and a liquid junction. The spray voltage was set to 2.15 kV. Solution A was composed of 0.5% acetic acid, and solution B was composed of 0.5% acetic acid and 80% acetonitrile. Peptides were loaded for 17 minutes at 300 nL/min in 95% solution A and 5% solution B, equilibrated for 5 minutes in the same mixture (17-22 minutes), and eluted by increasing solution B from 5% to 15% (22-87 minutes) and 15% to 38% (87-147 minutes), followed by a 10-minute wash to 90% and a 5-minute regeneration to 5%. Survey scans of peptide precursors from 120 to 2000 m/z were performed at 30 K resolution (at 200 m/z) with a 1.5 × 105 ion count target. Tandem MS was performed by isolation with the quadrupole with isolation window 2.0, higher-energy collision dissociation (HCD) fragmentation with normalized collision energy of 30, and rapid scan MS analysis in the ion trap. The MS2 ion count target was set to 1 × 105 and the maximal injection time was 250 ms. Only those precursors with charge state 2 to 8 were sampled for MS2. The dynamic exclusion duration was set to 60 seconds with a 10-ppm tolerance around the selected precursor and its isotopes. Monoisotopic precursor selection was turned on. The Filter Product Ion Trigger was selected, and a mass list corresponding to the masses of HexNAc (204.0867 m/z), HexNAcfragment (138.0545 m/z), and HexNAcHex (366.1396 m/z) was compiled. Electron transfer dissociation (ETD) was triggered when these masses were detected within the top 20 of the identified fragments. Tandem MS was performed on these ions employing a quadrupole isolation with isolation window of 3.0 ppm and an ETD reaction time of 70 milliseconds. The instrument was run in top-speed mode with 3-second cycles. All data were acquired with Xcalibur software (Thermo Scientific).
Data analysis
Data files from the Orbitrap Fusion were first analyzed using the Preview software (Protein Metrics, San Carlos, CA). The Preview software allows for recalibration of the m/z measurements and screen for peptide modifications; static (carbamidomethylation of cysteine residues [C] + 57.0214 Da) and dynamic modifications (oxidation of methionine residues [M] + 15.994 Da, deamination of asparagine residues [N] or glutamine residues [Q] + 0.9840 Da, addition of dithiothreitol to cysteine residues + 159.657 Da). The resulting data were subsequently analyzed employing Byonic software (Protein Metrics) and screened against the Uniprot_organism_9606_And_kw_0181 database. Additional parameters to identify C-mannosylation (hexose + 162 Da attached to a tryptophan [W]) and O-fucosylation (deoxyhexose + 146 Da or glucose-fucose + 308 Da attached to a serine [S] or threonine [T]) were added manually. Byonic software was used to screen the fragments against the following glycan databases: N-glycan 182 human no multiple fucose, O-glycan 70 human, N-glycan 57 human plasma, or a combination of the 3 databases (databases included in the Byonic software package). Spectra were further analyzed manually, adding additional peptide and glycan fragments to identified m/z peaks.
Results
We used tandem MS employing HCD and ETD to determine the glycosylation profile of pADAMTS13. The HCD fragmentation of glycan-modified peptides allowed for determining the glycan composition. Subsequent ETD modus was used to identify the location of glycans within a peptide sequence. Supplemental Figure 1, available on the Blood Web site, shows the sequence coverage for both trypsin (supplemental Figure 1A) and chymotrypsin digestion (supplemental Figure 1B). To increase the total sequence coverage, the flow through from the ZIC-HILIC purification was further purified using C18 stage tips and analyzed subsequently. Peptide coverage from the ZIC-HILIC purification was 88.7% and 85.9% for trypsin or chymotrypsin digestion, respectively. Combining the ZIC-HILIC and the C18-resin enrichments, the total coverage increased to 90.1% for trypsin and to 86.2% for chymotrypsin. Combining data from trypsin and chymotrypsin digestion, 94.8% of ADAMTS13 sequence was covered. Taking into account that the signal peptide (aa 1-29) and the pro-peptide (aa 30-74) are both removed prior to secretion, we identified 99.9% of the mature ADAMTS13 amino acid sequence.
Identification and determination of the N-linked glycan structures
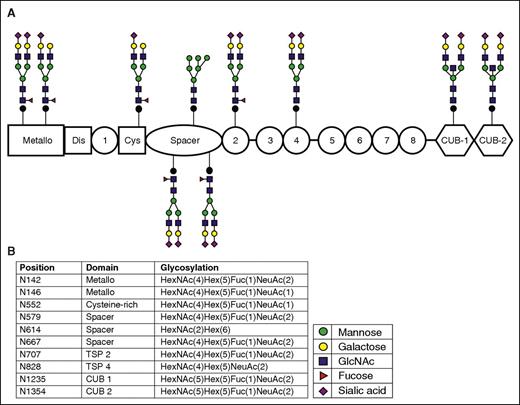
The biosynthetic pathways involved in the biosynthesis of N-linked glycans are well characterized (see supplemental Figure 2).16,17 To determine the glycan structures on ADAMTS13, we used the Byonic software package, which analyzes the fragmentation patterns obtained for each peptide. Peaks resulting from the fragmentation of attached glycans were scanned for fragment masses of 146 Da, 162 Da, 203 Da, and 291 Da corresponding to inclusion of fucose, mannose/glucose/galactose, GlcNAc/GalNAc, and sialic acid residues, respectively. Figure 1A shows the spectrum that was used to identify the N-glycan linked to Asn667, which includes the b- and y-ions from the fragmentation of the peptide backbone and the annotations for the glycan moieties. Byonic software identifies the glycan moieties as being either Hex, HexNAc, NeuAc, HexNAcHex, HexNAcHexNeuAc, but does not distinguish between galactose, mannose, and glucose or between GlcNAc and GalNAc. In Figure 1B, the corresponding cartoon symbols have been added manually to the spectrum. The total mass of the added glycan, the glycan moieties recovered from the spectrum, and our knowledge on the biosynthesis of complex glycans were used to derive the glycan structure attached to Asn667. The same procedure was followed for the other N-linked glycosylation sites. Figure 2A provides an overview of the N-linked glycans as detected by MS. Figure 2B displays the location of these glycans and the exact glycan fragment as identified by Byonic software. The glycan at position Asn828 in TSP 4 was newly identified; however, it was previously predicted to be a possible N-linked glycosylation site.18 Nine of the 10 N-linked glycans contained complex carbohydrate structures with a terminal sialic acid. In addition, the glycans at these N-linked sites were identified both with and without a fucose on the primary GlcNAc (Figure 2; Table 1). We were unable to identify a third GlcNAc residue in the glycan linked to Asn614 in the spacer domain (supplemental Figure 3). The absence of a third GlcNAc suggests that the glycan on Asn614 consists primarily of high mannose structures (see supplemental Figure 4 for all the N-linked glycan spectra and reconstructed glycans). Most glycan-containing Asn contained multiple glycan structures. Table 1 provides an overview of the glycans that were detected less frequently at the specified positions. The majority of the glycans identified were of the complex structure and contain terminal sialic acid, and ∼50% of the glycans were also fucosylated. Only high-mannose N-linked glycans were identified on Asn614 (Figure 2; Table 1). High-mannose N-linked glycans were also identified on Asn142, Asn146, and Asn1235, although spectra corresponding to these glycans were observed less frequently.
Reconstruction of a N-linked glycan at Asn667. (A) Spectrum as provided by Byonic software. In blue, the b-ions, and in red, the y-ions are depicted. In green, the glycan moieties are indicated as identified by Byonic software. (B) Corresponding glycan moieties depicted in the commonly used cartoons: green circles, blue squares, purple diamonds, and yellow circles for mannose, GlcNAc, sialic acid, and galactose, respectively. From this spectrum, we can determine the glycan structure attached to this asparagine. Starting with the core structure 2-GlcNAc-3-mannose, from the spectrum, we can conclude that this is a complex type glycan; a GlcNAc-galactose-sialic acid fragment is present. The presence of a fucose moiety suggests that this glycan is fucosylated. The fragment corresponding to the mass of the peptide + HexNAc + fucose suggests that the first GlcNAc is fucosylated. Combining these individual fragments results in the final structure of a fucosylated biantennary complex glycan structure attached to Asn667.
Reconstruction of a N-linked glycan at Asn667. (A) Spectrum as provided by Byonic software. In blue, the b-ions, and in red, the y-ions are depicted. In green, the glycan moieties are indicated as identified by Byonic software. (B) Corresponding glycan moieties depicted in the commonly used cartoons: green circles, blue squares, purple diamonds, and yellow circles for mannose, GlcNAc, sialic acid, and galactose, respectively. From this spectrum, we can determine the glycan structure attached to this asparagine. Starting with the core structure 2-GlcNAc-3-mannose, from the spectrum, we can conclude that this is a complex type glycan; a GlcNAc-galactose-sialic acid fragment is present. The presence of a fucose moiety suggests that this glycan is fucosylated. The fragment corresponding to the mass of the peptide + HexNAc + fucose suggests that the first GlcNAc is fucosylated. Combining these individual fragments results in the final structure of a fucosylated biantennary complex glycan structure attached to Asn667.
Schematic representation of the N-linked glycosylation sites and the most frequently identified glycans attached to each site. The glycan structures were determined using the workflow described in supplemental Figure 2. (A) Schematic representation of the glycans identified on ADAMTS13. Each glycan is attached to the corresponding domain. (B) Glycan composition, amino acid position, and corresponding domain of ADAMTS13. There is one glycan on pADAMTS13 that was only identified with a high mannose structure; the remaining glycosylation sites were predominantly identified with complex structures terminating in sialic acid; 6 of 10 glycans were fucosylated.
Schematic representation of the N-linked glycosylation sites and the most frequently identified glycans attached to each site. The glycan structures were determined using the workflow described in supplemental Figure 2. (A) Schematic representation of the glycans identified on ADAMTS13. Each glycan is attached to the corresponding domain. (B) Glycan composition, amino acid position, and corresponding domain of ADAMTS13. There is one glycan on pADAMTS13 that was only identified with a high mannose structure; the remaining glycosylation sites were predominantly identified with complex structures terminating in sialic acid; 6 of 10 glycans were fucosylated.
Identification and determination of the O-linked glycan structures and sites
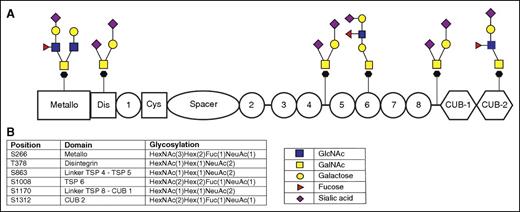
O-Linked glycosylation starts with the linkage of a GalNAc to a Ser/Thr residue by a family of glycosyltransferases known as UDP-GalNAC:polypeptide N-acetylgalactosaminyltransferases in the Golgi apparatus.19,20 This GalNAc linkage to a serine/threonine is also known as the Tn antigen. The Tn antigen is usually extended by different glycosyltransferases yielding a wide variety of complex O-glycans, which are commonly divided into 8 different O-linked core structures (supplemental Figure 5).17,19 Six O-linked glycans were identified on ADAMTS13; 5 O-linked glycans were attached to a Ser and 1 to a Thr (Figure 3B). Figure 3B shows the exact glycan fragment as identified by the Byonic software. Our MS approach is not able to distinguish between different hexoses (such as mannose, glucose, or galactose) or HexNAcs (such as GlcNAc or GalNAc). We used our current knowledge on the assembly of O-linked glycans to propose the most likely structure for each of the O-linked glycans on ADAMTS13 using the individual mass chromatograms for each O-linked glycosylation site, as shown in supplemental Figure 6. All 6 O-glycans contained a terminal sialic acid. Three O-linked glycans were fucosylated on either GlcNAc or galactose. Based on the expression of fucosyltransferase VI and the synthesis of ADAMTS13 in the liver,21-23 we propose that the fucosylation of these glycans is likely to occur on GlcNAc.
O-linked glycosylation of ADAMTS13.O-linked glycosylation starts with a GalNAc attached to either a serine or a threonine; glycan moieties such as galactose, fucose, GlcNAc, GalNAc, or sialic acid are added subsequently. There is no consensus on the composition of these structures, and the presented structures are possibilities corresponding to the residues identified by the MS and Byonic software. (A) Possible glycan structures that correspond to the glycan moieties identified using the Byonic software. Our MS approach is not able to distinguish between different hexoses (such as mannose, glucose, or galactose) or HexNAcs (such as GlcNAc and GalNAc); the presented structures are based on our current knowledge of the biosynthetic pathway of O-glycans. (B) Amino acid position (serine or threonine) and the corresponding domain of ADAMTS13 and glycan elements are identified on this position as determined using Byonic software.
O-linked glycosylation of ADAMTS13.O-linked glycosylation starts with a GalNAc attached to either a serine or a threonine; glycan moieties such as galactose, fucose, GlcNAc, GalNAc, or sialic acid are added subsequently. There is no consensus on the composition of these structures, and the presented structures are possibilities corresponding to the residues identified by the MS and Byonic software. (A) Possible glycan structures that correspond to the glycan moieties identified using the Byonic software. Our MS approach is not able to distinguish between different hexoses (such as mannose, glucose, or galactose) or HexNAcs (such as GlcNAc and GalNAc); the presented structures are based on our current knowledge of the biosynthetic pathway of O-glycans. (B) Amino acid position (serine or threonine) and the corresponding domain of ADAMTS13 and glycan elements are identified on this position as determined using Byonic software.
O-fucosylation and C-mannosylation of ADAMTS13
A common posttranslational modification of TSP1 repeats is O-fucosylation. Previously, O-fucosylation sites have been described in the TSP1, 2, 3, 5, 6, 7, and 8 in recombinant ADAMTS13.24 O-fucosylation of TSP2, 5, 7, and 8 was shown to contribute to the secretion of recombinant ADAMTS13.24 Our current analysis confirms the presence of O-linked fucose residues in TSP1, 2, 3, 5, 6, 7, and 8 of pADAMTS13. Unexpectedly, we identified 1 O-linked fucosylation sites in the disintegrin domain at Ser336 (Figure 4). This O-fucosylation site did not fulfill the proposed consensus sequence CSX(S/T)CG.25 Mass spectra identifying O-fucosylation of Ser336 are displayed in supplemental Figure 7. Unmodified, nonfucosylated peptides were identified at all 8 sites (data not shown).
O-fucosylation and C-mannosylation sites on ADAMTS13. (A) Schematic representation of the ADAMTS13 molecule with the O-fucosylation sites depicted as red triangles and blue circles (fucose and glucose residues); green circles represent the mannose residues. In total, 3 C-mannosylation sites were identified by the Byonic software. Eight O-fucosylation sites, all of them modified by a glucose-fucose group attached to either a serine or a threonine, were identified. (B) Amino acid position and corresponding domain of ADAMTS13 and attached glycan structures (Hex either being a mannose or a glucose, and dHex being a fucose moiety) are depicted.
O-fucosylation and C-mannosylation sites on ADAMTS13. (A) Schematic representation of the ADAMTS13 molecule with the O-fucosylation sites depicted as red triangles and blue circles (fucose and glucose residues); green circles represent the mannose residues. In total, 3 C-mannosylation sites were identified by the Byonic software. Eight O-fucosylation sites, all of them modified by a glucose-fucose group attached to either a serine or a threonine, were identified. (B) Amino acid position and corresponding domain of ADAMTS13 and attached glycan structures (Hex either being a mannose or a glucose, and dHex being a fucose moiety) are depicted.
We previously reported that Trp390 (TSP1) and Trp884 (linker TSP4-TSP5) were modified by C-mannosylation.14 In this study, we confirm that Trp884 is indeed modified by C-mannosylation (Figure 4; supplemental Figure 8). Peptides containing a mannose moiety attached to Trp390 were not identified in this study. We found evidence for a mannose moiety attached to Trp387 in accordance with the proposed consensus sequences for C-mannosylation (supplemental Figure 8).25,26 C-mannosylation of this residue was also reported by Akiyama and coworkers.27 We identified C-mannosylation of Trp1016 (TSP7) (supplemental Figure 8). Non-C-mannosylated peptides were identified spanning Trp387, Trp884, and Trp1016 (data not shown).
Discussion
In this study, we determined the glycan structures attached to pADAMTS13, employing glycopeptide enrichment followed by tandem MS. We identified glycans on all of the 10 predicted N-linked sites. As expected, individual sites contained several different N-linked glycans. Based on the limitation of MS being dependent on the efficiency of ionization and the fragmentation efficiency of peptides by high-energy collision dissociation and ETD, we can only discuss glycans that we identified; we cannot exclude that additional glycan structures are present, which escaped detection by the methods employed in this paper. We identified a single high-mannose–ending glycan at Asn614 in the spacer domain. In addition, 3 mannose-ending glycans were identified in other domains of ADAMTS13 (Table 1). We speculate that high-mannose glycans identified in this study are potentially involved in the binding of ADAMTS13 to the mannose receptor on dendritic cells.13
The glycan positioned at Asn1354 is closely located to the peptide “ASYILIRDTHSLRTTA” (residues 1355-1369), previously identified to be present on HLA-DRB1*03.11,12,14 The glycan at Asn1354 may modulate presentation of peptide 1355-1369 on major histocompatibility complex class II.12,14 Aberrant glycosylation has previously been implicated in initiation of autoimmune disorders.28 In normal individuals, type II collagen contains an O-linked glycan attached to a modified l-hydroxylysine at position 264.29 In patients with rheumatoid arthritis, the lysine at position 264 does not contain an O-linked glycan.29 In a murine model of collagen-induced arthritis, the deglycosylated peptide is recognized as a foreign entity by pathogenic CD4+ T cells.29
In this study, we show that ADAMTS13 is extensively modified by O-linked glycans. No consensus sequences for O-linked glycosylation have been defined; nevertheless, high-energy collision dissociation and ETD allowed for the identification of 6 O-linked glycans on ADAMTS13. The diversity of O-linked glycans provides a major challenge for their identification employing MS. The biosynthesis of O-linked glycans is less well defined when compared with that of N-linked glycans.30 The composition of the O-linked glycans can be accurately determined employing the methods used in this study (Figure 3). However, it is not possible to distinguish between GlcNAc or GalNAc due to their identical mass. We identified 3 O-linked glycosylation sites (Thr378, Ser863, and Ser1170) that were modified with a HexNAc(1)Hex(1)NeuAc(2) glycan structure corresponding to a disialyl-T antigen. The other 3 O-linked glycosylation sites (Ser266, Ser1008, and Ser1312) were more complex, and we propose the structures as shown in Figure 3 based on our current knowledge on the assembly and composition of the most prevalent O-linked glycans.17,19 The fucosylation of the O-linked glycosylation sites Ser266, Ser1008, and Ser1312 has been assigned to the GlcNAc residue within the glycan structure, based on our knowledge that ADAMTS13 is synthesized in the liver22,23 and the expression of fucosyltransferase VI in the liver.21 Fucosyltransferase VI adds a fucose moiety to a GlcNAc via an α-1-3 linkage.21 N- and O-linked glycans are hydrophilic and negatively charged and can have a significant effect on protein conformation. Small-angle X-ray scattering has demonstrated that the carboxyterminal TSP8 and CUB1-2 domains interact with the aminoterminal disintegrin, TSP1, Cys-rich, and spacer domains.31 In a separate study, South and coworkers showed that the CUB1-2 domains of ADAMTS13 interacted with the Arg568, Phe592, Arg660, Tyr661, and Tyr665 in the spacer domain.32 Both CUB domains and the spacer domain of ADAMTS13 contain a number of N- and O-linked glycans, and these may contribute to the interaction between the spacer and CUB domains. In this respect, it is interesting to note that the glycan attached to Asn614 is primarily composed of nonfucosylated, high-mannose–ending sugars, which suggests that this site may be poorly accessible for mannosidases, galactosyltransferases, and sialyltransferases due to its interaction with the CUB1-2 domains.31,32
We identified 8 O-fucosylation sites, of which 7 were previously reported by Ricketts and coworkers.14,24 These 7 O-fucosylations occur at serines in agreement with the consensus sequence for O-fucosylation established for the TSP domains. A glucose-fucose moiety was identified on all the O-fucosylation sites.31,32 TSP-derived peptides containing a single fucose moiety were also identified (data not shown). O-fucosylation has so far been reported exclusively in TSP domains and epidermal growth factor (EGF) domains.24,25,33 Interestingly, the EGF domains of several blood coagulation factors such as factor VII, factor IX, and factor XII contain O-fucosylated residues. A consensus sequence for O-fucosylation of EGF domains have been defined: Cys-X-X-X-X-(Ser/Thr)-Cys.34 O-fucose-linked glycans on EGF domains appeared to be quite heterogenous comprising O-fucose monosaccharides but also tetrasaccharide modifications as observed for factor IX.34 Also, EGF domains in Notch and the Notch-ligand Δ-like 1 (DLL1) are modified by O-fucosylation.34-36 O-fucosylation of TSP repeats has been demonstrated for TSP1.25 Identification of this modification on TSP of other proteins, which include properdin and ADAMTS13, has led to the proposal of the following consensus sequence: CX2-3(S/T)CX2G.24,33 In this study, we show that O-fucosylation is not limited to TSP repeats or EGF domains; O-fucosylated peptides derived from the disintegrin domain of ADAMTS13 were also identified (Figure 4). In the consensus sequence, the Ser/Thr residue that is modified by O-fucosylation is immediately followed by a cysteine residue.24,33 Ser336 (disintegrin) is also followed directly by a cysteine residue. Two O-fucosyltransferases have been identified so far. Protein O-fucosyltransferase 1 (Pofut1) has been implicated in fucosylation of EGF domains,37 whereas protein O-fucosyltransferase 2 (Pofut2) mediates fucosylation of TSPs.36 Following initial fucosylation, β1-3 glycosyltransferases can add an additional glucose moiety to fucosylated TSPs.36 More complex sugars are added to O-fucosylated residues within EGF domains, which are elongated by β3-N-acetylglucosaminyltransferases.38 We did not find evidence for the addition of complex O-linked sugars to O-fucosylated sites within ADAMTS13. The exclusive presence of disaccharide glucose-β(1,3)fucose suggests that the sequential action of Pofut2 and β1,3 glycosyltransferase is responsible for the attachment of the disaccharide to the non-TSP sites within pADAMTS13, as previously described for recombinant ADAMTS13.24 Both Pofut1 and Pofut2 are involved in quality control in the endoplasmic reticulum. Modification of O-fucosylation sites in TSP2, 5, 7, and 8 of recombinant ADAMTS13 resulted in reduced secretion,24 which suggests that O-fucosylation serves as a quality control for proper protein folding of the TSPs of ADAMTS13.
We identified 3 C-mannosylation sites at Trp387 (TSP1), Trp884 (linker TSP4-TSP5), and Trp1016 (TSP8) in this study. Trp387, Trp884, and Trp1016 were not fully C-mannosylated; we also identified non-C-mannosylated peptides (data not shown). In a previous study, we reported that Trp390 was C-mannosylated; however, in that particular study, we also observed C-mannosylated peptides that could not be unambiguously assigned to either Trp387 or Trp390.14 The assignment of Trp387 as a prominent site for C-mannosylation in the current study is based on the analysis of spectra derived of 34 peptides, of which 16 were found to contain a C-mannosylation at Trp387; on 6 peptides, a C-mannosylation was detected, but the modification could not be assigned to either Trp387 or Trp390. The remainder of the peptides spanning Trp387 and Trp390 were not modified by C-mannosylation (data not shown). No evidence of C-mannosylation of Trp390 was therefore obtained in our current analysis (data not shown). Nevertheless, we cannot fully exclude that a small proportion of pADAMTS13 is indeed C-mannosylated on Trp390 as suggested by our previous work.14 The NetCGlyc 1.0 prediction tool identifies Trp387 and Trp390 as potential targets for C-mannosylation.25,26 C-mannosylation of Trp387 was also suggested by electron density linked to the side chain of Trp387 in the crystal structure of a recombinant ADAMTS13 fragment containing a P475S substitution that was expressed in CHO Lec 3.2.8.1 cells.27 Based on the currently available data, we therefore feel it is likely that Trp387 is the preferred site of C-mannosylation in the TSP1 domain of ADAMTS13. Interestingly, we identified peptides containing C-mannosylated Trp387 that lacked the presence of an O-glycan at Thr378 (supplemental Figures 6 and 8) suggesting that these modifications are mutually exclusive. We speculate that following addition of an O-glycan at Thr378, Trp387 is not accessible for C-mannosylation. Conversely, C-mannosylation at Trp387 apparently also prevents addition of an O-glycan to Thr378.
Overall, our findings illustrate the complexity and heterogeneity of N-linked, O-linked, and C-linked glycans on ADAMTS13. We show that O-fucosylated residues are not exclusively present in the TSP repeats of ADAMTS13 and demonstrated that not all sites are fully occupied by glycans. In future studies, we will explore whether the observed glycan heterogeneity plays a role in the onset of acquired TTP.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Marshall Bern, Eric Carlson, and Pierre Allemand of Byonic for valuable input. The authors acknowledge Y. Fujimura for providing the A10 antibody used for purification of plasma-derived ADAMTS13. The authors also thank Ivan Peyron and Eelke Béquin for valuable input and critical comments.
This work was supported by a grant from the Landsteiner Foundation for Blood Transfusion Research.
Authorship
Contribution: F.C.V. designed and carried out the experiments, performed the analysis, prepared the figures, and wrote the manuscript; E.S. set up the mass spectroscopy protocol; P.H.P.K. purified the pADAMTS13; F.v.A. performed the C18 enrichment and operated the MS; and J.V. and A.B.M. contributed to the design and supervision of the experiments. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jan Voorberg, Department of Plasma Proteins, Sanquin Research, Plesmanlaan 125, 1066 CX Amsterdam, The Netherlands; e-mail: j.voorberg@sanquin.nl.
References
Author notes
F.C.V. and E.S. contributed equally to this study.