Abstract
Protease signaling in cells elicits multiple physiologically important responses via protease-activated receptors (PARs). There are 4 members of this family of G-protein–coupled receptors (PAR1-4). PARs are activated by proteolysis of the N terminus to reveal a tethered ligand. The rate-limiting step of PAR signaling is determined by the efficiency of proteolysis of the N terminus, which is regulated by allosteric binding sites, cofactors, membrane localization, and receptor dimerization. This ultimately controls the initiation of PAR signaling. In addition, these factors also control the cellular response by directing signaling toward G-protein or β-arrestin pathways. PAR1 signaling on endothelial cells is controlled by the activating protease and heterodimerization with PAR2 or PAR3. As a consequence, the genetic and epigenetic control of PARs and their cofactors in physiologic and pathophysiologic conditions have the potential to influence cellular behavior. Recent studies have uncovered polymorphisms that result in PAR4 sequence variants with altered reactivity that interact to influence platelet response. This further demonstrates how interactions within the plasma membrane can control the physiological output. Understanding the structural rearrangement following PAR activation and how PARs are allosterically controlled within the plasma membrane will determine how best to target this family of receptors therapeutically. The purpose of this article is to review how signaling from PARs is influenced by alternative cleavage sites and the physical interactions within the membrane. Going forward, it will be important to relate the altered signaling to the molecular arrangement of PARs in the cell membrane and to determine how these may be influenced genetically.
Plasma proteases have multiple physiological roles, which include generating bioactive peptides, converting zymogens to proteases, and initiating cellular signaling through protease-activated receptors (PARs). PARs are a family of G-protein–coupled receptors (GPCRs) that couple to multiple G-proteins depending on the cell type, activating protease, and cellular cofactors. The prototypical receptor, PAR1, was identified 25 years ago as the thrombin receptor.1 Subsequently, 3 other family members were identified (PAR2, PAR3, and PAR4) by homology screening of complementary DNA libraries.2-6 Since the original reports, numerous studies in cells, knockout mice, and preclinical animal models of disease have demonstrated the importance of PARs in a wide range of physiological and pathophysiological conditions. The role of PARs in hemostasis is well documented in several detailed reviews.2,7-9 The focus of this review is how co-receptors influence activation and signaling specificity. The goal is to integrate what is known about the molecular organization of PARs in the plasma membrane, how this influences both activation and downstream events, and how these may be regulated genetically. The specific signaling events emanating from each of the family members have been reviewed extensively and will only briefly be discussed here.
Proteolytic activation of PARs
The rate-limiting step of protease signaling via PARs is the proteolysis of the N terminus to expose the tethered ligand, which interacts with the extracellular loops of the receptor (Figure 1).10,11 This key step is determined by the nature of the enzyme-substrate interaction. Proteases use a variety of strategies such as exosites and cofactors to increase the specificity and efficiency of substrate hydrolysis. The wide panel of proteases capable of activating PARs in a variety of cell types has been comprehensively reviewed.7,12,13 Each protease has its distinct requirements for PAR activation including cofactors and noncanonical cleavage sites that may initiate specific signaling pathways.
PAR signaling is initiated by proteolysis of the N terminus to irreversibly activate the receptor by exposing the tethered ligand. (A-B) Proteases have distinct cleavage sites that generate unique tethered ligands that can direct the downstream signaling toward specific pathways. The PAR1 cleavage sites for thrombin, APC, MMP1, and MMP13 are shown as a representative example.
PAR signaling is initiated by proteolysis of the N terminus to irreversibly activate the receptor by exposing the tethered ligand. (A-B) Proteases have distinct cleavage sites that generate unique tethered ligands that can direct the downstream signaling toward specific pathways. The PAR1 cleavage sites for thrombin, APC, MMP1, and MMP13 are shown as a representative example.
The most well-characterized activator of PARs is thrombin. Thrombin is a multisubstrate enzyme that uses exosites to drive substrate specificity and reaction efficiency.14 Further, thrombin’s exosites are allosterically linked to its active site, and interactions at exosite I induce thrombin into a protease conformation.15 PAR1 is an excellent substrate due to a hirudin-like sequence that interacts with thrombin’s exosite I and greatly enhances cleavage efficiency.16-18 In contrast, structural and biochemical studies show that PAR4 does not bind to thrombin’s exosite I.19-21 This fundamental difference in enzyme-substrate interaction contributes to a 200-fold decrease in the first order rate constant (kcat) compared with PAR1 (17 s−1 vs 340 s−1).18 Mechanistically, PAR4 does not have the advantage of a second binding site that allosterically induces thrombin into the protease conformation, which contributes to the overall slower activation rate. PAR4 activation kinetics on the cells is consistent with the cleavage studies using purified exodomains.19,20
In vivo proteins are not expressed in isolation and the complexities at the cell surface dramatically influence PAR activation. The initial studies defining the roles of PARs in knockout mice uncovered a surprising dependence on PAR3 for efficient thrombin activation of PAR4 on mouse platelets.22 Nakanishi-Matsui et al proposed that PAR3 is a cofactor that recruits thrombin to the surface for efficient PAR4 cleavage.22 Leger et al subsequently demonstrated a similar mechanism for efficient PAR4 activation on human platelets where PAR1 serves as the cofactor for PAR4 (Figure 2A).23 The enhancement of PAR4 cleavage by PAR1 is exosite I dependent.19,23 Taken together, a plausible hypothesis is that, in addition to recruiting thrombin to the surface of cells, the PAR1-exosite I interaction is holding thrombin in the protease conformation for efficient cleavage of PAR4.15 On platelets, the glycoprotein Ib-IX (GPIb-IX) complex also binds thrombin, however, it was unclear what additional contributions thrombin binding to GPIb had beyond thrombin-induced PAR signaling.24 Early studies suggested that GPIb enhanced PAR1, but not PAR4, activation by thrombin.25,26 Estevez et al have recently shown that GPIb-IX signaling cooperates with PAR signaling via the 14-3-3, Rac1, LIMK1 axis using cell culture, human platelets, and mouse models.27 These studies demonstrate that signaling systems may also interact downstream of the point of activation and do not necessarily need to physically associate.
PARs use cofactors to regulate the specificity and rates of cleavage. (A) The current model of PAR4 activation on platelets. PAR1 serves as a cofactor to enhance the rate of PAR4 cleavage by thrombin. The PAR1-PAR4 cooperation is based on the model originally proposed for PAR3-PAR4 on mouse platelets by Nakanishi-Matsui et al.22 (B) PAR1 cleavage by APC requires the EPCR as a cofactor.
PARs use cofactors to regulate the specificity and rates of cleavage. (A) The current model of PAR4 activation on platelets. PAR1 serves as a cofactor to enhance the rate of PAR4 cleavage by thrombin. The PAR1-PAR4 cooperation is based on the model originally proposed for PAR3-PAR4 on mouse platelets by Nakanishi-Matsui et al.22 (B) PAR1 cleavage by APC requires the EPCR as a cofactor.
The first report of activated protein C (APC) cleaving PAR1 mechanistically linked PAR1 to APC’s cytoprotective effects in sepsis.28 Several studies have since shown that the APC–endothelial protein C receptor (EPCR)–PAR1 pathway plays an essential cytoprotective role in sepsis and endotoxemia (Figure 2B).9,29-31 Thrombin is required for generating APC, which initially made it unclear how APC could signal through PAR1 in the presence of thrombin because APC cleaves PAR1 10 000-fold less efficiently.32 A combination of later reports showed that low levels of APC were sufficient to confer barrier protection in endothelial cells and that limited PAR1 cleavage by APC could elicit PAR1 signaling through β-arrestin.33-36 These studies underscore the importance of co-receptors and membrane localization for signaling specificity.37-39 Another key development was demonstrating that the anticoagulant properties of APC could be separated from its signaling role by mutating key residues.40,41 Alanine substitution of residues in the Ca2+-binding site or in loop 37 resulted in APC mutants with a decreased ability to inactivate factor Va (FVa), but retained the ability to activate PAR1.40 These mutations were later combined to make APC-5A, which has <0.1% anticoagulant activity.41
Recent studies show that APC can also cleave PAR1 and Arg 46 and lead to cytoprotective signaling (Figure 1).42,43 The noncanonical cleavage site was demonstrated by analyzing the proteolytic fragments of peptides derived from the PAR1 exodomain. Further, PAR1 constructs with mutations at Arg 41, Arg 46, or both were overexpressed in cells and incubated with APC or thrombin. The mutations at Arg 41 or Arg 46 had reduced cleavage by APC compared with wild-type PAR1; cleavage was completely abrogated in the double mutant.42,43 As expected, mutating Arg 46 had no influence on thrombin cleavage. Finally, agonist peptides derived from the Arg 46 cleavage site (NPNDKYEP) were able to generate cytoprotective signaling in cells. A potential issue is the high concentrations of enzyme-extended incubation times required for cleavage at Arg 46, which needs to be further examined. However, Soh and Trejo clearly show that limited cleavage of PAR1 is sufficient for a cellular response,36 which will make it difficult to determine the precise localization of proteolysis. Most studies to date have focused on PAR1-mediated effects, however, alternative cleavage sites on PAR3 by APC and FXa have been reported to influence endothelial cell function.43,44
Matrix metalloproteinases (MMPs) are a family of zinc-dependent endopeptidases that degrade extracellular matrix proteins during tissue repair and cancer invasion.45 MMP-1 and MMP-13 both activate PAR1 on cells through noncanonical sites.46-48 MMP-1 cleaves at Asp 39 and MMP-13 cleaves at Ser 42 (Figure 1). The alternative site at Asp 39 directs signaling that is distinct from the canonical cleavage site of thrombin at Arg 41.46,47 For example, in human platelets, MMP-1 induces G12/13-Rho, p38 MAPK pathways, and shape change. However, MMP1 has a lower efficacy in stimulating intracellular Ca2+ mobilization and platelet aggregation than thrombin.46,47 To date, distinct signaling pathways downstream of MMP-13 cleavage of PAR1 have not been described. Finally, the MMPs do not appear to require specific membrane localization or cofactors to activate PAR1.
As more proteases are identified as PAR activators at noncanonical sites, there are key questions that must be kept in mind as the details are uncovered. First, how is specificity determined? The studies characterizing PAR1 cleavage by APC demonstrate the requirement of EPCR as a cofactor and proper membrane localization. These underscore the importance of the cellular context in which the receptor is expressed. Second, we need to be cautious when interpreting data from in vitro studies using high concentrations (nonphysiological) of proteases on cultured cells for long periods of time. Now that gene editing has become commonplace, it will be interesting to see the effects of mutating potential cleavage sites on endogenous PARs. Nonetheless, there are a multitude of possibilities for regulated protease signaling via PARs that will certainly have specific physiologic roles.
Biased signaling
GPCRs can signal through G-protein and β-arrestin pathways. Functional selectivity or biased agonism describes agonists that preferentially activate specific pathways downstream of the receptor.49 The precise mechanism of biased agonism is still under investigation. Structural studies with a prototypical GPCR, the β-adrenergic receptor, reveal partial changes in the structure that correlate with different states of activation using partial agonists.50,51 The multiple agonists used show that not all of the molecular switches need to be tripped in order for the receptor to activate signaling. These intermediate steps may allow for signaling to proceed down specific pathways and provide a structural basis for functional selectivity. Biased signaling downstream of PARs can be mediated by posttranslational modifications, cofactors, alternative cleavage sites, or pharmacologically.7-9,52-54
PAR1 signaling downstream of APC and thrombin have opposing effects in endothelial cells.9 The first report to mechanistically describe the biased signaling mediating this cellular response was Soh and Trejo in 2011.36 Prior to this work, it was understood that localization of EPCR and PAR1 into specialized membrane microdomains was a key contributor to the cytoprotective signaling of APC through PAR1.12,37 However, Bae et al have also shown that engagement of EPCR with a catalytically inactive protein C modulates the cellular response.38,39 Soh and Trejo built on this work to demonstrate that APC signals through Rac1 within 5 minutes in a β-arrestin-2–dependent manner. In contrast, thrombin signaling through RhoA was independent of β-arrestin.36 Since this original report, several studies have focused on biased signaling from PARs. The reader is directed to recent reviews of the literature that specifically focus on biased signaling from PARs in detail.7,12,13
A question that remains is how do PARs activate distinct signaling events under specific conditions. The association of receptors with downstream effectors varies; some complexes are preassembled whereas others form following activation.55-57 For example, there appears to be a population of PAR1 in preformed complexes with Gαi.55 There is a second population that associates with Gα12 and β-arrestin following stimulation; these interactions were detected with a t1/2 of 8.8 and 7.5 minutes, respectively.55 The same pattern occurred with PAR2; Gαi was found in preformed complexes and Gα12 and β-arrestin associated with PAR2 minutes after stimulation. The PAR2 study also included Gαo and it was in preformed complexes similar to Gαi.57 These data are in contrast with Soh and Trejo who observed that PAR1 was in complex with β-arrestin in endothelial cells under basal conditions.36 These differences may be due to multiple PAR1 populations in specific cell types. The detailed studies by Ayoub and colleagues focused primarily on thrombin-activated PAR1.55,56 More light will be shed on the populations of receptors as these dynamics are examined with other activating proteases that use alternative cleavage sites or with allosteric modulators (parmodulins) that select for Gα12/13 over Gαq.52
The arrangement and stoichiometry of PARs in cell membranes
We can draw from studies that have examined the arrangement of other GPCRs in cells and synthetic membranes to provide a context for the molecular arrangement of PARs.58-62 There is compelling evidence for both monomeric and dimeric/oligomeric GPCRs as a functional unit, as such, the topic remains highly debated.63-67 Experimental systems consisting of purified monomers in nanodiscs show that rhodopsin and β2-adrenergic receptors can mediate agonist-dependent nucleotide exchange from G-proteins.58,59 In constrast, Jastrzebska and colleagues demonstrated with cryoelectron microscopy that bovine rhodopsin purified from native tissues forms a heteropentamer consisting of 2 receptors in complex with a heterotrimeric G-protein.68,69 The matter becomes more complicated because the oligomeric status can be dynamic depending on the receptor and activation state. For example, the M2-muscarinic receptor forms stable tetramers, whereas the N-formyl peptide receptor is in equilibrium between monomers and dimers.70,71 In other cases, agonists and antagonists influence the organization of the receptors within the plasma membrane. The serotonin 5-HT-2C receptors are primarily dimers that become disrupted when treated with an antagonist and the M3-muscarinic receptors are found as dimers and tetramers.72-74 These studies highlight the potential dynamic nature of GPCR organization within the membrane. PARs are no exception as evidence exists for both constitutive and agonist-induced dimers.8,23,75-78
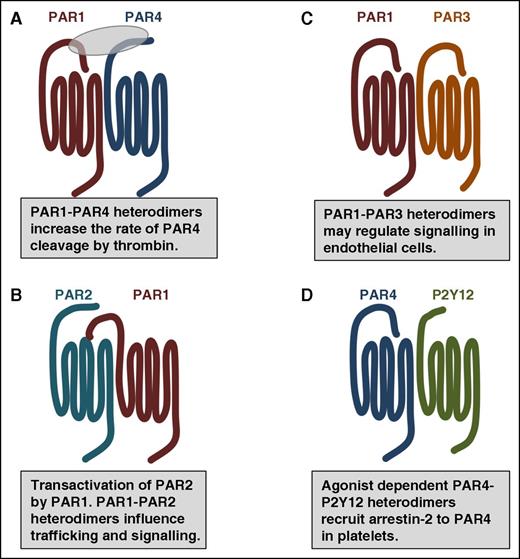
The major point of contention regarding the organization of GPCRs is defining a reliable readout for oligomerization of receptors in native tissues. PARs have a distinct advantage over other receptor families because they cooperate in specific ways that are consistent with dimerization or oligomerization. There are examples of PARs interacting to influence their rate of activation, downstream signaling, and trafficking in platelets, smooth muscle cells, endothelial cells, and podocytes (Figure 3).8 As described in “Proteolytic activation of PARs,” coordinated activity between PARs was first identified for PAR3 and PAR4 on mouse platelets and later for PAR1 and PAR4 on human platelets.22,23 In each case, the enhanced rate of PAR4 cleavage by thrombin is dependent on PAR4 heterodimerization.23,78,79 In platelets, PAR4 also forms agonist dependent heterodimers with the ADP receptor, P2Y12.76,77 Li et al showed that simultaneous stimulation of PAR4 and P2Y12 resulted in arrestin-2 recruitment to PAR4.77 Following recruitment, arrestin-2 served as a scaffold to support Akt signaling. There is a growing body of literature suggesting that heterodimers of GPCRs may influence signaling by acting as allosteric modulators.80 For years the signaling functions of PAR3 remained elusive. There are now emerging roles for PAR3 as an allosteric regulator of PAR1 on endothelial cells and PAR4 on mouse platelets by forming heterodimers and influencing signaling through specific G-proteins.44,75,79,81
PAR family members interact with one another and other GPCRs as homodimers and heterodimers. (A) In addition to homodimers, PAR1 and PAR4 form heterodimers that influence the rate of PAR4 activation by thrombin. (B) Thrombin-cleaved PAR1 can transactivate an adjacent PAR2. The PAR1-PAR2 heterodimer traffic within the cell as a unit and activate Rac1 pathways. (C) PAR1-PAR3 heterodimers may regulate cytoprotective signaling on endothelial cells. (D) In platelets, PAR4-P2Y12 form heterodimers that recruit β-arrestin-2 to PAR4 where it serves as a scaffold for AKT signaling.
PAR family members interact with one another and other GPCRs as homodimers and heterodimers. (A) In addition to homodimers, PAR1 and PAR4 form heterodimers that influence the rate of PAR4 activation by thrombin. (B) Thrombin-cleaved PAR1 can transactivate an adjacent PAR2. The PAR1-PAR2 heterodimer traffic within the cell as a unit and activate Rac1 pathways. (C) PAR1-PAR3 heterodimers may regulate cytoprotective signaling on endothelial cells. (D) In platelets, PAR4-P2Y12 form heterodimers that recruit β-arrestin-2 to PAR4 where it serves as a scaffold for AKT signaling.
A unique interaction between PAR family members is PAR1’s transactivation of PAR2 that was first described on endothelial cells by O’Brien and colleagues (Figure 3).82 In this model, the PAR1-tethered ligand generated by thrombin binds to the ligand-binding pocket of an adjacent PAR2; PAR2 is not cleaved by thrombin. These initial studies were followed up by Kaneider et al who demonstrated that thrombin signaling in endothelial cells switched from RhoA to Rac1 concomitantly with PAR1-PAR2 dimer formation.83 More recently, Lin and Trejo identified that PAR1 and PAR2 traffic together as heterodimers that are directed by PAR1 localization signals.84 They went on to show that thrombin stimulation induced internalization of the PAR1-PAR2 heterodimer. Finally, the PAR1-PAR2 heterodimer created an interface on PAR2 that bound β-arrestin that was distinct from the interface on PAR2 when it was expressed on its own.84 These studies provide a mechanism for thrombin-mediated β-arrestin signaling through the PAR1-PAR2 heterodimer. These new data suggest that the change in signaling observed by Kaneider and colleagues may due to β-arrestin signaling; this needs to be formally tested.
There are several lines of evidence for a functional interaction between PAR family members. However, the precise stoichiometry and geometric arrangement PARs are still under investigation. To date, all studies examining PAR dimerization have relied on fluorescence resonance energy transfer, bioluminescence energy transfer, or coimmunoprecipitation and cannot distinguish between dimers and higher-order oligomers such as tetramers. Future studies that use advanced microscopy such as spatial intensity distribution analysis and spectrally resolved 2-photon microscopy are necessary to shed light on the stoichiometry and dynamics of PAR homo- and hetero-oligomerization as they have for other GPCRs.72-74 The point remains that these high-powered techniques are, for the most part, limited to exogenously expressed receptors and are no substitution for ultimately analyzing receptors in native tissues. Naturally occurring mutations or polymorphisms may provide further evidence of cooperation between PARs.
Regulation of PAR expression and polymorphisms
There are multiple genetic risk factors for cardiovascular disease. The genetic and epigenetic control of PAR expression has the potential to define the sensitivity and signaling outcomes in response to proteases in a variety of physiological and pathophysiological settings. For example, alterations in flow patterns influence PAR2 expression in endothelial cells via Kruppel-like factor 2 (KLF2) and PAR4 expression is increased in vascular smooth muscle cells in diabetes.85-87 There have been relatively few studies examining the regulation of PAR promoters in cells that influence hemostasis. However, studies from the cancer literature show that the PAR1 promoter is regulated by p53, the androgen receptor, and activator protein 2 (AP2).88-90 Although PAR expression has not been directly connected to epigenetic factors, recent studies have correlated hypomethylation of f2rl3 (the PAR4 gene) to smoking in multiple populations.91-94 The decreased methylation of f2rl3 is linked to an increase in mortality due to coronary heart disease when other factors are controlled and is a potential biomarker.95-97 Future studies that examine PAR4 expression and function empirically in these populations may provide novel insights to disease progression.
Two transcription factors regulate multiple PARs in endothelial cells and vascular smooth muscle cells. KLF2 is thromboprotective in endothelial cells and its expression is upregulated by laminar flow.85 PAR1 and PAR2 are downregulated by KLF2; as a consequence, the endothelial cells no longer respond to thrombin. These results suggest that PAR1 and PAR2 are differentially regulated across the vasculature as the flow patterns change and that specific cellular responses will be dependent on the local environment.85,98 In vascular smooth muscle cells, PAR1 and PAR3 expression is controlled by nuclear factor of activated T cells.99,100 Interestingly, PAR3 expression is dynamic and thrombin-dependent. The apparent coordinated regulation of PAR expression in endothelial cells (PAR1 and PAR2) and vascular smooth muscle cells (PAR1 and PAR3) provides an attractive model for fine-tuning receptor regulation through heterodimerization and warrants further investigation (Figure 3). Linking the flow-mediated changes in expression to the detailed signaling studies in cultured endothelial cells will be a major challenge.
Genetic polymorphisms that alter the coding sequence, control expression levels, or regulate downstream signaling have been described for PAR1, PAR2, and PAR4.101-106 The challenge is establishing a direct link from the identified polymorphisms to receptor expression or function, and ultimately to a physiological output. For example, a polymorphism in an intron of the PAR1 gene (f2r) results in a lower density of PAR1 on the platelet surface and decreased aggregation in response to PAR1 agonists.103 However, in a clinical study with 660 patients who underwent percutaneous coronary intervention, there was no evidence of increased major adverse cardiovascular events or bleeding risk correlated with the polymorphism.107
The recently described difference in reactivity between PAR4 sequence variants provides the most compelling evidence that single nucleotide polymorphisms can influence a physiological response downstream of PARs.104,105 Edelstein and colleagues identified 2 single nucleotide polymorphisms (rs773902 and rs2227346) that result in sequence variants at positions 120 (Ala/Thr) in transmembrane helix 2 (TM2) and 296 (Phe/Val) in TM6, respectively (Figure 4A).104 The PAR4 variants correlate with platelet reactivity and are distributed by race. The G allele of rs773902 encodes for Ala at 120 and is found in 37% of black subjects compared with 81% of white subjects. In contrast, the A allele encodes for Thr at 120 and is found in 63% of blacks subjects and 19% of white subjects. In all cases, regardless of race, PAR4-120T is hyperreactive compared with PAR4-120A. The differences in platelet response were specific to PAR4 and could not be accounted for by changes in PAR4 expression. The reactivity of each variant was recapitulated when expressed in HEK293 cells demonstrating that it is an intrinsic property of the receptor. The PAR4 variants also have implications pharmacologically as PAR4-120T is resistant to the antagonist YD-3. Taken together, these data suggest that the amino acid change from Ala to Thr in TM2 likely alters ligand-binding site either directly or in an allosteric manner. The T allele of rs2227346 is common and encodes for Phe at 296, whereas the G allele is rare and encodes for Val at 296. It is of interest that PAR4-296V has low reactivity regardless of the amino acid at 120, which suggests a different mechanism by which it affects PAR4 function. Interestingly, individuals that are heterozygous for PAR4-296V have an overall low platelet reactivity even if they are homozygous for PAR4-120T. This suggests that PAR4-296V is acting as a dominant-negative receptor, potentially through homodimerization (Figure 4B).108 The PAR4 variants may provide a means to study the consequences of PAR4 homodimerization in native tissues.
Polymorphisms in the PAR4 gene (f2rl3) give rise to sequence variants that have altered reactivities. (A) PAR4-120T is hyperreactive compared with PAR4-120A. The rare PAR4 variant, PAR4-296V, has low reactivity regardless of the amino acid at 120. (B) Individuals heterozygous for PAR4-296V have low PAR4 reactivity and subsequent low platelet response to PAR4 agonists. A potential mechanism is that the hyporeactive allele sequesters the hyperreactive allele as a dominant-negative receptor.
Polymorphisms in the PAR4 gene (f2rl3) give rise to sequence variants that have altered reactivities. (A) PAR4-120T is hyperreactive compared with PAR4-120A. The rare PAR4 variant, PAR4-296V, has low reactivity regardless of the amino acid at 120. (B) Individuals heterozygous for PAR4-296V have low PAR4 reactivity and subsequent low platelet response to PAR4 agonists. A potential mechanism is that the hyporeactive allele sequesters the hyperreactive allele as a dominant-negative receptor.
These studies directly link PAR4 to the heritable interindividual variation in platelet reactivity, which is one of the genetic risk factors for cardiovascular disease and is greater in black than white individuals.109 The mechanism by which the PAR4 variants elicit their distinct effects on platelet function is not known. The polymorphisms may change the interactions of PAR4 within the membrane, allosterically alter ligand binding, or influence the transition of the receptor to an active state.110 Structural studies examining the differences between the PAR4 sequence variants are necessary to determine the molecular basis for the differences in reactivity.
Structural studies on PARs
Defining how the multiple ligands generated by alternative cleavage sites influence the overall rearrangement of PARs following activation at the atomic level would significantly move the field forward. The recent advancements of automated technology and protein engineering have facilitated great progress in the structural and biophysical data available for GPCRs since the first structure was solved 15 years ago.111,112 A high-resolution crystal structure of PAR1 bound to the antagonist vorapaxar was recently solved.113 Vorapaxar bound PAR1 within the transmembrane helices near the extracellular surface. However, it was also covered by residues from extracellular loop 2 (ECL2). It is unclear how vorapaxar gains access to the closed ligand-binding pocket. Zhang et al hypothesize that the unbound receptor may have a more open ligand-binding pocket.113
The insights gained from the high-resolution structure of PAR1 reveals how vorapaxar binds to PAR1. However, there was little structural information gleaned regarding how the native ligand interacts with the ligand-binding site. This is experimentally challenging because of the modifications required for crystallization. However, molecular simulations based on the PAR1 structure suggest the ligand interacts on the surface of the receptor rather than deep within the transmembrane bundle.113 These modeling studies are consistent with earlier studies mapping the ligand-binding site to specific residues at the extracellular surface (Leu 96, Asp 256, Glu 260, and Glu 347).10,113 Recently, Alsteens and colleagues developed a modification of atomic force microscopy to probe the ligand-binding site of PAR1.114 Based on these studies, the authors proposed a 2-step binding mechanism where the ligand first interacts in a low-affinity mode, then progresses to a high-affinity mode. It would be interesting to use this novel technique to examine how the alternative tethered ligands that are generated by noncanonical cleavage sites interact with the ligand-binding site. The specific structural rearrangements that occur following activation by the tethered ligand and how cofactors and heterodimers influence the receptor allostery are remaining questions that will require alternative techniques such as structural mass spectrometry.115,116
Targeting PARs for therapies
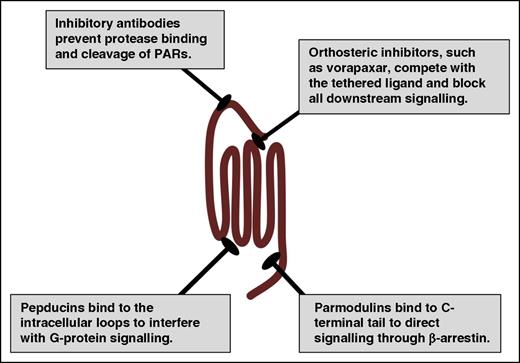
A primary goal of structural studies is to gain insights on the mechanisms by which receptors operate at the molecular level and how they interact with potential therapeutics. These details can provide the tools required for rational drug design and high-throughput screening of chemical libraries that can be the basis for lead compound development. The major hurdle with targeting PARs is that orthosteric compounds must compete with the nondiffusible tethered ligand, which has an estimated concentration of 400 μM.113 The only US Food and Drug Administration (FDA)-approved drug targeting PARs is vorapaxar (Zontivity), which has a black box warning for intracranial hemorrhage.117-119 Vorapaxar binds to PAR1 with high affinity and is essentially irreversible. This leads to challenging clinical management due to its pharmacokinetic and pharmacodynamics properties. There have been other approaches to targeting PARs therapeutically by interfering with the intracellular loops, blocking activation with antibodies, or directing signaling via allosteric modulators (Figure 5).52,53,120-122 These alternative strategies may provide a means to deal with the tethered ligand and potentially target a subset of PAR signaling events on specific cell types.
Therapeutic strategies for targeting PARs. PARs have been targeted with traditional orthosteric antagonists, such as vorapaxar for PAR1. In addition, PAR signaling may be inhibited at the initiation step by preventing the protease from cleaving the N terminus with blocking antibodies. Alternatively, pepducins target the intracellular face of the receptor to interfere with G-protein signaling. Parmodulins target the C-terminal eighth helix and have been selected to direct signaling to the cytoprotective pathways (β-arrestin) while blocking Gαq signaling.
Therapeutic strategies for targeting PARs. PARs have been targeted with traditional orthosteric antagonists, such as vorapaxar for PAR1. In addition, PAR signaling may be inhibited at the initiation step by preventing the protease from cleaving the N terminus with blocking antibodies. Alternatively, pepducins target the intracellular face of the receptor to interfere with G-protein signaling. Parmodulins target the C-terminal eighth helix and have been selected to direct signaling to the cytoprotective pathways (β-arrestin) while blocking Gαq signaling.
Future perspectives
The emerging picture shows that PAR signaling is fine-tuned through alternative cleavage sites, cofactors, and receptor oligomerization, which may provide innovative strategies to target PARs therapeutically. Further, as we continue to understand the subtle genetic and epigenetic factors that control PAR expression and activity, we may be able to target specific populations or diseases according to genotype. The key to furthering these developments is a balanced approach that determines: (1) the biophysical and structural requirements for PAR activation, (2) how the initiation of specific signaling pathways is controlled, and (3) the genetics that control expression and function of PARs or cofactors. Together, these will determine which of the PARs make the most sense to go after therapeutically and provide a strategy to move forward.
Acknowledgments
The author regrets that work from some colleagues could not be referenced or discussed due to space limitations.
This work was supported by the American Heart Association Grant-in-Aid (15GRNT25090222) and the National Institutes of Health National Heart, Lung, and Blood Institute (HL098217).
Authorship
Contribution: M.T.N. wrote the manuscript.
Conflict-of-interest disclosure: The author declares no competing financial interests.
Correspondence: Marvin T. Nieman, Department of Pharmacology, Case Western Reserve University, 2109 Adelbert Rd, Wood Building W305C, Cleveland, OH 44106; e-mail: nieman@case.edu.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal