Key Points
ENG regulatory elements target hemogenic mesoderm and hemogenic endothelium.
Hemogenic progenitors can be enriched using these elements as molecular probes to discover novel regulators of hematopoiesis.
Abstract
Enhancers are the primary determinants of cell identity, and specific promoter/enhancer combinations of Endoglin (ENG) have been shown to target blood and endothelium in the embryo. Here, we generated a series of embryonic stem cell lines, each targeted with reporter constructs driven by specific promoter/enhancer combinations of ENG, to evaluate their discriminative potential and value as molecular probes of the corresponding transcriptome. The Eng promoter (P) in combination with the −8/+7/+9-kb enhancers, targeted cells in FLK1 mesoderm that were enriched for blast colony forming potential, whereas the P/−8-kb enhancer targeted TIE2+/c-KIT+/CD41− endothelial cells that were enriched for hematopoietic potential. These fractions were isolated using reporter expression and their transcriptomes profiled by RNA-seq. There was high concordance between our signatures and those from embryos with defects at corresponding stages of hematopoiesis. Of the 6 genes that were upregulated in both hemogenic mesoderm and hemogenic endothelial fractions targeted by the reporters, LRP2, a multiligand receptor, was the only gene that had not previously been associated with hematopoiesis. We show that LRP2 is indeed involved in definitive hematopoiesis and by doing so validate the use of reporter gene–coupled enhancers as probes to gain insights into transcriptional changes that facilitate cell fate transitions.
Introduction
With advances in microscopy and histology, different cell types can now readily be distinguished from one another. However, the molecular characteristics that make each cell type unique and help distinguish stem cells from their more differentiated progeny in a tissue are still obscure. Harvesting pure populations of stem cells is a prerequisite to probing their molecular identity. Over the years, protocols combining flow cytometry with single-cell serial transplantation assays have been progressively refined to purify mouse and human adult hematopoietic stem cells (HSCs).1,2
One of the utilitarian benefits of determining the molecular fingerprint of an HSC is that it could serve as a measurable goal when developing protocols aimed at generating HSCs from differentiated cells.3 The failure of current protocols to generate long-term repopulating HSCs from embryonic stem/induced pluripotent stem (ES/iPS) cells is attributed in part to our incomplete understanding of the developmental journey that mesodermal progenitors traverse in the embryo when generating the complement of HSCs that are resident in the bone marrow of a newborn.4 Determining the molecular identities of embryonic HSC precursors is complicated by the lack of consensus regarding the precise HSC intermediates in the embryo, functional assays that are less than ideal for assessment of these intermediates and knowledge that these intermediates are transitory cell populations that are present in very small numbers.5 FLK1 expressing mesodermal cells in the posterior primitive streak when isolated from the embryo and cultured in vitro generate blast colonies that have blood, endothelial, and vascular smooth muscle potential.6 Blast colony forming cell (BL-CFC) potential in FLK1+ mesoderm has been estimated to be ∼1:300.7 Hemogenic potential in TIE2+c-KIT+ hemogenic endothelium (HE) or VE-CAD+CD45−CD41− pre-HSC cells in the dorsal aorta that transit to hematopoietic cells range from 1:100 to 1:300.8-10 These functional estimates are too low to probe the molecular identities of either the early hemangioblast or HE cell populations in the developing embryo using currently available protocols.
Cell identity is encoded within the sequences of tissue-specific gene regulatory elements (GREs) that direct and coordinate gene expression in a cell.11 A number of regulatory elements of hematopoietic transcription factors (TFs) have previously been shown to direct reporter expression to developing blood cells in the mouse embryo and include enhancers of Scl, Runx1, Gata2, Erg, Fli1, Lmo2, and Lyl1, which also form a recursive circuit in human adult HSCs.12 The Runx1+23 enhancer marks a population of early HE cells that transit to HSCs and has been used to isolate cells from different embryonic stages for transcriptomic analysis.13 Ly6a/Sca1 and Eng (CD105) serve as useful cell surface markers for isolation of murine HSC fractions.14,15 The promoter of Ly6a and promoter/enhancer combinations of Eng also target embryonic hematopoiesis and in the case of the former have been used in conjunction with a reporter to isolate HE cells and HSCs from early embryos.16-18
Endoglin (ENG) is an accessory receptor and modulator of TGF-β superfamily signaling.19 ENG is expressed on FLK1+ mesoderm and is required for normal BL-CFC development, and its expression facilitates the hematopoietic program in these cells.10,20 ENG null mice die at E9.5 with vascular defects due to abnormal endothelial and pericyte development.21 It is also a marker of adult murine HSCs that was identified using a Scl+19-driven fluorescent reporter coupled with transcriptomic and proteomic assessment of purified cells.15 An emerging concept of developmental hematopoiesis posits that HSC development from the dorsal aorta at E10 reflects maturation of cells that were fated earlier during embryogenesis toward the hematopoietic lineage.13 As such, we rationalized that transcriptional regulation of ENG, which is functionally important for the development of hematopoietic intermediates, could be instructive in helping elucidate the transcriptional environment of these cells. We have previously shown that sequence information within the promoter and hematoendothelial enhancers of Eng determine how reporter genes are targeted to either endothelial or blood and endothelial tissues in the embryo.17,22 Given the spectrum of cell types that are involved in the developmental journey of embryonic HSCs and the deterministic role that ENG plays in their development, we hypothesized that distinct combinations of promoter/enhancers of this gene are used by different hematopoietic intermediates to regulate ENG expression. We rationalized that if distinct promoter/enhancer constructs indeed targeted functionally distinct hematopoietic intermediates, they could be used as molecular probes to profile the transcriptional environment of these cells.
Here, we show using ES cells with single-copy reporter-coupled transgenes targeted to the constitutively active HPRT locus that distinct promoter/enhancer combinations of ENG are used by FLK1+ mesoderm and HE that are enriched for BL-CFC and hematopoietic potential, respectively. Using these reporter-coupled transgenes as probes to harvest cell populations from embryonic stem cell (ESC) differentiation assays, we performed RNA-seq to identify gene sets that were associated with functional enrichment of hematopoietic potential and show their complementarity with primary mouse tissues at matching stages of development.
Of the 6 genes that were upregulated in both hemogenic mesoderm (HB) and HE fractions targeted by the reporters, LRP2, a multiligand receptor, was the only gene that had not previously been associated with hematopoiesis. Here, we show that LRP2 is indeed involved in aorta-gonad-mesonephros (AGM) hematopoiesis and by doing so validate the use of reporter gene–coupled enhancers as a discovery tool.
Materials and methods
Murine ES cell culture and Hprt targeting
ES cell differentiation into embryoid bodies (EBs) and lacZ staining
To generate EBs, ES cells were collected and cultured as detailed in the supplemental data.
Flow cytometry and cell sorting
Cells were collected from EBs and liquid blast cultures and dissociated into a single-cell suspension. Details of the procedure and antibodies are listed in the supplemental data.
Methylcellulose blast BL-CFC assay and liquid cultures
Details of the BL-CFC assay and liquid cultures are listed in the supplemental data.
Hematopoietic methylcellulose colony-forming assay
Cells isolated from day 2 or 4 of liquid blast cultures and seeded for colony-forming unit–cell (CFU-C) assays as detailed in the supplemental data.
Reverse transcription polymerase chain reaction (RT-PCR)
A list of primers and methods are supplied in the supplemental data.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed as detailed previously.22 See the supplemental data for a list of primers and experimental details.
Mouse embryo immunostaining and imaging
Details of the procedure and antibodies are listed in the supplemental data.
Generating zebrafish morpholinos and analysis
Details of generating zebrafish morpholinos and the analysis are listed in the supplemental data.
Statistical analysis
RT-PCR data, BL-CFCs, and hematopoietic colony counts were statistically analyzed using Student t test or paired Student t test.
RNA sequencing and analysis
The data have been deposited in the Gene Expression Omnibus under the accession number GSE77390. The Ingenuity IPA Core Analysis Tool (version 17199142) and the GSEA Java Desktop tool (v 2.0.13) were used for analysis. See the supplemental data for details.
Study approval
All experiments were approved by the institutional biological committee and animal care and ethics committee of the University of New South Wales, Manchester University, and Harvard University.
Results
Mesoderm to hemangioblast transition is accompanied by increased Eng expression and chromatin accessibility at hematoendothelial regulatory elements
The promoter of ENG when coupled with −8-kb, +7-kb, and +9-kb enhancers have previously been shown to direct reporter expression to either endothelial or blood and endothelial tissues in the embryo (Figure 1A17,22 ). The Bry-GFP ESC line has been used extensively to investigate the developmental progression of premesoderm (GFP−/FLK1−) to prehemangioblast mesoderm (GFP+/FLK1−; G+/F−) to the hemangioblast (GFP+/FLK1+; G+F+)7 (Figure 1B). We used this cell line to first evaluate expression of Eng and chromatin accessibility at hematoendothelial regulatory elements of Eng17,22 because cells progressed from prehemangioblast mesoderm to hemangioblast mesoderm. Eng expression increased by ∼3-fold (Figure 1C) and enrichments of H3K9 acetylation (an active chromatin mark) increased ∼10- to 20-fold at the Eng promoter and −8-kb, +7-kb, and +9-kb Eng enhancers (Figure 1D). There was no change in H3K9Ac at −4-kb, a region that is highly conserved across species but shows no enhancer activity.22
Mesoderm to hemangioblast transition is accompanied by increased Eng expression and chromatin accessibility at hematoendothelial regulatory elements. (A) Schematic representation of the ENG locus. The transcription start site is marked with an arrow. The −8-kb, +7-kb, and +9-kb enhancers and the promoter (P) are marked in orange; exons are marked in brown, and the 5′untranslated region is marked in cyan. (B) Schematic representation of Bry-GFP ES cell differentiation. At day 3 of EB differentiation, Bry-GFP+/FLK1− (G+F−) and Bry-GFP+/FLK1+ (G+F+) cells were sorted and analyzed by RT-PCR and ChIP. (C) Bar graph shows Eng mRNA expression levels in sorted FLK1+ve and –ve mesodermal cell populations in day 3 EBs generated from Bry-GFP ES cells. (D) Bar graph shows levels of enrichment of the active chromatin mark, H3K9Ac at Eng −8, P, +7, and +9 hematoendothelial enhancers relative to immunoglobulin G in prehemangioblast mesoderm (G+F−; black) and in hemangioblast mesoderm (G+F+; gray). Eng −4 was included as a negative control region. **P < .01, ***P < .001.
Mesoderm to hemangioblast transition is accompanied by increased Eng expression and chromatin accessibility at hematoendothelial regulatory elements. (A) Schematic representation of the ENG locus. The transcription start site is marked with an arrow. The −8-kb, +7-kb, and +9-kb enhancers and the promoter (P) are marked in orange; exons are marked in brown, and the 5′untranslated region is marked in cyan. (B) Schematic representation of Bry-GFP ES cell differentiation. At day 3 of EB differentiation, Bry-GFP+/FLK1− (G+F−) and Bry-GFP+/FLK1+ (G+F+) cells were sorted and analyzed by RT-PCR and ChIP. (C) Bar graph shows Eng mRNA expression levels in sorted FLK1+ve and –ve mesodermal cell populations in day 3 EBs generated from Bry-GFP ES cells. (D) Bar graph shows levels of enrichment of the active chromatin mark, H3K9Ac at Eng −8, P, +7, and +9 hematoendothelial enhancers relative to immunoglobulin G in prehemangioblast mesoderm (G+F−; black) and in hemangioblast mesoderm (G+F+; gray). Eng −4 was included as a negative control region. **P < .01, ***P < .001.
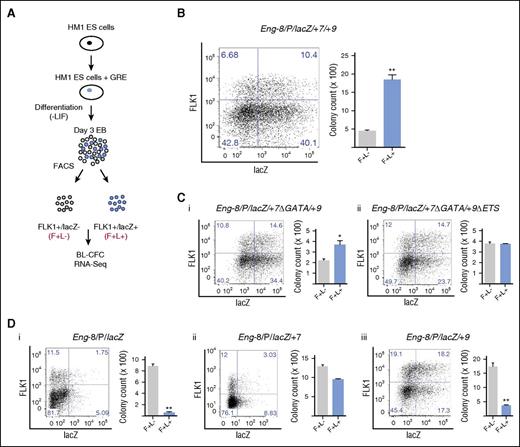
The Eng promoter, when combined with the −8, +7, and +9 hematoendothelial enhancers, targets FLK1+ mesodermal cells enriched for BL-CFC potential
HM1 ESCs have a disrupted Hprt locus that can be reconstituted by homologous recombination of a targeting vector.23 They serve as a useful tool to evaluate reporter activity of single copies of GREs at a constitutively active locus at different stages of ESC differentiation. We took advantage of this system to introduce combinations of Eng regulatory elements with blood and endothelial activity in in vivo transgenic assays (supplemental Figure 1). Successful recombination and generation of ESC lines with −8/P/lacZ, −8/P/lacZ/+7, −8/P/lacZ/+9, −8/P/lacZ/+7/+9, −8/P/lacZ/+7Δ (GATA)/+9, and −8/P/lacZ/+7Δ (GATA)/+9 Δ (ETS), were confirmed by RT-PCR and southern blotting (supplemental Figure 2). We used these ESC lines as a toolkit with which to track, evaluate, and compare the activity of each of these GREs during different stages of hematopoietic development and to fractionate cells for functional validation and transcriptomic analysis.
To identify which, if any, of the Eng GREs targeted FLK1+ mesoderm enriched for hemangioblast potential, we generated EBs from each ESC line and fractionated FLK1+lacZ− (F+L−) and FLK1+lacZ+ (F+L+) cells and performed BL-CFC assays (Figure 2A). The Eng −8/P/lacZ/+7/+9 construct, which showed robust blood and endothelial staining in vivo,17 targeted a fraction of the FLK1+ mesoderm that showed increased (∼4-fold) BL-CFC potential (Figure 2B; supplemental Figure 3A). We have previously shown that mutating the GATA binding motifs in +7 and ETS binding motifs in +9 diminished endothelial activity and extinguished hematopoietic activity of the Eng −8/P/lacZ/+7/+9 construct in transgenic assays.17 There was a corresponding reduction or failure of the mutant constructs to preferentially target cells with BL-CFC potential (Figure 2Ci,ii; supplemental Figure 3B). The −8P/lacZ construct showed strong endothelial but no hematopoietic activity in transgenic assays.17 FLK1+ cells targeted by this construct (F+L+) showed significantly lower BL-CFC potential than F+L− cells (Figure 2Di; supplemental Figure 3A). For the −8P/lacZ/+7 and −8P/lacZ/+9 constructs, which showed strong endothelial and low to moderate hematopoietic activity in in vivo transgenic assays,17 FLK1+ mesoderm (F+L+) had either lower or equivalent BL-CFC activity than FLK1+ (F+L−) cells (Figure 2Dii,iii; supplemental Figure 3A). It is important to note that the total number of BL-CFCs generated by FLK1+ mesoderm will vary from clone to clone but that comparisons are of BL-CFC potential of L+ and L− sorted FLK1+ cells from each clone.
The Eng promoter when combined with the −8, +7, and +9 hematoendothelial enhancers targets FLK1+ mesodermal cells enriched for BL-CFC potential. (A) Schematic representation of the experimental procedure. The Eng −8/P/LacZ, Eng −8/P/LacZ/+7, Eng −8/P/LacZ/+9, Eng −8/P/LacZ/+7/+9, Eng −8/P/LacZ/+7Δ/+9, and Eng −8/P/LacZ/+7Δ/+9Δ reporter constructs were introduced by homologous recombination into the HPRT locus of HM1 ES cells. Recombinant clones were differentiated into day 3 EBs and stained for FLK1 expression and β-galactosidase activity. FLK1+/LacZ− (F+/L−; gray) and FLK1+/LacZ+ (F+/L+; blue) cells were sorted and seeded into BL-CFC assays. Fractions sorted from the Eng −8/P/LacZ/+7/+9 were further analyzed by RNA sequencing. (B) Flow cytometry profiles of Eng −8/P/LacZ/+7/+9 day 3 EBs (left). BL-CFCs from sorted F+/L− (gray) and F+/L+ (blue) fractions. (C) (i) Flow cytometry profile of day 3 EBs derived from ES cells targeted with Eng −8/P/LacZ/+7Δ/+9 (mutated GATA motifs in the +7 enhancer) is shown to the left with corresponding BL-CFCs from sorted F+/L− (gray) and F+/L+ (blue) fractions shown to the right. (ii) Flow cytometry profile of day 3 EBs derived from ES cells targeted with Eng −8/P/LacZ/+7Δ/+9Δ (mutated GATA motifs in the +7 enhancer and mutated ETS motifs in the +9 enhancer) and corresponding BL-CFCs from sorted F+/L− (gray) and F+/L+ (blue) fractions. (D) Flow cytometry profiles of day 3 EBs and BL-CFCs from sorted F+/L− (gray) and F+/L+ (blue) fractions are shown for ES cells targeted with (i) Eng −8/P/LacZ, (ii) Eng −8/P/LacZ/+9, and (iii) Eng −8/P/LacZ/+7. BL-CFC counts are the total number of blast colonies generated from 2 × 104 seeded cells. Statistical analysis was conducted using Student t test, *P < .05, **P < .01.
The Eng promoter when combined with the −8, +7, and +9 hematoendothelial enhancers targets FLK1+ mesodermal cells enriched for BL-CFC potential. (A) Schematic representation of the experimental procedure. The Eng −8/P/LacZ, Eng −8/P/LacZ/+7, Eng −8/P/LacZ/+9, Eng −8/P/LacZ/+7/+9, Eng −8/P/LacZ/+7Δ/+9, and Eng −8/P/LacZ/+7Δ/+9Δ reporter constructs were introduced by homologous recombination into the HPRT locus of HM1 ES cells. Recombinant clones were differentiated into day 3 EBs and stained for FLK1 expression and β-galactosidase activity. FLK1+/LacZ− (F+/L−; gray) and FLK1+/LacZ+ (F+/L+; blue) cells were sorted and seeded into BL-CFC assays. Fractions sorted from the Eng −8/P/LacZ/+7/+9 were further analyzed by RNA sequencing. (B) Flow cytometry profiles of Eng −8/P/LacZ/+7/+9 day 3 EBs (left). BL-CFCs from sorted F+/L− (gray) and F+/L+ (blue) fractions. (C) (i) Flow cytometry profile of day 3 EBs derived from ES cells targeted with Eng −8/P/LacZ/+7Δ/+9 (mutated GATA motifs in the +7 enhancer) is shown to the left with corresponding BL-CFCs from sorted F+/L− (gray) and F+/L+ (blue) fractions shown to the right. (ii) Flow cytometry profile of day 3 EBs derived from ES cells targeted with Eng −8/P/LacZ/+7Δ/+9Δ (mutated GATA motifs in the +7 enhancer and mutated ETS motifs in the +9 enhancer) and corresponding BL-CFCs from sorted F+/L− (gray) and F+/L+ (blue) fractions. (D) Flow cytometry profiles of day 3 EBs and BL-CFCs from sorted F+/L− (gray) and F+/L+ (blue) fractions are shown for ES cells targeted with (i) Eng −8/P/LacZ, (ii) Eng −8/P/LacZ/+9, and (iii) Eng −8/P/LacZ/+7. BL-CFC counts are the total number of blast colonies generated from 2 × 104 seeded cells. Statistical analysis was conducted using Student t test, *P < .05, **P < .01.
Taken together, these data show that the GREs of ENG that showed increased chromatin accessibility as prehemangioblast mesoderm progressed to hemangioblast mesoderm (ie, −8/P/+7/+9; Figure 1D) act collectively to target reporter gene expression to BL-CFCs in FLK1+ mesoderm. It is noteworthy that in in vivo transgenic assays, it was also this construct (−8/P/lacZ/+7/+9) that had the strongest and most specific activity in blood and endothelium in the developing embryo.17
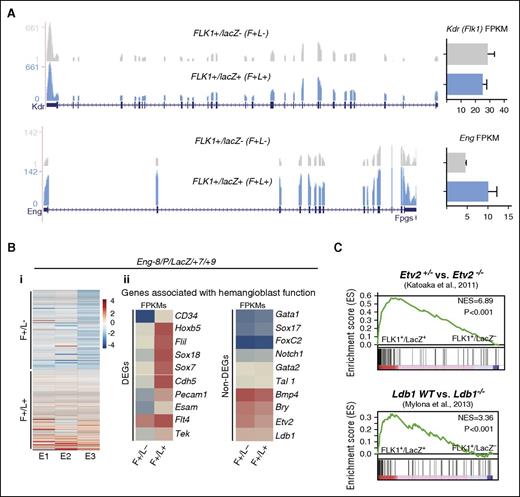
Global transcriptomic analysis of FLK1 mesoderm targeted by Eng −8/P/LacZ/+7/+9 identifies genes associated with hemangioblast activity
To discover genes associated with the activation of these GREs and increased activity of the reporters, we performed RNA sequencing on sorted lacZ+ and lacZ− cell fractions from 3 independent experiments. As expected, Kdr (Flk1) expression was comparable in both fractions and Eng transcripts were increased in the lacZ+ fraction consistent with Eng GRE-driven reporter activity (Figure 3A). There was also a shared set of genes that was consistently differentially expressed between the lacZ+ and lacZ− cell fractions (Figure 3Bi) and included 107 upregulated and 101 downregulated genes. These genes included cell surface receptors (CSRs) and TFs known to be associated with blood and endothelium (fold change ≥2 and P value < .5; supplemental Table 1; supplemental Figure 4A) and genes that have been associated with hemangioblast development in the LifeMap Sciences embryonic development compendium (Figure 3Bii). Individually, the expression of many genes known to be associated with early mesoderm (eg, Bry/T and Bmp4), blood (eg, Gata1 and Tal1), and endothelial (eg, Foxc2 and Etv2) development did not vary significantly between these cell fractions (Figure 3Bii). Indeed, as hemangioblasts are a subpopulation of FLK1+ mesoderm with multilineage differentiation potential, it would have been unusual to see significant differences in expression of individual genes that are strongly associated with commitment to a specific lineage. However, ingenuity pathway analysis (IPA) revealed that when differentially expressed genes were considered as a collective, there were strong associations with blood and blood vessel development for genes in the FLK1+/lacZ+ set (supplemental Figure 4B). Consistent with these biological functions, this gene set also showed significant associations with signaling pathways that govern endothelial development and endothelial nitric oxide synthase signaling (supplemental Figure 4C-F).
RNA sequencing of FLK1 mesoderm targeted by Eng −8/P/LacZ/+7/+9 identifies genes associated with hemangioblast activity. (A) RNA-sequencing profiles shows Kdr (Flk1) transcripts (top) and Eng transcripts (bottom) in the F+/L− and F+/L+ fractions. FPKM expression values are shown to the right. (B) (i) Heat map representation of up- and downregulated genes in FLK1+/LacZ− (F+/L−) and FLK1+/LacZ+ (F+/L+) fractions in 3 independent experiments. (ii) Expression (FPKM values) levels of genes that have previously been associated with hemangioblast function. The left panel shows a subset of genes that are differentially expressed between F+/L− and F+/L+ fractions, and the right panel shows a subset of genes that are not. (C) GSEA profiles shows the correspondence of genes that are differentially expressed between F+/L− and F+/L+ fractions and those that are differentially expressed in ETV2+/− vs ETV2−/− (top) and LDB wt vs LDB−/− gene sets. DEG, differentially expressed genes.
RNA sequencing of FLK1 mesoderm targeted by Eng −8/P/LacZ/+7/+9 identifies genes associated with hemangioblast activity. (A) RNA-sequencing profiles shows Kdr (Flk1) transcripts (top) and Eng transcripts (bottom) in the F+/L− and F+/L+ fractions. FPKM expression values are shown to the right. (B) (i) Heat map representation of up- and downregulated genes in FLK1+/LacZ− (F+/L−) and FLK1+/LacZ+ (F+/L+) fractions in 3 independent experiments. (ii) Expression (FPKM values) levels of genes that have previously been associated with hemangioblast function. The left panel shows a subset of genes that are differentially expressed between F+/L− and F+/L+ fractions, and the right panel shows a subset of genes that are not. (C) GSEA profiles shows the correspondence of genes that are differentially expressed between F+/L− and F+/L+ fractions and those that are differentially expressed in ETV2+/− vs ETV2−/− (top) and LDB wt vs LDB−/− gene sets. DEG, differentially expressed genes.
To investigate the in vivo relevance of our gene set, we used GSEA analysis to compare expression overlaps with gene expression data from FLK1+ cells in ETV224,25 and LDB126 knockout embryos, both of which are defective in hemangioblasts (Figure 3C). There were strong overlaps between genes expressed in FLK1+ mesoderm targeted by ENG −8/P/lacZ/+7/+9 and genes expressed in hemangioblast competent wild-type ETV2 and LDB1 embryos compared with ETV2+/− or LDB1−/− embryos, respectively. Therefore, the molecular signature of BL-CFC-enriched FLK1+ mesoderm that was identified using differential reporter activity of ENG GREs is consistent with in vivo functional capacity.
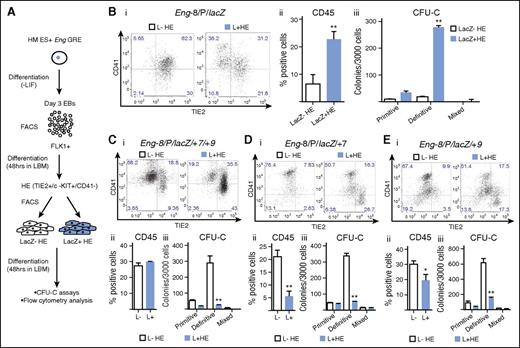
The Eng promoter in combination with the −8 endothelial enhancer targets HE cells enriched for hematopoietic potential
Definitive hematopoiesis in the embryo progresses through a TIE2+/c-KIT+/CD41− HE intermediate.9 We used a cell culture system that mirrors this in vivo transition to investigate whether any of the reporter ESC lines preferentially targeted HE cells and whether they could be used to isolate cell fractions that were enriched for hematopoietic potential.9 To this end, FLK1+ cells were sorted from day 3 EBs and seeded into liquid blast culture media (Figure 4A). At day 2 of culture, lacZ+ and lacZ− HE cells were isolated by fluorescence-activated cell sorter and reseeded into liquid blast media (LBM) for 2 further days followed by flow cytometry and CFU-C assays to evaluate the hematopoietic potential of each fraction. Of the reporter ESC lines, Eng −8/P/lacZ was unique in that it was active in a fraction of HE cells that generated more TIE2−/CD41+ and CD45+ cells after 48 hours in culture (Figure 4Bi),ii) and contained almost all CFU-C potential (Figure 4Biii). Whereas Eng −8/P/lacZ/+7/+9 targeted FLK1+ mesoderm with increased BL-CFC potential (Figure 2), it did not target HE cells with increased hematopoietic potential (Figure 4Ci-iii). Indeed, this and each of the other constructs targeted HE cells that had lower hematopoietic potential (Figure 4D,E; supplemental Figure 5). Taken together, these data showed that not only was there a specific combination of ENG GREs that targeted HE cells but also the combination was distinct from that which targeted BL-CFCs in FLK1+ mesoderm.
The Eng promoter in combination with the −8 endothelial enhancer targets HECs enriched for hematopoietic potential. (A) Schematic diagram outlining the experimental procedure. Recombinant ES cells generated using the Eng reporter constructs were differentiated into day 3 EBs. FLK1+ mesodermal cells were sorted from representative clones for each recombinant ES cell line and cultured in LBM. At 48 hours, CD41−/TIE2+/c-KIT+ (HE) cells were sorted into lacZ+ and lacZ– fractions. The sorted fractions were recultured in LBM for a further 48 hours followed by flow cytometry and CFU-C assays. (B) (i) CD41 and TIE2 expression in sorted c-KIT+ HE LacZ− (white) and LacZ+ (blue) fractions (after 2 days of reculture) derived from Eng −8/P/LacZ ES cells. (ii) Percentage of CD45+ cells generated from LacZ− and LacZ+ HE fractions. (iii) Bar chart shows the number and type of hematopoietic colonies generated by each fraction. (C) (i-iii) Corresponding data to (B) generated from Eng −8/P/LacZ/+7/+9 ES cells. (D) (i-iii) Corresponding data to (B) generated from Eng −8/P/LacZ/+7 ES cells. (E) (i-iii) Corresponding data to (B) generated from Eng −8/P/LacZ/+9 ES cells. Primitive and definitive colonies were scored after 4 and 9 days, respectively. Statistical analysis was conducted using Student t test, *P < .05, **P < .01.
The Eng promoter in combination with the −8 endothelial enhancer targets HECs enriched for hematopoietic potential. (A) Schematic diagram outlining the experimental procedure. Recombinant ES cells generated using the Eng reporter constructs were differentiated into day 3 EBs. FLK1+ mesodermal cells were sorted from representative clones for each recombinant ES cell line and cultured in LBM. At 48 hours, CD41−/TIE2+/c-KIT+ (HE) cells were sorted into lacZ+ and lacZ– fractions. The sorted fractions were recultured in LBM for a further 48 hours followed by flow cytometry and CFU-C assays. (B) (i) CD41 and TIE2 expression in sorted c-KIT+ HE LacZ− (white) and LacZ+ (blue) fractions (after 2 days of reculture) derived from Eng −8/P/LacZ ES cells. (ii) Percentage of CD45+ cells generated from LacZ− and LacZ+ HE fractions. (iii) Bar chart shows the number and type of hematopoietic colonies generated by each fraction. (C) (i-iii) Corresponding data to (B) generated from Eng −8/P/LacZ/+7/+9 ES cells. (D) (i-iii) Corresponding data to (B) generated from Eng −8/P/LacZ/+7 ES cells. (E) (i-iii) Corresponding data to (B) generated from Eng −8/P/LacZ/+9 ES cells. Primitive and definitive colonies were scored after 4 and 9 days, respectively. Statistical analysis was conducted using Student t test, *P < .05, **P < .01.
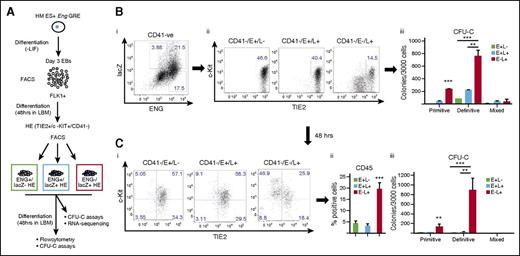
Hematopoietic potential is highest in Eng −8/P/lacZ targeted HE cells that do not as yet express surface ENG
Cell fate transitions are dynamic, and our purpose was to use these reporter constructs to capture HE cells that were intrinsically fated toward the hematopoietic lineage at the earliest possible time point in culture. Based on the assumption that there would be a delay between transactivating the Eng GRE reporter and surface expression of ENG, we repeated the experiments described in Figure 4 using the Eng −8/P/lacZ ES cell line but here also incorporating surface ENG expression to isolate TIE2+/C-KIT+/CD41− HE fractions that were ENG+/lacZ−, ENG+/lacZ+, or ENG−/lacZ+ (Figure 5A). Interestingly, CFU-C potential within the lacZ+ fraction was highest in ENG−/lacZ+ HE cells (Figure 5Bi,ii). ENG+/lacZ−, ENG+/lacZ+, and ENG−/lacZ+ HE cells were reseeded in LBM and analyzed by flow cytometry and CFU-C assays after 2 further days of culture. The proportions of TIE2−/CD41+ (Figure 5Ci) and CD45+ (Figure 5Cii) cells and CFU-C potential (Figure 5Ciii) were highest for cultured ENG−/lacZ+ HE cells. Taken together, these data show that the hematopoietic potential within HE cells can be targeted by Eng −8/P/lacZ and that these ESCs could be used to interrogate the earliest transcriptional changes associated with this cell fate decision.
Hematopoietic potential is highest in Eng −8/P/lacZ targeted HE cells that do not express surface ENG. (A) Schematic diagram outlining the experimental procedure. FLK1+ mesodermal cells were sorted from day 3 EBs generated from the Eng −8/P/LacZ recombinant ES cell line and cultured in liquid blast culture media. At 48 hours, CD41−/TIE2+/c-KIT+ (HE) cells were sorted into ENG+/LacZ−, ENG+/LacZ+, and ENG−/LacZ+ fractions. These fractions were either directly seeded into CFU-C assays (B) or recultured in LBM for a further 48 hours and analyzed by flow cytometry and CFU-C assays (C). (B) (i-ii) Flow cytometry shows the frequencies of CD41−/TIE2+/c-KIT+ (HE) cells in ENG+/lacZ+, ENG+/lacZ−, and ENG-lacZ+ fractions. (iii) CFU-C potential of each sorted fractions in (i). (C) (i) Flow cytometry analysis of CD41 and TIE2 expression in sorted HE cell fractions after 2 days of reculture in LBM. (ii) Bar chart shows the percentage of CD45− positive cells in sorted fraction. (iii) Bar chart shows hematopoietic colony numbers from each fraction. Primitive and definitive colonies were scored after 4 and 9 days, respectively. Statistical analysis was conducted using Student t test, **P < .01, and ***P < .001.
Hematopoietic potential is highest in Eng −8/P/lacZ targeted HE cells that do not express surface ENG. (A) Schematic diagram outlining the experimental procedure. FLK1+ mesodermal cells were sorted from day 3 EBs generated from the Eng −8/P/LacZ recombinant ES cell line and cultured in liquid blast culture media. At 48 hours, CD41−/TIE2+/c-KIT+ (HE) cells were sorted into ENG+/LacZ−, ENG+/LacZ+, and ENG−/LacZ+ fractions. These fractions were either directly seeded into CFU-C assays (B) or recultured in LBM for a further 48 hours and analyzed by flow cytometry and CFU-C assays (C). (B) (i-ii) Flow cytometry shows the frequencies of CD41−/TIE2+/c-KIT+ (HE) cells in ENG+/lacZ+, ENG+/lacZ−, and ENG-lacZ+ fractions. (iii) CFU-C potential of each sorted fractions in (i). (C) (i) Flow cytometry analysis of CD41 and TIE2 expression in sorted HE cell fractions after 2 days of reculture in LBM. (ii) Bar chart shows the percentage of CD45− positive cells in sorted fraction. (iii) Bar chart shows hematopoietic colony numbers from each fraction. Primitive and definitive colonies were scored after 4 and 9 days, respectively. Statistical analysis was conducted using Student t test, **P < .01, and ***P < .001.
Transcriptomic analysis of HE fractions identifies genes associated with HE to hematopoietic transition
To discover genes that act on Eng −8/P/lacZ and drive reporter gene activity and by extension are associated with hemogenic potential in TIE2+/C-KIT+/CD41− HE cells, we performed RNA sequencing on sorted ENG+/lacZ−, ENG+/lacZ+, and ENG−/lacZ+ HE cell fractions from 3 independent experiments. As expected, the fractions, which expressed surface ENG, had abundant Eng transcripts, which were still comparatively low in ENG−/lacZ+ HE cells (Figure 6A). Consistent with its role as a major determinant of endothelial-to-hematopoietic transition (EHT),9,27,28 Runx1 transcripts were abundant in HE cells that were enriched with functional hemogenic cells (ENG−/lacZ+ HE) and relatively low in those (ENG+/lacZ− and ENG+/lacZ+ HE) that were not. In total, there were 707 upregulated and 981 downregulated genes in ENG−/lacZ+ HE cells compared with ENG+/lacZ− and ENG+/lacZ+ HE cells (Figure 6Bi; supplemental Table 2). It was interesting to note that only a subset of genes that have previously been attributed to mark HE cells based on cell surface protein expression were16 differentially expressed between these functionally distinct HE subpopulations (Figure 6Bii). This observation does not imply that these genes are not important but that their higher or lower expression is not associated with these early subtle transitions.
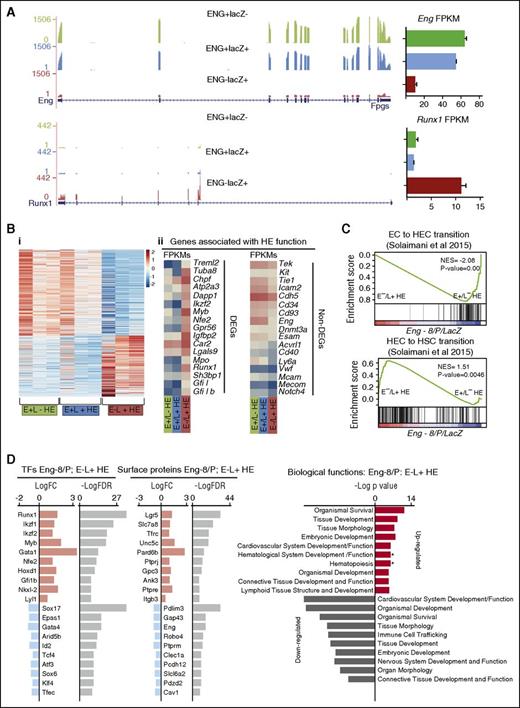
Transcriptomic analysis of HE fractions identifies genes associated with HE to hematopoietic transition. (A) RNA-sequencing profiles show Eng transcripts (top) and Runx1 transcripts (bottom) in the E+/L−, E+/L+, and E−/L+ fractions. FPKM expression values are shown to the right. (B) (i) Heat map representation of up- and downregulated genes in ENG+/LacZ− (E+/L−) HE, ENG+/LacZ+ (E+/L+) HE, and ENG−/LacZ+ (E−/L+) HE fractions in 3 independent experiments. (ii) Expression (FPKM values) levels of genes that have previously been associated with HE. The top panel shows a subset of genes that are differentially expressed between E+/L−, E+/L+, and E−/L+ fractions, and the bottom panel shows a subset of genes that are not. (C) GSEA profiles show the correspondence of genes that are differentially expressed between the E+/L−, E+/L+, and E−/L+ fractions and those that are differentially expressed in EC vs HECs (top) and HECs vs HSC gene sets. (D) TFs and CSRs that are up- and downregulated in the Eng −8/P E−/L+ HE fraction. The log fold changes (logFC) and log false discovery rates (logFDR) are listed for each gene.
Transcriptomic analysis of HE fractions identifies genes associated with HE to hematopoietic transition. (A) RNA-sequencing profiles show Eng transcripts (top) and Runx1 transcripts (bottom) in the E+/L−, E+/L+, and E−/L+ fractions. FPKM expression values are shown to the right. (B) (i) Heat map representation of up- and downregulated genes in ENG+/LacZ− (E+/L−) HE, ENG+/LacZ+ (E+/L+) HE, and ENG−/LacZ+ (E−/L+) HE fractions in 3 independent experiments. (ii) Expression (FPKM values) levels of genes that have previously been associated with HE. The top panel shows a subset of genes that are differentially expressed between E+/L−, E+/L+, and E−/L+ fractions, and the bottom panel shows a subset of genes that are not. (C) GSEA profiles show the correspondence of genes that are differentially expressed between the E+/L−, E+/L+, and E−/L+ fractions and those that are differentially expressed in EC vs HECs (top) and HECs vs HSC gene sets. (D) TFs and CSRs that are up- and downregulated in the Eng −8/P E−/L+ HE fraction. The log fold changes (logFC) and log false discovery rates (logFDR) are listed for each gene.
To investigate the in vivo relevance of our gene set, we used GSEA analysis to compare expression overlaps between ENG−/lacZ+ HE vs ENG+/lacZ− HE and gene sets generated from primary embryonic endothelial cell (EC), hemogenic endothelial cells (HECs), and HSCs.16 Consistent with our functional data, the gene sets associated with EC to HE transition (Figure 6Ci) and HE to HSC transition (Figure 6Cii) showed strong overlaps with genes expressed in ENG−/lacZ+ HE. Gene sets associated with HIF1a and DNA replication also showed strong overlaps with genes expressed in ENG−/lacZ+ HE cells (supplemental Figure 6A). Genes that were UP in ENG−/lacZ+ HE compared with ENG+/lacZ− HE cells feature prominently in IPA reconstructions of gene networks governing hematopoietic development (supplemental Figure 6B). Whereas genes that were UP in ENG−/lacZ+ HE cells compared with either ENG+/lacZ− HE or ENG+/lacZ+ HE cells were associated with biological processes relating to blood development, genes that were DOWN in ENG−/lacZ+ HE cells relative to the other 2 fractions were associated more with angiogenesis or vasculogenesis (supplemental Figure 6C-E). Interrogation of differentially expressed TFs and CSRs in the more functionally hemogenic ENG−/lacZ+ HE fraction relative to the ENG+/lacZ− HE fraction showed upregulation of a number of TFs (eg, Runx1,29 Myb,30 Gfi1b,31 ) and CSRs (Lgr532 ) that are known to play a role in HSC development and downregulation of others (eg, Sox1733 ), which are important for HE to HSC transition (Figure 6D).
Lrp2 is required for normal blood emergence in the zebrafish aorta
We then overlapped our gene sets to visualize associations between genes that were UP or DOWN in HB and/or HE (supplemental Figures 7 and 8; supplemental Tables 1 and 2) to interrogate their function. Six genes were shared between the upregulated groups (Figure 7A; supplemental Figure 7C) and 8 genes between the downregulated groups (supplemental Figure 8A). Genes that were DOWN in both HB and HE cells included several with no known association with hematopoiesis (supplemental Figure 8B). However, we focused on genes that were UP in both HB and HE cells (Figure 7A; supplemental Figure 7C) for practical considerations given that their expression and functional role would be easier to validate. This group included hematopoietic TFs (Gfi131 and Lyl134 ), a platelet protein kinase C substrate (Plek35 ) and granulocyte lysosomal and lysosomal membrane proteins (Mpo36 and Laptm537 ), all of which have known functions in the hematopoietic system. It also included a multifunctional ligand (Lrp2) with no previously described role in blood or blood development. Lrp2/Megalin is a member of an endocytic receptor complex that is involved in maternal-fetal transport of folate and other nutrients, lipids, and morphogens such as sonic hedgehog (Shh) and retinoids.38 Given these associations, we postulated that Lrp2 upregulation in blood precursors was likely to be of functional significance.
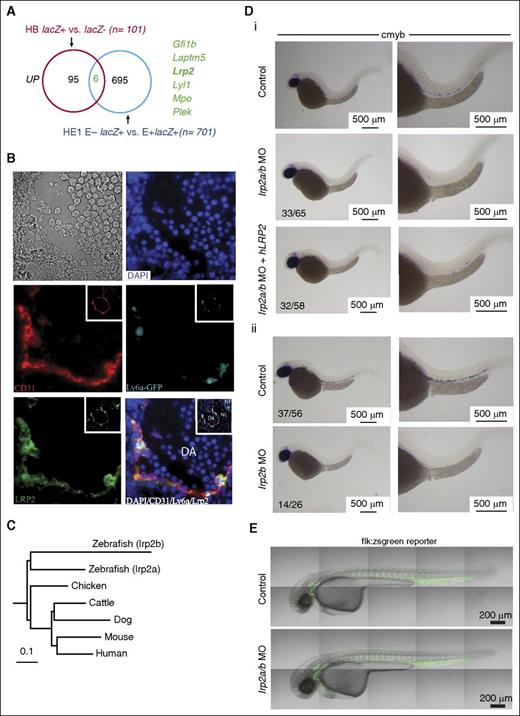
Lrp2 is required for normal definitive hematopoiesis. (A) Venn diagram shows the overlap of genes that are UP in FLK1 mesoderm enriched for BL-CFCs and/or HE cells enriched for hemogenic potential. (B) Immunohistochemistry of E10.5 Ly6aGFP AGM shows coexpression of GFP and LRP2 in ECs and hematopoietic clusters. The insets show the same sections at low magnification. (C) Homology relationships of zebrafish lrp2a and lrp2b coding sequences with that of Lrp2 in different vertebrate species. (D) ISH for the HSC marker cmyb in zebrafish at 36 hpf. (i) Low (left) and high (right) magnification images of control zebrafish (top row), lrp2 a/b morpholinos (middle row), and lrp2a/b morpholinos coinjected with hLRP2 mRNA (bottom row) zebrafish. (ii) Low (left) and high (right) magnification images of control zebrafish (top row) and lrp2b morpholinos. (E) Confocal images of flk:zsgreen reporter embryos show an intact vasculature in both control (top) and lrp2a/b morphant (bottom) embryos. DA, dorsal aorta; NC, notochord; NT, neural tube.
Lrp2 is required for normal definitive hematopoiesis. (A) Venn diagram shows the overlap of genes that are UP in FLK1 mesoderm enriched for BL-CFCs and/or HE cells enriched for hemogenic potential. (B) Immunohistochemistry of E10.5 Ly6aGFP AGM shows coexpression of GFP and LRP2 in ECs and hematopoietic clusters. The insets show the same sections at low magnification. (C) Homology relationships of zebrafish lrp2a and lrp2b coding sequences with that of Lrp2 in different vertebrate species. (D) ISH for the HSC marker cmyb in zebrafish at 36 hpf. (i) Low (left) and high (right) magnification images of control zebrafish (top row), lrp2 a/b morpholinos (middle row), and lrp2a/b morpholinos coinjected with hLRP2 mRNA (bottom row) zebrafish. (ii) Low (left) and high (right) magnification images of control zebrafish (top row) and lrp2b morpholinos. (E) Confocal images of flk:zsgreen reporter embryos show an intact vasculature in both control (top) and lrp2a/b morphant (bottom) embryos. DA, dorsal aorta; NC, notochord; NT, neural tube.
The Ly6aGFP (Sca1) mouse model, in which all HSCs throughout development are GFP+,14,39 has facilitated the study of EHT. There mice were used to show in real time the transition of morphologically flat endothelial GFP+ cells in the E10.5 aorta to round GFP+ cells that coexpress other HSC markers.40 Given that LRP2 was upregulated in HE cells, we evaluated LRP2 expression in Ly6aGFP E10.5 AGM. LRP2 shows specific expression in ECs with strong expression in Ly6aGFP+ ECs and hematopoietic clusters (Figure 7B).
EHT is an evolutionarily conserved process in vertebrates, and real-time imaging of transgenic zebrafish embryos has also shown the transition of aortic ECs to hematopoietic cells.41,42 Lrp2 is highly conserved across different vertebrate species (Figure 7C; average sequence identity across all species shown = 70%). The zebrafish genome has 2 closely related protein-coding genes, lrp2a on chromosome 9 and lrp2b on chromosome 12, both of which are expressed at 24 to 72 hpf.43 To validate the involvement of LRP2 in HSC generation, we used a zebrafish morpholino oligo (MO) knockdown approach targeting both lrp2a and 2b together and each alone. At 36 hours postfertilization (hpf), morphants were assayed by ISH for cmyb and runx1, markers for emerging blood progenitors in the aorta.44 Wild-type embryos showed robust cmyb and runx1 expressing cells along the dorsal aorta in contrast to lrp2a/b morphants that showed severe reductions (Figure 7Di; supplemental Figure 9A). There was partial rescue of AGM blood progenitors when lrp2a/b morphants were coinjected with hLRP2 mRNA. The partial rescue was probably due to only partial homology of protein sequences between humans and fish (∼65%) and quality of in vitro transcribed mRNA given the large size of LRP2 cDNA (∼14 kb). To exclude nonspecific toxicity-related loss of cmyb and runx1 expressing cells, we coinjected lrp2a/b MO with tp53 MO and saw no restitution of cmyb expressing cells in the morphants (supplemental Figure 9B). Injection of lrp2bMO, but not lrp2aMO, reduced the numbers of cmyb expressing AGM blood progenitors (lrp2bMO; Figure 7Dii; lrp2a; data not shown). To establish that this defect in blood cell production was not secondary to loss of vascular integrity, we injected lrp2a/b MO into flk:zsgreen transgenic embryos and saw no difference between morphants and controls at 32 hpf (Figure 7E). In addition to vascular integrity, we also assessed blood flow in morphants. Both heart function and blood flow was indistinguishable from control embryos (data not shown). Taken together, these data support a role for LRP2 during AGM hematopoiesis.
Discussion
Regulatory elements of genes that demonstrate tissue-specific expression have previously been used to target and characterize various cell populations in ESC systems.45-48 When initiating these experiments, we did not envisage that distinct combinations of ENG promoter/enhancers would target hemangioblast potential in FLK1 mesoderm and hemogenic potential in TIE2+/C-KIT+/CD41− hemogenic endothelium. In retrospect, given the distinct transcriptomes and functional properties of hemangioblasts and hemogenic endothelium, this should not have come as a surprise, nor did we predict that hemogenic potential would be enriched in ENG− HE1 cells targeted by the ENG −8PlacZ transgene. Given that F+L+ cells expressed higher levels of ENG (Figure 3A), these data raise the question whether HE1 cells emerge from F+L− ENG low cells, which are less able than their F+L+ counterparts to generate BL-CFCs or whether F+L+ ENG high cells subsequently shutdown ENG expression to facilitate their hemogenic potential in HE1 cells. As these populations were targeted by different transgenes (F+L+; Eng-8/P/lacZ/+7/+9 and ENG-L+ HE; Eng-8/P/lacZ), this could not be directly tested. However, ES cells targeted with dual reporters each driven by either Eng-8/P/+7/+9 or Eng-8/P may assist in addressing this specific question.
Lrp2, a gene that encodes megalin, a multiligand uptake receptor that regulates circulating levels of diverse compounds,49 emerged as a novel regulator of hematopoiesis. Mutations in LRP2 result in impaired neuroepithelial development and are causative of Donnai-Barrow and facio-oculo-acoustico-renal syndromes.50 It has been implicated in balancing BMP4 and SHH signaling in neuroepithelium by acting as a clearance receptor for BMP4 and by concentrating or depleting SHH by ligand recycling or clearance, respectively, in a cell-type and context-dependent manner.51 This control mechanism is of interest as the BMP4-SHH gradient between the neural tube and dorsal aorta has also been implicated in the induction of the HSC developmental program in the ventral wall of the dorsal aorta.52 On a C57Bl/6N background, the LRP2 mutation causes lethality in mice around the time of birth, and there are no mutant pups, although embryo collections at all embryonic stages to E18.5 show expected Mendelian ratios. LRP2 mutations on 129 or CD1 backgrounds also do not yield survivors (A.H. and T.W., unpublished data). On a FVB/N background, however, LRP2 null mice are viable with neural tube defects, and this receptor has previously been implicated in folate endocytosis in the developing neural tube.53 However, peripheral blood and bone marrow hematopoietic stem and progenitor cell numbers were comparable in FVB/N wild-type and mutant adult mice at 6 to 9 months of age (supplemental Figure 10). A more detailed analysis of embryonic hematopoiesis in mutant mice on both C57Bl/6N and FVB/N backgrounds will be required to establish whether the numbers of emergent HSCs differ at various time points and the identity of any modifier genes in FVB/N that compensate for the loss of LRP2; these investigations are ongoing. However, taken together with the zebrafish data, which show reduction rather than loss of HSCs, LRP2 is likely to facilitate rather than be absolutely required for EHT. Indeed, it is important to keep in mind that Lrp2 transcripts were higher in HB cells with greater BL-CFC potential and HE cells with greater CFU-C potential, but cells with lower numbers of transcripts were also able to generate BL-CFCs and CFU-Cs.
Deficiency of dietary folate also results in impaired neural tube development and megaloblastic anemia.54 Targeted inactivation of the reduced folate carrier, which facilitates folate delivery into cells, results in embryonic lethality at E10.5 due to neural and hematopoietic defects,55 and components of the Megalin complex are among the most significantly disrupted genes in null embryos.38 Coordinated upregulation of a receptor that facilitates folate uptake in HECs would be consistent with demand for an essential hematinic in cells that are on the threshold of a replicative phase.
Although we focused our attention on Lrp2, as a gene without a described role in hematopoiesis, from a list of 6 that were upregulated in both hemangioblasts and HE cells, there were other genes that were UP in only one or the other cell fraction. Given the overlap of these gene sets with those generated from gene knockout embryos with specific developmental defects, they will serve as a rich resource to explore and manipulate the emergence of hemangioblasts from FLK1+ mesoderm or hematopoiesis in HE cells. Insights gained from these manipulations will in turn inform tissue regeneration protocols that aim to generate functional HSCs.
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE77390).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the National Health and Medical Research Council of Australia, Australian Research Council, and the Tom Bee Stem Cell Research Fund (J.E.P.), Cancer Research UK (V.K. and G.L.), the Biotechnology and Biological Sciences Research Council, Leukaemia and Lymphoma Research, The Leukaemia and Lymphoma Society, and core support grants by the Wellcome Trust to the Cambridge Institute for Medical Research and Wellcome Trust–Medical Research Council Cambridge Stem Cell Institute (B.G.). This work was supported by grants from the National Institutes of Health National Heart, Lung, and Blood Institute (R01 HL04880, P015PO1HL32262-32, and 5U01 HL10001-05) and National Institute of Diabetes and Digestive and Kidney Diseases (5P30 DK49216, 5R01 DK53298, and R24 DK092760) (L.I.Z.) and by European Research Council Advanced Grant 341096. E.M.F. is a Fellow of the Leukemia and Lymphoma Society.
Authorship
Contribution: R.N., E.M.F., P.S., K.K., R.T., M.S., F.P., J.K., A.U., J.T., D.B., C.S.V., A.E., and R.P. performed research and analyzed data. A.S., J.W., A.H., D.K., R.R., T.W., B.G., E.D., and L.I.Z. contributed essential reagents and advice with data analysis and interpretation. R.N., G.L., V.K., and J.E.P. contributed to study design, data interpretation, and manuscript preparation. All authors read and approve the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for P.S. is BeDataDriven BV, The Hague, The Netherlands.
The current affiliation for A.S. is GSK Pharmaceutical Company, London, United Kingdom.
Correspondence: John E. Pimanda, Lowy Cancer Research Centre and the Prince of Wales Clinical School, UNSW Australia, Sydney, NSW 2052, Australia; e-mail: jpimanda@unsw.edu.au; Valerie Kouskoff, Cancer Research United Kingdom Manchester Institute, The University of Manchester, 553 B5093, Manchester M20 4BX, United Kingdom; e-mail: valerie.kouskoff@cruk.manchester.ac.uk; Georges Lacaud, Cancer Research United Kingdom Manchester Institute, The University of Manchester, 553 B5093, Manchester M20 4BX, United Kingdom; e-mail: georges.lacaud@cruk.manchester.ac.uk.