Key Points
Autophagy is required for maintenance of AML-initiating cells and peripheral myeloblast survival.
Loss of autophagy potentiates the therapeutic effects of AraC in vivo.
Abstract
Despite advances in the treatment of acute myeloid leukemia (AML), relapse and drug resistance frequently occur. Therefore, detailed mechanisms of refractoriness, including leukemia-initiating cell (LIC) biology, should be elucidated to treat AML. The self-degradative property of cytosolic macromolecules is central to autophagy and can contribute to homeostasis and stress response. Recent reports suggest the importance of autophagy in hematopoietic stem cells and various tumors. Thus, this study investigated the functional role of autophagy in AML maintenance and drug resistance using tamoxifen-inducible conditional knockout mice of Atg5 or Atg7, which are essential genes for autophagy, combined with an mixed lineage leukemia–eleven nineteen leukemia–induced murine AML model. Inactivation of autophagy by deletion of Atg5 or Atg7 prolonged survival in leukemic mice and reduced functional LICs. Atg7-deficient LICs displayed enhanced mitochondrial activity and reactive oxygen species production together with increased cell death. In addition, Atg7 deletion markedly decreased peripheral blood leukemia cells, concurrent with increased apoptosis, suggesting a higher dependency on autophagy compared with bone marrow leukemia cells. Finally, cytarabine (AraC) treatment activated autophagy in LICs, and Atg7 deletion potentiated the therapeutic effects of AraC, which included decreased LICs and prolonged survival, suggesting that autophagy contributes to AraC resistance. Our results highlight the intratumoral heterogeneity related to autophagy in AML and the unique role of autophagy in leukemia development and drug resistance.
Introduction
The prognosis of acute myeloid leukemia (AML) remains dismal in the majority of patients even after recent advances in the pathophysiology in AML. One reason for this is the acquisition of resistance to key chemotherapeutic drugs such as cytarabine (AraC) and anthracyclines, resulting in relapse, which is mediated by a small subset of self-renewing cells called leukemia-initiating cells (LICs). Thus, further studies on the biological features of LICs and their mechanisms of drug resistance are essential to treat AML patients.
Macroautophagy (hereafter referred to as autophagy) is a process used for the degradation of cytosolic macromolecules by lysosomal enzymes.1,2 Autophagy has various physiological roles through the maintenance of cellular homeostasis by degrading damaged or excess cytosolic components and supplying nutrients to starved cells.3,4 Through autophagic processes, double membrane vesicles, called autophagosomes, engulf cytosolic components and transport their cargo to lysosomes. A group of proteins including autophagy–related 5 (ATG5) and ATG7 are indispensable for autophagy.1,5,6
Based on the complicated relationship between autophagy and tumors, it is unknown if inhibition of autophagy could be a beneficial approach to treat cancer.7,8 In general, autophagy contributes to cell survival in established tumors.9,10 Accordingly, inactivation of autophagy, through Atg3 deletion, in breakpoint cluster region–Abelson murine leukemia viral oncogene homolog 1 (BCR-ABL1)–transduced murine bone marrow (BM) cells diminishes their capacity to produce chronic myeloid leukemia (CML) in transplanted mice because of p53-dependent growth arrest.11 As such, the efficacy of the autophagy inhibitor chloroquine and its derivatives is currently being tested in clinical trials for the treatment of various types of cancer.12 In contrast, it has also been reported that autophagy genes function as tumor suppressors in normal tissues.13 Additionally, even in established tumors, it has been reported that inhibition of autophagy promotes cancer progression in a murine TP53-deficient pancreatic cancer model and in EGFR-mutated lung cancer.14,15 Considering these facts, the dependency on autophagy could be strongly affected by biological context such as gene mutations and physiological conditions of the tumor microenvironment.
In addition, its association with drug resistance makes autophagy an attractive drug target. Autophagy is recognized as a nonapoptotic mechanism of programmed cell death and is required for cell death after chemotherapy in some cases.16,17 However, combination treatment using an autophagy inhibitor with chemotherapy, irradiation, or targeted therapy has demonstrated efficacy in various cancers including hematological malignancies.8,18,19 Considering the fact that the function of autophagy is context-dependent, it is worthwhile to evaluate the impact of inhibition of autophagy in vivo in different types of tumors, using animal models that can recapitulate physiological conditions and tumor microenvironments.
The role of autophagy in AML progression, including LIC maintenance, is not yet fully understood. Moreover, the exact function of autophagy in drug resistance in vivo needs to be elucidated; however, some studies have revealed that autophagy is required for drug resistance.20-24 In the present study, to investigate the role of autophagy in AML progression and drug resistance in vivo, we inhibited autophagy in murine mixed lineage leukemia–eleven nineteen leukemia (MLL-ENL) leukemia and CML blast crisis (CML-BC).
Methods
Mice
Atg5flox/flox mice,25 Atg7flox/flox mice,5 and GFP-LC3 mice26 were obtained from RIKEN BRC (Tsukuba, Japan). Cre-ERT2 mice27 were obtained from JAX (Bar Harbor, ME). Atg5flox/flox or Atg7flox/flox mice were crossed with Cre-ERT2 mice. Genotyping was performed with primers shown in supplemental Table 1 (available on the Blood Web site). Wild-type C57BL/6J (8- to 12-week-old) mice were purchased from Sankyo Laboratory Service (Tokyo, Japan). These mice were kept at the Center for Disease Biology and Integrative Medicine at the University of Tokyo, according to institutional guidelines. All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of Tokyo.
Generation of murine leukemia model
The procedure for developing murine bone marrow transplantation (BMT) leukemia models was derived from previous reports.28,29 For producing retrovirus, MSCV-MLL-ENL-IRES-GFP, MSCV-MLL-ENL-IRES-Puro,28 pGCDNsam-BCR-ABL1-IRES-GFP, or pGCDNsam-NUP98-HOXA9-IRES-KusabiraOrange were transfected into Plat-E packaging cells30 using polyethylenimine.31 The detailed protocol is described in the supplemental Methods.
Drug administration
For excising the loxP-flanked region in vivo, 1 mg/mouse tamoxifen (TAM; Cayman Chemical Company, Ann Arbor, MI) dissolved in peanut oil (Sigma, St. Louis, MO) was intraperitoneally administrated once daily for 5 consecutive days. In leukemia mouse models, TAM treatment started from day 7 after BMT. AraC (Tokyo Chemical Industry, Tokyo, Japan) was dissolved in phosphate-buffered saline (PBS) and intraperitoneally administrated to leukemic mice at a dose of 1 mg/mouse once daily.
Flow cytometry analyses and sorting
For staining, isolated mononulcear cells were incubated with 3% fetal calf serum/PBS containing diluted antibody as listed in supplemental Table 2. Stained cells were washed once with 3% fetal calf serum/PBS and analyzed. For apoptosis analysis, stained cells were washed once and subsequently incubated with annexin-V-allophycocyanin (BioLegend, San Diego, CA) and 4′,6-diamidino-2-phenylindole (DAPI; 1 µg/mL) in accordance with the manufacturers’ protocols. Mitochondrial activity was detected by MitoTrackerOrange CMTMRos (Life Technologies, Waltham, MA), and reactive oxygen species (ROS) levels were determined by CellRox DeepRed (Life Technologies). Cells (2 × 105) that were stained with antibodies against surface markers were washed once and incubated with 1× Hanks balanced salt solution containing a 1:2000 dilution of MitoTracker or a 1:500 dilution of CellRox at 37°C for 15 minutes. Stained cells were analyzed using an LSRII (BD Biosciences, San Jose, CA) and sorted with a BD FACSAria (BD Biosciences). Data were analyzed by FlowJo software (TreeStar, Ashland, OR).
Electron micrograph
Sorted c-Kit−CD11b+ BM or peripheral blood (PB) cells from MLL-ENL leukemic mice were fixed with 2% paraformaldehyde and 2% glutaraldehyde in 50 mM phosphate buffer (pH 7.4) for 10 minutes at 4°C. The buffer was then replaced with 2% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4) and incubated at 4°C overnight. Thereafter, sample preparation and imaging were conducted at Tokai Electron Microscopy Inc. (Nagoya, Japan).
Statistics
P values, for comparison of 2 different groups, were calculated using an unpaired 2-tailed Student t test unless otherwise mentioned. LIC frequency was assessed using the Poisson distribution. To analyze the survival curves, a log-rank test was used. P values <.05 were considered significant. Data analysis was performed using R software (http://www.R-project.org).
Results
Autophagy deficiency in MLL-ENL AML mice retarded leukemia progression
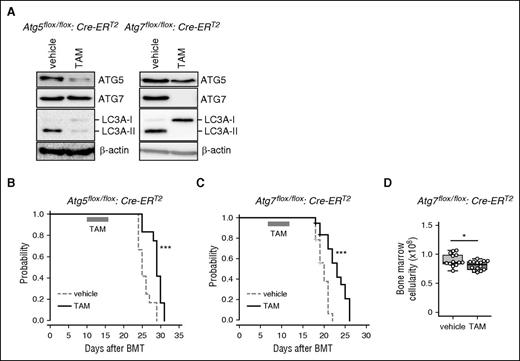
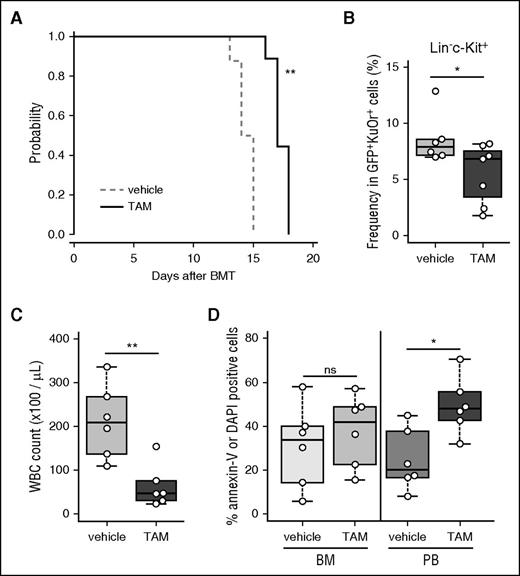
To investigate the importance of autophagy in AML progression, we developed a leukemia mouse model through BMT with Atg5- or Atg7-floxed BM cells transduced with MLL-ENL. To delete the Atg5 or Atg7 gene in vivo, Atg5flox/flox or Atg7flox/flox mice were crossed with TAM-inducible Cre-ERT2 mice.27 Green fluorescent protein (GFP)-labeled MLL-ENL-driven AML cells, from primary recipients, were serially transplanted into secondary recipients, and secondary recipients received TAM treatment to delete Atg5 or Atg7 (supplemental Figure 1A). Genomic polymerase chain reaction and western blots of BM cells from TAM-treated, Atg5flox/flox:Cre-ERT2 or Atg7flox/flox:Cre-ERT2 MLL-ENL leukemic mice confirmed the in vivo deletion of Atg5 or Atg7 (supplemental Figure 2A; Figure 1A). Unconjugated microtubule-associated protein 1A/1B-light chain 3A (LC3A, or LC3A-I) covalently binds to phosphatidylethanolamine to form LC3A-II to induce autophagy through associations with several ATG molecules including ATG5 and ATG7.1 Thus, LC3A-II is commonly used as an indicator of autophagy. We confirmed diminished conversion of LC3A-I to LC3A-II in TAM-treated Atg5Δ/Δ and Atg7Δ/Δ leukemic cells, revealing that genetic deletion of Atg5 or Atg7 can efficiently suppress autophagy in MLL-ENL leukemic cells (Figure 1A). In terms of disease progression in MLL-ENL mice, Atg5Δ/Δ and Atg7Δ/Δ animals displayed significantly longer survival than Atg5flox/flox and Atg7flox/flox controls (Atg5Δ/Δ: median = 29 days vs Atg5flox/flox: median = 25 days; Atg7Δ/Δ: median = 23 days vs Atg7flox/flox: median = 20 days) (Figure 1B-C). Atg7Δ/Δ MLL-ENL mice showed lower BM cellularity than control mice (Figure 1D), although the frequency of GFP+ MLL-ENL cells in BM and spleen weight were not different between vehicle- and TAM-treated mice (supplemental Figure 2B-C).
Retarded progression of MLL-ENL–induced murine leukemia caused by autophagy deficiency. (A) Protein levels of ATG5, ATG7, and LC3A were assessed by western blot analysis in BM cells isolated from MLL-ENL leukemic mice at day 18 post-BMT. β-actin was used as a loading control. (B-C) Kaplan-Meier curves represent vehicle- or TAM-treated Atg5flox/flox:Cre-ERT2 (n = 12 each) and Atg7flox/flox:Cre-ERT2 (n = 15 each) MLL-ENL leukemic mice. The P value was calculated by a log-rank test. (D) BM cellularities of vehicle- and TAM-treated Atg7flox/flox:Cre-ERT2 leukemic mice are shown (n = 13). The box depicted in the upper and lower quartiles, and the median, are signified by thick lines within the box. The whiskers represent maximum or minimum data within the 1.5× interquartile range from the top or the bottom of the box, respectively. Each circle represents an individual animal. *P < .05; ***P < .001. All results except for western blotting are pooled data from at least 3 independent experiments with 1 to 4 animals per experiment.
Retarded progression of MLL-ENL–induced murine leukemia caused by autophagy deficiency. (A) Protein levels of ATG5, ATG7, and LC3A were assessed by western blot analysis in BM cells isolated from MLL-ENL leukemic mice at day 18 post-BMT. β-actin was used as a loading control. (B-C) Kaplan-Meier curves represent vehicle- or TAM-treated Atg5flox/flox:Cre-ERT2 (n = 12 each) and Atg7flox/flox:Cre-ERT2 (n = 15 each) MLL-ENL leukemic mice. The P value was calculated by a log-rank test. (D) BM cellularities of vehicle- and TAM-treated Atg7flox/flox:Cre-ERT2 leukemic mice are shown (n = 13). The box depicted in the upper and lower quartiles, and the median, are signified by thick lines within the box. The whiskers represent maximum or minimum data within the 1.5× interquartile range from the top or the bottom of the box, respectively. Each circle represents an individual animal. *P < .05; ***P < .001. All results except for western blotting are pooled data from at least 3 independent experiments with 1 to 4 animals per experiment.
Atg7 deletion in MLL-ENL leukemic mice decreased LIC frequency via induction of cell death
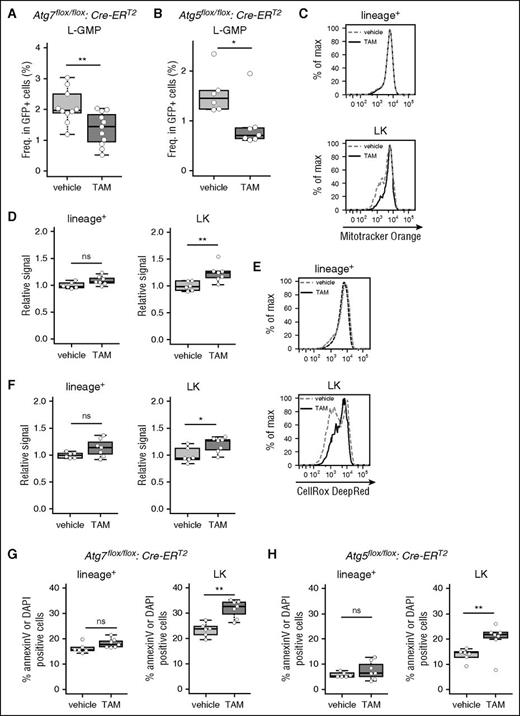
To further examine the role of autophagy in MLL-ENL leukemic mice, LIC frequency was evaluated in Atg7-deficient leukemic mice; MLL-ENL LICs are enriched in lineage−c-Kit+Sca1−CD34+CD16/32+ (L-GMP) or lineage−c-Kit+ (LK) cells.32,33 Flow cytometric analysis revealed that LK and L-GMP frequency were decreased in Atg7-deleted leukemic mice (Figure 2A; supplemental Figure 3A-B). Additionally, a similar reduction in LICs was observed in Atg5Δ/Δ leukemic mice, suggesting that the decreased frequency of LICs was caused by autophagy-related mechanisms (Figure 2B; supplemental Figure 3C). Hereafter, we mainly focused on the Atg7 conditional knockout model to study the role of autophagy, as phenotypes were identical after knocking out Atg5 or Atg7. In normal hematopoietic stem cells (HSCs), an Atg7 deletion causes accumulation and increased activity of mitochondria, resulting in increased ROS production.34,35 Similarly, Atg7Δ/Δ MLL-ENL LK cells demonstrated enhanced mitochondrial activity and increased ROS production compared with those of their Atg7flox/flox counterpart (Figure 2C-F). In lineage+ cells, a population rich in differentiated cells, the Atg7 deletion had no effect on mitochondrial activity or ROS production (Figure 2C-F). When we assessed the apoptotic status of Atg7Δ/Δ and Atg7flox/flox MLL-ENL mice, annexin-V+/DAPI+ dead cells increased only in the Atg7Δ/Δ LK fraction (Figure 2G; supplemental Figure 3D). We confirmed that cell death in the LK fraction was also induced in TAM-treated Atg5flox/flox MLL-ENL leukemic mice (Figure 2H).
Induction of cell death, associated with activated mitochondria and ROS production, is reduced in LICs by Atg7 deletion. Collective data indicating the frequencies of L-GMP cells from vehicle-treated (n = 10) or TAM-treated (n = 11) Atg7flox/flox:Cre-ERT2 MLL-ENL leukemic mice (A) and vehicle-treated (n = 6) or TAM-treated (n = 7) Atg5flox/flox:Cre-ERT2 MLL-ENL leukemic mice (B). (C) Representative flow cytometric analysis of mitochondrial activity in lineage+ and LK cells from vehicle- or TAM-treated Atg7flox/flox:Cre-ERT2 MLL-ENL leukemic mice. (D) Collective data of relative signal level of Mitotracker Orange in lineage+ and LK cells from vehicle-treated (n = 6) or TAM-treated (n = 7) Atg7flox/flox:Cre-ERT2 MLL-ENL leukemic mice. Relative signal represents geometric mean fluorescent intensity (MFI) of each sample divided by the average MFI from all vehicle samples. (E) Representative flow cytometric analysis of ROS levels in lineage+ and LK cells from vehicle- or TAM-treated Atg7flox/flox:Cre-ERT2 MLL-ENL leukemic mice. (F) Collective data of relative signal levels of CellRox DeepRed in lineage+ and LK cells from vehicle-treated (n = 6) or TAM-treated (n = 7) Atg7flox/flox:Cre-ERT2 MLL-ENL leukemic mice. (G) Boxplot of the frequencies of annexin-V+ or DAPI+ cells from vehicle-treated (n = 6) or TAM-treated (n = 7) Atg7flox/flox:Cre-ERT2 MLL-ENL leukemic mice. (H) Boxplot of the frequencies of annexin-V+ or DAPI+ cells from vehicle-treated (n = 6) or TAM-treated (n = 7) Atg5flox/flox:Cre-ERT2 MLL-ENL leukemic mice. *P < .05; **P < .01. All results represent pooled data from at least 3 independent experiments with 2 to 4 animals per experiment.
Induction of cell death, associated with activated mitochondria and ROS production, is reduced in LICs by Atg7 deletion. Collective data indicating the frequencies of L-GMP cells from vehicle-treated (n = 10) or TAM-treated (n = 11) Atg7flox/flox:Cre-ERT2 MLL-ENL leukemic mice (A) and vehicle-treated (n = 6) or TAM-treated (n = 7) Atg5flox/flox:Cre-ERT2 MLL-ENL leukemic mice (B). (C) Representative flow cytometric analysis of mitochondrial activity in lineage+ and LK cells from vehicle- or TAM-treated Atg7flox/flox:Cre-ERT2 MLL-ENL leukemic mice. (D) Collective data of relative signal level of Mitotracker Orange in lineage+ and LK cells from vehicle-treated (n = 6) or TAM-treated (n = 7) Atg7flox/flox:Cre-ERT2 MLL-ENL leukemic mice. Relative signal represents geometric mean fluorescent intensity (MFI) of each sample divided by the average MFI from all vehicle samples. (E) Representative flow cytometric analysis of ROS levels in lineage+ and LK cells from vehicle- or TAM-treated Atg7flox/flox:Cre-ERT2 MLL-ENL leukemic mice. (F) Collective data of relative signal levels of CellRox DeepRed in lineage+ and LK cells from vehicle-treated (n = 6) or TAM-treated (n = 7) Atg7flox/flox:Cre-ERT2 MLL-ENL leukemic mice. (G) Boxplot of the frequencies of annexin-V+ or DAPI+ cells from vehicle-treated (n = 6) or TAM-treated (n = 7) Atg7flox/flox:Cre-ERT2 MLL-ENL leukemic mice. (H) Boxplot of the frequencies of annexin-V+ or DAPI+ cells from vehicle-treated (n = 6) or TAM-treated (n = 7) Atg5flox/flox:Cre-ERT2 MLL-ENL leukemic mice. *P < .05; **P < .01. All results represent pooled data from at least 3 independent experiments with 2 to 4 animals per experiment.
To exclude the possibility that TAM treatment alone or activation of Cre-estrogen receptor fusion protein (Cre-ER) could cause artificial effects in hematopoietic cells,36,37 we used Cre-ER negative (Cre−) Atg7flox/flox leukemic mice and Cre-ER wild-type leukemic mice as controls. TAM treatment did not prolong survival in Atg5flox/flox:Cre− or Atg7flox/flox:Cre− leukemic mice and did not reduce the frequency of L-GMPs in Atg7flox/flox:Cre− leukemic mice (supplemental Figure 4A-C). Moreover, the frequency of L-GMP cells in Cre-ER wild-type leukemic mice did not change after TAM treatment (supplemental Figure 4D).
These results suggest that autophagy is required for LIC maintenance by preventing cell death associated with accumulation of mitochondria and induced ROS production.
Atg7-deficient MLL-ENL LICs possessed decreased tumorigenic capacity
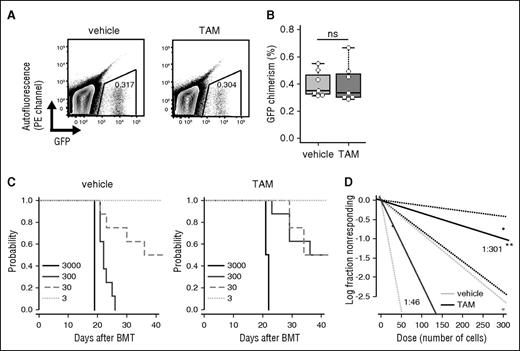
To characterize the defects of LICs in autophagy-deficient leukemic mice, functional analyses of Atg7Δ/Δ leukemic cells were performed. GFP+Atg7Δ/Δ BM cells had decreased in vitro colony-forming capacity compared with GFP+Atg7flox/flox BM cells (supplemental Figure 5A-B). When sorted GFP+ BM cells from Atg7flox/flox or Atg7Δ/Δ MLL-ENL leukemic mice were serially transplanted into tertiary recipient mice, it was confirmed that the Atg7 deletion had no impact on homing ability (Figure 3A-B; supplemental Figure 1B). Limiting dilution transplantation analysis revealed that the frequency of functional LICs decreased in autophagy-inactivated Atg7Δ/Δ mice (Atg7flox/flox vs Atg7Δ/Δ, 1:46 vs 1:301, respectively) (Figure 3C-D). Taken together, Atg7 is required for maintenance of functional LICs in MLL-ENL leukemia.
Decreased frequency of functional LICs in Atg7-deleted MLL-ENL mice. (A) Representative flow cytometric plot of donor cell engraftment 16 hours after BMT with 1 × 106 GFP+Atg7flox/flox or Atg7Δ/Δ cells. (B) Collective data of donor cell engraftment at 16 hours after BMT with 1 × 106 GFP+Atg7flox/flox or Atg7Δ/Δ cells (n = 7 per group). (C) Kaplan-Meier curves represent survival of tertiary recipients with 3, 30, 300, or 3000 GFP+ BM cells from vehicle- or TAM-treated leukemic mice (n = 4-8 per group). (D) Poisson statistical analysis of limiting dilution transplantation assay. Plots were generated for estimation of the frequency of functional LICs in GFP+ BM cells from vehicle- or TAM-treated leukemic mice (n = 4-8 at each cell dose). The plot represents the logarithm of the frequency of living mice at 42 days after injection for the indicated number of cells. LIC frequency was 1:46 for vehicle-treated (95% confidence interval, 1:18-115) and 1:301 for TAM-treated (95% confidence interval, 1:126-718) leukemic mice. P = .0002.
Decreased frequency of functional LICs in Atg7-deleted MLL-ENL mice. (A) Representative flow cytometric plot of donor cell engraftment 16 hours after BMT with 1 × 106 GFP+Atg7flox/flox or Atg7Δ/Δ cells. (B) Collective data of donor cell engraftment at 16 hours after BMT with 1 × 106 GFP+Atg7flox/flox or Atg7Δ/Δ cells (n = 7 per group). (C) Kaplan-Meier curves represent survival of tertiary recipients with 3, 30, 300, or 3000 GFP+ BM cells from vehicle- or TAM-treated leukemic mice (n = 4-8 per group). (D) Poisson statistical analysis of limiting dilution transplantation assay. Plots were generated for estimation of the frequency of functional LICs in GFP+ BM cells from vehicle- or TAM-treated leukemic mice (n = 4-8 at each cell dose). The plot represents the logarithm of the frequency of living mice at 42 days after injection for the indicated number of cells. LIC frequency was 1:46 for vehicle-treated (95% confidence interval, 1:18-115) and 1:301 for TAM-treated (95% confidence interval, 1:126-718) leukemic mice. P = .0002.
PB MLL-ENL leukemia cells had a high dependency on autophagy for survival
Intriguingly, both Atg5Δ/Δ and Atg7Δ/Δ MLL-ENL leukemic mice had significantly lower peripheral white blood cell (WBC) counts compared with those of their respective counterparts (Figure 4A-B). This contrasted the minimal contribution of Atg5/Atg7 deletion in reducing AML burden in BM (Figure 1D; supplemental Figure 2B-C). These results led us to hypothesize that various extracellular factors in PB can result in autophagy dependence in peripheral leukemia cells. We thus evaluated the apoptotic status of PB Atg5Δ/Δ and Atg7Δ/Δ MLL-ENL leukemia cells. To minimize possible bias derived from differential frequencies of c-Kit+ cells between BM and PB, CD11b+ cells, in which MLL-ENL (GFP+) leukemia cells accumulate in both BM and PB, were subdivided into c-Kit− and c-Kit+ fractions for further analysis (supplemental Figure 6A). Strikingly, a TAM-induced Atg5/Atg7 deletion caused increased apoptosis only in PB CD11b+ leukemia cells and not in similar cells of BM origin, irrespective of c-Kit expression (Figure 4B-C; supplemental Figure 6B).
Dependence of PB leukemia cells on autophagy. (A) WBC counts of PB from Atg5flox/flox:Cre-ERT2 or Atg7flox/flox:Cre-ERT2 MLL-ENL leukemic mice were measured at indicated days after BMT (>7 mice per group, mean ± standard deviation [SD]). P values were calculated by a Welch t test. Collective data of frequencies of annexin-V+ or DAPI+ cells in c-Kit−CD11b+ or c-Kit+CD11b+ cells obtained from vehicle- or TAM-treated Atg7flox/flox:Cre-ERT2 (B) and Atg5flox/flox:Cre-ERT2 (C) mice (n = 6-9 per group). (D) Relative colony numbers from GFP+ PB cells sorted from vehicle- or TAM-treated Atg7flox/flox:Cre-ERT2 MLL-ENL leukemic mice are shown (n = 4 per group, mean ± SD). (E) Kaplan-Meier curves representing survival of tertiary recipients of GFP+ PB cells from vehicle- or TAM-treated Atg7flox/flox:Cre-ERT2 MLL-ENL leukemic mice (n = 9 per group). The P value was calculated by a log-rank test. *P < .05; **P < .01. All results represent pooled data from at least 3 independent experiments with 1 to 4 animals per experiment.
Dependence of PB leukemia cells on autophagy. (A) WBC counts of PB from Atg5flox/flox:Cre-ERT2 or Atg7flox/flox:Cre-ERT2 MLL-ENL leukemic mice were measured at indicated days after BMT (>7 mice per group, mean ± standard deviation [SD]). P values were calculated by a Welch t test. Collective data of frequencies of annexin-V+ or DAPI+ cells in c-Kit−CD11b+ or c-Kit+CD11b+ cells obtained from vehicle- or TAM-treated Atg7flox/flox:Cre-ERT2 (B) and Atg5flox/flox:Cre-ERT2 (C) mice (n = 6-9 per group). (D) Relative colony numbers from GFP+ PB cells sorted from vehicle- or TAM-treated Atg7flox/flox:Cre-ERT2 MLL-ENL leukemic mice are shown (n = 4 per group, mean ± SD). (E) Kaplan-Meier curves representing survival of tertiary recipients of GFP+ PB cells from vehicle- or TAM-treated Atg7flox/flox:Cre-ERT2 MLL-ENL leukemic mice (n = 9 per group). The P value was calculated by a log-rank test. *P < .05; **P < .01. All results represent pooled data from at least 3 independent experiments with 1 to 4 animals per experiment.
To further analyze autophagy-related functions in PB MLL-ENL leukemia cells, in vitro colony formation assays and in vivo serial BMT assays with PB leukemia cells were performed. PB Atg7Δ/Δ leukemia cells showed decreased colony forming capacity in vitro (Figure 4D), and recipients of PB Atg7Δ/Δ leukemia cells displayed marginal but significantly prolonged survival in vivo (Figure 4E; supplemental Figure 1C). This demonstrated that PB MLL-ENL leukemia cells also had leukemia-initiating capacity, which was partly regulated by autophagy.
Additionally, we evaluated the apoptotic status of PB cells from Atg7flox/flox:Cre− or Cre-ER leukemic mice with wild-type Atg genes as controls. TAM treatment did not cause reduction in WBCs or induction of apoptosis in PB. Therefore, the possibility of a cytotoxic effect on PB leukemic cells mediated by the Cre-ER system was excluded (supplemental Figure 4E-I).
Taken together, autophagy protects PB leukemia cells from apoptosis irrespective of their immunophenotypic maturity.
PB MLL-ENL leukemia cells had an increased number of autophagosomes
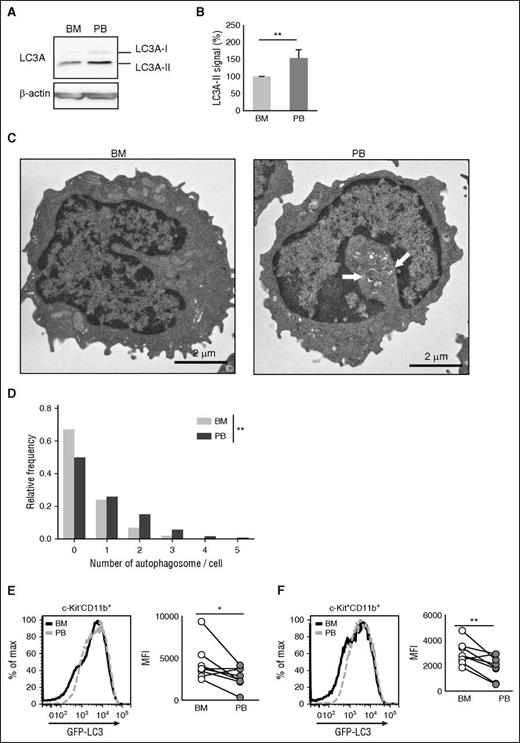
We next investigated whether PB leukemia cells have enhanced autophagic activity. For this, BM and PB c-Kit−CD11b+ MLL-ENL leukemia cells were subjected to western blotting to quantify LC3A conversion. Protein analysis revealed that PB leukemia cells had a higher level of LC3A-II than BM leukemia cells, indicating that PB leukemia cells were rich in autophagosomes (Figure 5A-B). In addition, electron microscopic analysis confirmed that PB leukemia cells contained an increased number of autophagosomes compared with BM cells (Figure 5C-D). We next used GFP-LC3 transgenic mice to evaluate autophagy.26 When autophagy is activated in GFP-LC3 mice, GFP-LC3 is degraded resulting in decreased GFP intensity.38 We developed GFP-LC3 transgenic MLL-ENL leukemic mice (supplemental Figure 1D), and GFP expression in BM and PB leukemia cells was measured by flow cytometry (supplemental Figure 7A). In both c-Kit−CD11b+ and c-Kit+CD11b+ fractions, GFP expression in PB leukemia cells was decreased compared with that in BM leukemia cells (Figure 5E-F). To examine if the accumulation of autophagosomes was cell autonomous, we performed ex vivo culture of BM and PB leukemia cells. The enhanced LC3A-II expression in PB cells that was detected in vivo immediately disappeared when BM and PB c-Kit−CD11b+ cells from MLL-ENL leukemic mice were cultured (supplemental Figure 8A). Moreover, the difference in GFP-LC3 expression between BM and PB was maintained in leukemic mice after BMT of PB leukemic cells that expressed lower levels of GFP-LC3 (supplemental Figure 8B-C). These results suggest that the PB environment might enhance autophagy in PB leukemia cells.
Increased number of autophagosomes and decreased GFP-LC3 expression were observed in PB leukemic cells. (A) Expression of LC3A-I and LC3A-II in BM and PB from GFP-LC3 MLL-ENL leukemic mice. (B) Relative signal intensities of LC3A-II in BM and PB from GFP-LC3 MLL-ENL leukemic mice are shown. Values were normalized to the intensity of LC3A-II/β-actin from BM samples (n = 5, mean ± SD). (C) Representative electron micrograph of c-Kit−CD11b+ cells from BM or PB of MLL-ENL leukemic mice. The grids were observed by a transmission electron microscope (JEM-1400Plus; JEOL Ltd., Tokyo, Japan). Digital images were taken with a CCD camera (VELETA; Olympus Soft Imaging Solutions, Germany). White arrows indicate autophagosomes. (D) Frequency distribution of detectable autophagosomes per cell from electron micrograph analysis. The number of autophagosomes in 100 cells, derived from 2 independent mice, was counted. The P value was calculated with the Wilcoxon rank sum test. Representative histogram and collective data (n = 8) of GFP-LC3 expression in c-Kit−CD11b+ (E) or c-Kit+CD11b+ (F) cells in BM or PB from GFP-LC3 MLL-ENL leukemic mice. Geometric MFIs were plotted. P values were calculated by a Student paired t test. *P < .05; **P < .01. All results are pooled data from at least 3 independent experiments with 1 to 3 animals per experiment.
Increased number of autophagosomes and decreased GFP-LC3 expression were observed in PB leukemic cells. (A) Expression of LC3A-I and LC3A-II in BM and PB from GFP-LC3 MLL-ENL leukemic mice. (B) Relative signal intensities of LC3A-II in BM and PB from GFP-LC3 MLL-ENL leukemic mice are shown. Values were normalized to the intensity of LC3A-II/β-actin from BM samples (n = 5, mean ± SD). (C) Representative electron micrograph of c-Kit−CD11b+ cells from BM or PB of MLL-ENL leukemic mice. The grids were observed by a transmission electron microscope (JEM-1400Plus; JEOL Ltd., Tokyo, Japan). Digital images were taken with a CCD camera (VELETA; Olympus Soft Imaging Solutions, Germany). White arrows indicate autophagosomes. (D) Frequency distribution of detectable autophagosomes per cell from electron micrograph analysis. The number of autophagosomes in 100 cells, derived from 2 independent mice, was counted. The P value was calculated with the Wilcoxon rank sum test. Representative histogram and collective data (n = 8) of GFP-LC3 expression in c-Kit−CD11b+ (E) or c-Kit+CD11b+ (F) cells in BM or PB from GFP-LC3 MLL-ENL leukemic mice. Geometric MFIs were plotted. P values were calculated by a Student paired t test. *P < .05; **P < .01. All results are pooled data from at least 3 independent experiments with 1 to 3 animals per experiment.
Normal mature myeloid cells in PB did not depend on autophagy
The importance of autophagy in hematopoietic cells has been emphasized by several prior reports.39-42 Considering the enhanced dependency of PB MLL-ENL leukemia cells on autophagy, we next elucidated the role of autophagy in normal PB myeloid cells. For this, PB cells obtained from vehicle- or TAM-treated MLL-ENL-free Atg7flox/flox:Cre-ERT2 mice were analyzed. Atg7Δ/Δ mice showed no change in PB WBC counts even after TAM administration (supplemental Figure 9A-B). Additionally, an Atg7 deletion had no impact on apoptosis in PB c-Kit−CD11b+ or c-Kit+CD11b+ cells (supplemental Figure 9C). Moreover, normal c-Kit−CD11b+ cells of the PB exhibited lower expression of LC3A-II and displayed enhanced expression of GFP-LC3 compared with those of their counterparts in BM (supplemental Figure 9D-E). Therefore, normal BM or PB myeloid cells have no dependency on autophagy for at least short-term survival, suggesting that enhanced autophagy dependency in PB is specific for MLL-ENL leukemia cells.
Role of autophagy in a murine CML–blast phase model
To investigate the role of autophagy in other AML models, we used BCR-ABL1 and NUP98-HOXA9–driven murine CML-BC models (supplementary Figure 1E).43 Similar to the MLL-ENL model, Atg7Δ/Δ CML-BC mice survived longer with no reduction in tumor burden in the BM or spleen (Figure 6A; supplemental Figure 10A-B). Moreover, Atg7Δ/Δ CML-BC mice showed a significantly diminished LK fraction, which contains LICs in this model44 (Figure 6B; supplemental Figure 10C). In addition, deletion of Atg7 reduced the WBC count and induced apoptosis specifically in PB leukemia cells, which was similar to results observed for MLL-ENL leukemia (Figure 6C-D). The BM of deceased or moribund CML-BC mice was occupied by myeloblasts irrespective of Atg7 deletion, suggesting leukemia development in these mice (supplemental Figure 10D). These results suggest that the leukemia promoting and PB cell supportive roles of autophagy are not restricted to MLL-ENL leukemia but are also present in other types of myeloid leukemia.
Requirement for autophagy in leukemia progression, stem cell maintenance, and PB leukemia cell survival in a murine CML-BC model. (A) Kaplan-Meier curves representing vehicle-treated (n = 9) and TAM-treated (n = 9) Atg7flox/flox:Cre-ERT2 CML-BC mice. The P value was calculated by a log-rank test. (B) Frequencies of LIC fractions (GFP+KusabiraOrange+Lineage−c-Kit+) in BM from vehicle-treated (n = 6) or TAM-treated (n = 7) Atg7flox/flox:Cre-ERT2 CML-BC mice were determined by flow cytometric analysis. (C) WBC counts in PB from vehicle- or TAM-treated Atg7flox/flox:Cre-ERT2 CML-BC mice at day 14 after BMT were plotted (n = 6 per group). (D) Frequencies of annexin-V+ or DAPI+ cells in c-Kit−CD11b+ cells in BM or PB from vehicle- or TAM-treated Atg7flox/flox:Cre-ERT2 CML-BC mice were plotted (n = 6 per group). *P < .05; **P < .01. All results represent pooled data from at least 3 independent experiments with 2 to 3 animals per experiment.
Requirement for autophagy in leukemia progression, stem cell maintenance, and PB leukemia cell survival in a murine CML-BC model. (A) Kaplan-Meier curves representing vehicle-treated (n = 9) and TAM-treated (n = 9) Atg7flox/flox:Cre-ERT2 CML-BC mice. The P value was calculated by a log-rank test. (B) Frequencies of LIC fractions (GFP+KusabiraOrange+Lineage−c-Kit+) in BM from vehicle-treated (n = 6) or TAM-treated (n = 7) Atg7flox/flox:Cre-ERT2 CML-BC mice were determined by flow cytometric analysis. (C) WBC counts in PB from vehicle- or TAM-treated Atg7flox/flox:Cre-ERT2 CML-BC mice at day 14 after BMT were plotted (n = 6 per group). (D) Frequencies of annexin-V+ or DAPI+ cells in c-Kit−CD11b+ cells in BM or PB from vehicle- or TAM-treated Atg7flox/flox:Cre-ERT2 CML-BC mice were plotted (n = 6 per group). *P < .05; **P < .01. All results represent pooled data from at least 3 independent experiments with 2 to 3 animals per experiment.
Inactivation of autophagy potentiated the therapeutic effects of AraC in vivo
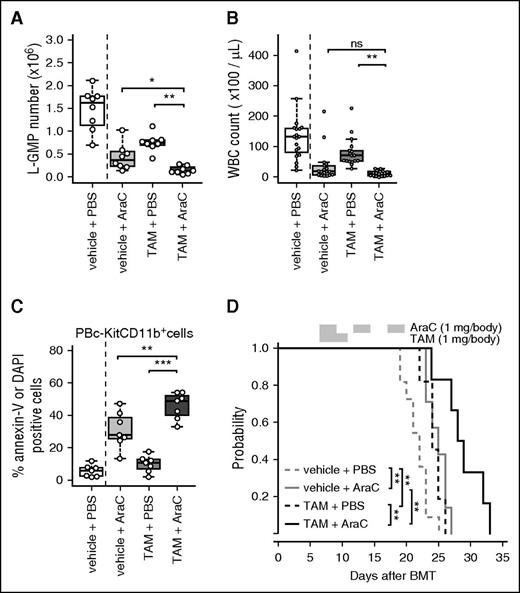
Although the contribution of autophagy to drug resistance in AML has been investigated by in vitro culture, the exact role of autophagy in drug resistance in vivo remains elusive. Hence, the effect of autophagy inactivation in combination with AraC, one of the most common clinically used chemotherapeutic agents for AML, was investigated in MLL-ENL leukemic mice. First, the expression of GFP-LC3 in MLL-ENL leukemia cells after AraC treatment was measured. AraC treatment reduced GFP-LC3 expression in L-GMP cells but not in lineage+ cells (supplemental Figure 11A-C). The overall survival of Atg7flox/flox:Cre-ERT2 MLL-ENL mice was enhanced in the cohort treated with TAM and AraC, indicating that Atg7 deletion potentiated the cytotoxic effect of AraC (Figure 7A). Atg7 deletion showed an additive effect with AraC treatment on BM cell reduction (supplemental Figure 11D). Furthermore, Atg7 deletion enhanced the efficacy of AraC in reducing the frequency and the absolute number of LICs (Figure 7B; supplemental Figure 11E). When the effect of TAM plus AraC therapy was assessed in PB from Atg7flox/flox:Cre-ERT2 MLL-ENL mice, WBC counts in MLL-ENL mice treated with TAM and AraC were prominently reduced and were comparable to those of mice treated with AraC alone (Figure 7C). Of note, AraC treatment combined with Atg7 deletion resulted in increased apoptosis in PB MLL-ENL leukemia cells compared with AraC treatment or Atg7 deletion alone; however, the combinatorial effect on apoptosis in BM leukemia cells was not significant (Figure 7D; supplemental Figure 11F). In deceased or moribund TAM- and AraC-treated mice, the BM was occupied by myeloblasts similar to that observed in control mice, suggesting that these mice developed leukemia before death (supplemental Figure 11G). Altogether, Atg7 deletion potentiated the efficacy of AraC treatment, and especially the capacity to target LICs, which highlights the important role of autophagy on AraC resistance in MLL-ENL leukemia.
Combinatorial effect of AraC treatment and Atg7 deletion in MLL-ENL leukemic mice. (A) Kaplan-Meier curves of Atg7flox/flox:Cre-ERT2 MLL-ENL leukemic mice treated with vehicle or TAM in combination with PBS or AraC (n = 6-11 mice per group). The P value was calculated by a log-rank test. (B) L-GMP numbers in GFP+ BM MLL-ENL cells were plotted (n = 8 per group). (C) WBC counts in PB from Atg7flox/flox:Cre-ERT2 MLL-ENL mice receiving vehicle + PBS (n = 19), vehicle + AraC (n = 18), TAM + PBS (n = 17), and TAM + AraC (n = 19) were plotted. (D) Frequencies of annexin-V+ or DAPI+ cells in c-Kit−CD11b+ PB from Atg7flox/flox:Cre-ERT2 MLL-ENL mice receiving vehicle + PBS (n = 6), vehicle + AraC (n = 7), TAM + PBS (n = 7), and TAM + AraC (n = 7) were plotted. Multiple comparisons were performed by analysis of variance followed by a Student t test, adjusting for multiple testing using Holm's step-down method. **P < .01; *** P < .001. All results represent pooled data from at least 2 independent experiments with 1 to 2 animals per experiment.
Combinatorial effect of AraC treatment and Atg7 deletion in MLL-ENL leukemic mice. (A) Kaplan-Meier curves of Atg7flox/flox:Cre-ERT2 MLL-ENL leukemic mice treated with vehicle or TAM in combination with PBS or AraC (n = 6-11 mice per group). The P value was calculated by a log-rank test. (B) L-GMP numbers in GFP+ BM MLL-ENL cells were plotted (n = 8 per group). (C) WBC counts in PB from Atg7flox/flox:Cre-ERT2 MLL-ENL mice receiving vehicle + PBS (n = 19), vehicle + AraC (n = 18), TAM + PBS (n = 17), and TAM + AraC (n = 19) were plotted. (D) Frequencies of annexin-V+ or DAPI+ cells in c-Kit−CD11b+ PB from Atg7flox/flox:Cre-ERT2 MLL-ENL mice receiving vehicle + PBS (n = 6), vehicle + AraC (n = 7), TAM + PBS (n = 7), and TAM + AraC (n = 7) were plotted. Multiple comparisons were performed by analysis of variance followed by a Student t test, adjusting for multiple testing using Holm's step-down method. **P < .01; *** P < .001. All results represent pooled data from at least 2 independent experiments with 1 to 2 animals per experiment.
Discussion
In this study, we evaluated the role of autophagy in leukemia progression, LIC maintenance, and drug resistance in vivo. Our results revealed that in the BM autophagy is specifically required for LICs, but not for differentiated leukemic blasts, to prevent oxidative stress; this should result in the maintenance of appropriate LIC frequency. In contrast, in the PB, autophagy supported survival of leukemia cells irrespective of their differentiation status. Furthermore, inactivation of autophagy potentiated the efficacy of AraC treatment in vivo.
The reason why in the BM LICs, but not other myeloblasts, required autophagy remains elusive. Our data support the hypothesis that accumulation of mitochondria and ROS production are possibly associated with the maintenance of LICs. In fact, normal HSCs have a fewer numbers of mitochondria than those of progenitor cells and use glycolytic metabolism in the hypoxic BM niche.45 Although leukemia cells contain more numbers of mitochondria than those of normal counterparts,46 fewer mitochondria might be required for maintenance of self-renewal capacity in LICs. These mechanisms are expected to be affected by the microenvironment, and this might account for inconsistencies in previous reports, specifically that pharmacological inhibition of autophagy did not inhibit proliferation in leukemic cell lines.20,23 For comprehensive understanding of the mechanisms of autophagy in LIC maintenance, further investigation is warranted.
Our study could also help to characterize the impact of autophagy on AraC resistance in vivo. The relationship between autophagy and drug resistance has been vigorously investigated; histone deacetylase inhibitors, mammalian target of rapamycin inhibitors, and AraC were all affected by autophagy in AML cell lines.20-24 However, considering that the dependency on autophagy in leukemia cells is expected to be highly affected the surroundings such as the BM or PB microenvironment, it is important to evaluate if autophagy has an essential role in vivo. As expected, we revealed that in vivo treatment of AraC activated autophagy specifically in LICs, and that Atg7 deletion potentiated the therapeutic efficacy of AraC.
An excess of leukemic blasts in the PB is known to be a sign of overflow from the BM. Our unique findings could suggest that PB leukemia cells also have leukemia-initiating capacity, which is partly regulated by autophagy. Possible explanations for the dependency on autophagy in PB leukemia cells can be derived from several environmental differences between BM and PB. These include oxygen partial pressure, cytokine concentrations, and adhesion to stromal cells, among others. These factors might render PB leukemia cells more dependent on autophagy. Taking into account the recent finding that granulocyte colony-stimulating factor–mobilized PB HSCs have enhanced autophagy and dependency on autophagy,47 it is suggested that undifferentiated cells, which normally reside in BM, might require autophagy to manage the cellular stress of the PB environment.
In this study, we detected an enhanced number of autophagosomes in PB leukemia cells through both electron microscopy and biochemical methods. At the same time, we confirmed that the degradation of autophagic substrates was induced in PB leukemia cells through measuring GFP-LC3 expression. These results support the notion that autophagy is activated in PB leukemia cells, although further experiments by using lysosome inhibitors are still required to confirm autophagic flux.48 Molecular mechanisms underlying the possible activation of autophagy in PB leukemia cells were not revealed by this study. Semicomprehensive reverse transcription polymerase chain reaction analysis revealed that a small number of genes were upregulated or downregulated in PB cells (greater than twofold), although expression of the majority of autophagy-associated genes was not altered (data not shown). More precise investigation of the mechanisms of altered autophagy would provide further understanding of the intratumor heterogeneity of AML.
Some issues remain before autophagy inhibitors can be applied to human AML. First, we revealed that survival in leukemic mice was slightly but significantly prolonged after Atg5/7 deletion. To determine the clinical importance of this finding, similar studies using models that have high clinical predictability, such as human AML-transplanted immunodeficient mice, are required. Second, the fact that normal HSCs also depend on autophagy (shown in previous reports) raises concern about possible toxicity of autophagy inhibition.34,40,49 However, defects in HSCs and myeloproliferation, specifically caused by autophagy deficiency, seem to be observed 6 to 9 weeks after autophagy inactivation. In contrast, the deficiencies in LICs were detectable 1 week after TAM treatment in our study, implying that LICs might be more susceptible to autophagy inhibition than HSCs. As our study did not evaluate the effects of systemic inhibition of autophagy, further investigation is required. Third, it has recently been reported that heterozygous deletion of Atg5 results in increased leukemogenic potential in a murine BMT model with transduction of MLL-ENL.50 In that study, it was also shown that homozygous deletion of Atg5 induces substantial cell death in a MLL-ENL induced leukemic cell line. These results suggest that complete ablation of autophagy could inhibit tumor survival or self-renewal activity; however, mild inactivation of autophagy might enhance the malignant phenotype of MLL-ENL leukemia. Thus, small-molecule inhibitors with high efficacy might be required for successful autophagy inhibition therapy.
Despite these concerns, autophagy inactivation by small molecules including chloroquine is a promising concept for leukemia therapy. As autophagy inhibition reduced LIC frequency, with little effect on total tumor burden (as observed in this study), autophagy inhibitors might be suitable for use in combination with other conventional drugs such as AraC or anthracyclines. Furthermore, based on the higher dependency on autophagy of PB leukemia cells, treatment with autophagy inhibitors combined with stem cell mobilizer drugs like plerixafor could be a promising for antileukemia therapy.
In summary, our findings demonstrate that leukemia cells use cytoprotective autophagy for LIC maintenance, survival in PB, and AraC resistance. Even considering that normal hematopoiesis requires autophagy, this function could be a possible therapeutic target in AML and other cancers.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank M. Komatsu for providing Atg7 conditional knockout mice; N. Mizushima for Atg5 conditional knockout mice and GFP-LC3 mice; T. Kitamura for Plat-E packaging cells; R. Ono and T. Nosaka for MLL-ENL cDNA; T. Nakamura for NUP98-HOXA9 cDNA; F. Ueki and R. Toyama for expert technical assistance; and Editage (http://www.editage.jp) for English-language editing.
This work was supported by the Japan Society for the Promotion of Science Grants-in-Aid for Scientific Research (KAKENHI) (24249055, 16H01196, and 15K09466).
Authorship
Contributions: Y.S., J.K., K.N., K.K., T.S., and M.K. designed the research; Y.S., T.T.-K, and K.M. performed the experiments; and Y.S., J.K., K.N., T.S., and M.K. wrote the manuscript.
Conflict-of-interest disclosure: Y.S. is an employee of Kyowa Hakko Kirin Co. Ltd. The remaining authors declare no competing financial interests.
Correspondence: Mineo Kurokawa, Department of Hematology & Oncology, Graduate School of Medicine, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-8655, Japan; e-mail: kurokawa-tky@umin.ac.jp.

![Figure 4. Dependence of PB leukemia cells on autophagy. (A) WBC counts of PB from Atg5flox/flox:Cre-ERT2 or Atg7flox/flox:Cre-ERT2 MLL-ENL leukemic mice were measured at indicated days after BMT (>7 mice per group, mean ± standard deviation [SD]). P values were calculated by a Welch t test. Collective data of frequencies of annexin-V+ or DAPI+ cells in c-Kit−CD11b+ or c-Kit+CD11b+ cells obtained from vehicle- or TAM-treated Atg7flox/flox:Cre-ERT2 (B) and Atg5flox/flox:Cre-ERT2 (C) mice (n = 6-9 per group). (D) Relative colony numbers from GFP+ PB cells sorted from vehicle- or TAM-treated Atg7flox/flox:Cre-ERT2 MLL-ENL leukemic mice are shown (n = 4 per group, mean ± SD). (E) Kaplan-Meier curves representing survival of tertiary recipients of GFP+ PB cells from vehicle- or TAM-treated Atg7flox/flox:Cre-ERT2 MLL-ENL leukemic mice (n = 9 per group). The P value was calculated by a log-rank test. *P < .05; **P < .01. All results represent pooled data from at least 3 independent experiments with 1 to 4 animals per experiment.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/128/12/10.1182_blood-2015-12-684696/4/m_1614f4.jpeg?Expires=1769795425&Signature=1stbPcOLkdizIr5poj3~lc1qTzt9GjIOnuuuOEm5ra-UseSuhN4dtBK87xnnvqzio36ILdz49lweXxg1gpcLn3-MtypeufKdejpJGxKBPOyffInR8NEzAnhC~7yK9gA3qv8G77WuKRPjjaaLtit9dOVmJQMT~LsE3O89tldZ~jtlOZ4zVm9yLZYVvC53zVL~6GVrNCliJuLZMHClwxNQPeBIrofYBaaxFtX8sOhJbDtEdHIGNAaDQERIGwbKj9rPXxNINbUKVtRsGprIIkRocb71IbivtreGlo9dEQ7OPUHGFIkVY~MaPxEZiQ0d6YmcUdsT5Sb53S1jcIdfP15jDA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal