Key Points
Nonpermissive mismatches associated with survival after HCT reflect FD between recipient-donor HLA-DPB1.
FD within HLA-DPB1 is determined by the combined impact of nonconservative peptide-binding AA substitutions.
Abstract
The role of HLA amino acid (AA) polymorphism for the outcome of hematopoietic cell transplantation (HCT) is controversial, in particular for HLA class II. Here, we investigated this question in nonpermissive HLA-DPB1 T-cell epitope (TCE) mismatches reflected by numerical functional distance (FD) scores, assignable to all HLA-DPB1 alleles based on the combined impact of 12 polymorphic AAs. We calculated the difference in FD scores (ΔFD) of mismatched HLA-DPB1 alleles in patients and their 10/10 HLA-matched unrelated donors of 379 HCTs performed at our center for acute leukemia or myelodysplastic syndrome. Receiver-operator curve-based stratification into 2 ΔFD subgroups showed a significantly higher percentage of nonpermissive TCE mismatches for ΔFD >2.665, compared with ΔFD ≤2.665 (88% vs 25%, P < .0001). In multivariate analysis, ΔFD >2.665 was significantly associated with overall survival (hazard ratio [HR], 1.40; 95% confidence interval [CI], 1.05-1.87; P < .021) and event-free survival (HR, 1.39; 95% CI, 1.05-1.82; P < .021), compared with ΔFD ≤2.665. These associations were stronger than those observed for TCE mismatches. There was a marked but not statistically significant increase in the hazards of relapse and nonrelapse mortality in the high ΔFD subgroup, whereas no differences were observed for acute and chronic graft-versus-host disease. Seven nonconservative AA substitutions in peptide-binding positions had a significantly stronger impact on ΔFD compared with 5 others (P = .0025), demonstrating qualitative differences in the relative impact of AA polymorphism in HLA-DPB1. The novel concept of ΔFD sheds new light onto nonpermissive HLA-DPB1 mismatches in unrelated HCT.
Introduction
Recipient-donor disparity for polymorphic HLA molecules is a frequent condition in unrelated hematopoietic cell transplantation (HCT), which is performed in 10% to 25% of cases across HLA-A, B, C, or DRB1 mismatches,1 and in more than 80% of cases across HLA-DPB1 mismatches.2-4 It is well established that HLA disparity increases the risks of adverse clinical outcome including overall and nonrelapse mortality (NRM), as well as graft-versus-host disease (GVHD), although it can also mediate a beneficial graft-versus-leukemia (GVL) effect.5-10 The identification of clinically permissive, ie, well-tolerated HLA mismatches is the subject of intensive research efforts that include the search for high-risk mismatch combinations,11-13 structural comparison of HLA molecules,14,15 and the identification of shared T-cell epitopes (TCEs) in mismatched HLA-DPB1 alleles.4,9,16,17 Considerable attention has also been given to the association between specific amino acid (AA) substitutions in mismatched HLA class I alleles and adverse outcome,11,18-20 resulting in a number of bio-informatic models for in silico outcome prediction.21-25 For HLA class II, structural variability has been extensively studied in the context of solid organ transplantation, with some in silico models predictive of antibody formation and/or kidney transplant outcome already entered into clinical use.26,27 These models did not prove equally valid for HCT outcome prediction,23,28 probably reflecting the more complex nature of HLA-peptide recognition by the T-cell receptor (TCR) compared with allo-antibodies.29,30
Here, we have addressed the functional role of AA polymorphism in HLA class II for clinical outcome of HCT in the context of nonpermissive TCE mismatches at HLA-DPB1, shown by some4,9 but not all31 multicenter studies to be associated with mortality after 10/10 HLA-matched unrelated HCT. We previously experimentally established a detailed landscape of the functional impact of single AA substitutions in HLA-DPB1 on in vitro T-cell allo-reactivity.32 This showed a correlation between the combined impact of AA substitutions in HLA-DPB1 alleles relative to HLA-DPB1*09:01, designated functional distance (FD) with HLA-DPB1 TCE groups, an observation that is since being used for updating the free online “DPB1 T-Cell Epitope Algorithm” for the assignment of nonpermissive TCE mismatches with newly described HLA-DPB1 alleles.33,34 We hypothesized that the difference between the FD (ΔFD) scores of HLA-DPB1 alleles from recipients and donors, could be a surrogate for nonpermissive TCE mismatches in unrelated HCT. This hypothesis was tested here in the clinical context of 10/10 HLA-matched unrelated HCT, and by in-depth analysis of the structural and biochemical characteristics of AA substitutions in relation to their functional impact.
Patients and methods
HLA typing
Genomic high resolution typing (second field) of HLA-A, B , C, DRB1, DQB1, and DPB1 was performed by sequence-specific oligonucleotide probing (LABType SSO; One Lambda, Canoga Park, CA), sequence-specific priming (Olerup SSP; Olerup SSP AB, Stockholm, Sweden), and sequence-based typing (CTS-Sequence; CTS, Heidelberg, Germany), according to previously described protocols35 under accreditation by the European Federation for Immunogenetics.
HLA-DPB1 TCE group matching
Patients, transplants, and outcome definitions
Patients who underwent a first unrelated HCT from a high-resolution (second field) 10/10 HLA-A, B, C, DRB1, and DQB1 identical unrelated donor for the treatment of acute myeloid leukemia (AML), myelodysplastic syndrome (MDS), or acute lymphoblastic leukemia (ALL) between 2005 and 2014 at the Department of Bone Marrow Transplantation of the University Hospital Essen (Essen, Germany) and for whom genomic DNA was available, were included in the study. Transplants were performed after written informed consent, under clinical protocols approved by the Ethical Review Board of the University Hospital Essen, in accordance with the Declaration of Helsinki.
Most patients received unmanipulated peripheral blood stem cells from, where possible, cytomegalovirus (CMV) serostatus-matched donors. GVHD prophylaxis consisted of standard short-course methotrexate and cyclosporine A in all patients, with or without anti–thymocyte-globulin (ATG). Details of patients, HLA-DPB1 mismatched transplants, and disease characteristics are listed in Table 1.
Patient and transplant characteristics
| . | HLA-DPB1 ΔFD ≤2.665 . | HLA-DPB1 ΔFD >2.665 . | . |
|---|---|---|---|
| . | N (%) . | N (%) . | P* . |
| Number of patients | 252 (67) | 127 (33) | — |
| Recipient age, median (range) | 53 (19-73) | 52 (19-71) | NS |
| Age at transplant | |||
| <Median age (53 y) | 122 (48) | 67 (53) | NS |
| ≥Median age (53 y) | 130 (52) | 60 (47) | — |
| Male sex | 130 (52) | 63 (50) | NS |
| Disease at transplant | |||
| AML | 177 (70) | 95 (75) | NS |
| ALL | 39 (15) | 19 (15) | — |
| MDS | 36 (14) | 13 (10) | — |
| Disease status at transplant | |||
| Early | 112 (44) | 53 (42) | NS |
| Intermediate | 60 (24) | 29 (23) | — |
| Advanced | 80 (32) | 45 (35) | — |
| Graft type | |||
| Bone marrow | 14 (6) | 6 (5) | NS |
| PBSC | 238 (94) | 121 (95) | — |
| Conditioning regimen | |||
| Myeloablative | 124 (49) | 58 (46) | NS |
| Reduced intensity | 128 (51) | 69 (54) | — |
| In vivo T-cell depletion (ATG) | |||
| Yes | 135 (64) | 56 (44) | NS |
| No | 117 (46) | 71 (56) | — |
| Donor/recipient sex match | |||
| Female/male | 15 (6) | 10 (8) | NS |
| Others | 237 (94) | 117 (92) | — |
| Donor/recipient CMV match | |||
| Negative/negative | 77 (30) | 36 (28) | NS |
| Positive/positive | 102 (40) | 46 (36) | NS |
| Positive/negative | 13 (5) | 9 (7) | — |
| Negative/positive | 60 (24) | 36 (28) | — |
| Donor age, median (range) | 35 (18-59) | 36 (18-58) | NS |
| Number of HLA-DPB1 allele mismatches | |||
| 1 | 194 (77) | 59 (46) | <.0001 |
| 2 | 58 (23) | 68 (54) | — |
| HLA-DPB1 TCE status | |||
| Matched (permissive) | 189 (75) | 15 (12) | <.0001 |
| Mismatched (nonpermissive) | 63 (25) | 112 (88) | — |
| HLA-DPB1 ΔFD, median (range) | 1.12 (0.01-2.66) | 3.47 (2.67-7.46) | <.0001 |
| . | HLA-DPB1 ΔFD ≤2.665 . | HLA-DPB1 ΔFD >2.665 . | . |
|---|---|---|---|
| . | N (%) . | N (%) . | P* . |
| Number of patients | 252 (67) | 127 (33) | — |
| Recipient age, median (range) | 53 (19-73) | 52 (19-71) | NS |
| Age at transplant | |||
| <Median age (53 y) | 122 (48) | 67 (53) | NS |
| ≥Median age (53 y) | 130 (52) | 60 (47) | — |
| Male sex | 130 (52) | 63 (50) | NS |
| Disease at transplant | |||
| AML | 177 (70) | 95 (75) | NS |
| ALL | 39 (15) | 19 (15) | — |
| MDS | 36 (14) | 13 (10) | — |
| Disease status at transplant | |||
| Early | 112 (44) | 53 (42) | NS |
| Intermediate | 60 (24) | 29 (23) | — |
| Advanced | 80 (32) | 45 (35) | — |
| Graft type | |||
| Bone marrow | 14 (6) | 6 (5) | NS |
| PBSC | 238 (94) | 121 (95) | — |
| Conditioning regimen | |||
| Myeloablative | 124 (49) | 58 (46) | NS |
| Reduced intensity | 128 (51) | 69 (54) | — |
| In vivo T-cell depletion (ATG) | |||
| Yes | 135 (64) | 56 (44) | NS |
| No | 117 (46) | 71 (56) | — |
| Donor/recipient sex match | |||
| Female/male | 15 (6) | 10 (8) | NS |
| Others | 237 (94) | 117 (92) | — |
| Donor/recipient CMV match | |||
| Negative/negative | 77 (30) | 36 (28) | NS |
| Positive/positive | 102 (40) | 46 (36) | NS |
| Positive/negative | 13 (5) | 9 (7) | — |
| Negative/positive | 60 (24) | 36 (28) | — |
| Donor age, median (range) | 35 (18-59) | 36 (18-58) | NS |
| Number of HLA-DPB1 allele mismatches | |||
| 1 | 194 (77) | 59 (46) | <.0001 |
| 2 | 58 (23) | 68 (54) | — |
| HLA-DPB1 TCE status | |||
| Matched (permissive) | 189 (75) | 15 (12) | <.0001 |
| Mismatched (nonpermissive) | 63 (25) | 112 (88) | — |
| HLA-DPB1 ΔFD, median (range) | 1.12 (0.01-2.66) | 3.47 (2.67-7.46) | <.0001 |
ΔFD, δ-functional distance; NS, not significant; PBSC, peripheral blood mobilized stem cell.
The Wilcoxon rank-sum test was used to test differences of continuous variables. Differences in frequencies of discrete variables were tested by the two-sided Fisher’s exact test.
Outcomes in this study were defined as follows: overall survival (OS) summarized the time interval between HCT and the last follow-up date of surviving patients within 5 years posttransplant; event-free survival (EFS) included the same time interval of patients surviving without hematologic disease recurrence after HCT; acute and chronic GVHD were diagnosed and clinically graded following the commonly accepted criteria36,37 ; hematologic disease relapse was defined by standard cytomorphologic blood and marrow criteria, or biopsy and/or analysis of cerebrospinal fluid in case of suspected isolated extramedullary or central nervous system relapse; and NRM was assumed as the time interval between HCT and death in all deceased patients without detectable disease recurrence or persistence after HCT.
Statistical methods
Differences in frequencies of discrete variables were tested by the 2-sided Fisher’s exact test. The Wilcoxon rank-sum test was used to test differences of continuous variables.
Receiver operating characteristic (ROC) analyses for the end point OS were performed for prediction of the best ΔFD cutoff values in terms of sensitivity and specificity, using the BIAS 10.02 software program (http://www.bias-online.de/).
For comparison of time-to-event end points without competing risks, ie, OS and EFS, the probabilities of events over time were calculated by the product-limit method and heterogeneity of time-to-event distribution functions was compared by the log-rank test with Šidák’s adjustment for multiple testing.38,39 To account for interactions of competing events on relapse (ie, death without relapse) and NRM (ie, relapse), the probabilities of events over time were estimated by cause-specific cumulative incidence rates.40,41 For the comparison of cumulative incidence rates between patient subsets, the time-to-event was compared by proportional hazards regression Cox models of the event-specific hazard functions using the 2-sided Wald test.42 In all multivariate proportional hazards general linear model (PHGLM) analyses on clinical end points, the stratified HLA-DPB1 ΔFD and TCE matching, pretransplant recipient-donor CMV serostatus, categorized patient age, disease stage, categorized disease duration (less or greater than 12 months), stem cell source, GVHD prophylaxis, and the European Society for Blood and Marrow Transplantation risk score were included as categorical covariates.43,44 All PHGLM analyses were performed using forward and backward selection steps. Only those covariates with a significance level below 5% were entered into the model building procedure. Further, only covariates with a significance level below 5% after adjustment for the other significant covariates selected in the forward and backward model building were regarded as significant in the final models. The hazard ratio (HR) and its 95% confidence limit were derived for each significant covariate included in the final PHGLM models. For those covariates not included in the final model, the HR and 95% confidence limits were derived after adjustment for all significant covariates in the final model. Statistical analysis and presentation was performed using Statistical Analysis Software (release 9.4, version number 7.100.1.2711, 2015) procedures and macros (SAS/STAT User’s Guide 14.1; Cary, NC). Date of the final analysis was March 18, 2016.
HLA-DP homology modeling
The crystal structure of HLA-DPB1*02:01 in association with HLA-DPA1*01:03 (PDB-ID: 3LQZ; http://www.rcsb.org)45,46 served as a template for homology modeling of HLA-DPB1*09:01 in association with HLA-DPA1*02:01 and the HLA-DP9 restricted peptide MP-10R13 derived from the streptococcal M12 protein,47 using the Swiss Model Workspace.48 The QMEAN z score was used to estimate the quality of the model.49 Single AA substitutions were modeled by exchanging AAs encoded by HLA-DPB1*09:01 with the corresponding residues encoded by other DPB1 alleles. Subsequently, few cycles of energy minimization were performed to release any internal structural constraints by using the GROningen MOlecular Simulation 43B1 force field.50,51 Model analysis and pictures were generated with the Swiss-PDB Viewer Software DeepView, version 4.1.52 Nonconservative AA substitutions were defined as those introducing different biochemical properties in terms of charge, hydrophobicity, and/or size.53
Results
Definitions of FD scores in HLA-DPB1
We previously defined FD in HLA-DPB1 as the combined impact of 10 polymorphic AAs encoded in exon 2 of HLA-DPB1 alleles, on recognition by T cells allo-reactive to wild-type (WT) HLA-DPB1*09:01.32 HLA-DPB1*09:01 was chosen as a reference because it is the prototype of immunogenic HLA-DPB1 alleles from the TCE model.16 FD is present at 3 interdependent levels: the AA level, the HLA-DPB1 allele level, and the HCT patient-donor pair level (Figure 1).54-56
HLA-DPB1 FD scores. (A) Schematic representation of the peptide antigen-binding groove encoded by HLA-DPB1*09:01 and definition of FDAA, FDAllele, and ΔFD scores. In the HLA-DPB1*09:01 molecule, the positions and side chains of 10 polymorphic AA residues used for the determination of FDAA scores are listed in boldface. The primary data for the development of FDAA and FDAllele scores were published previously.32 Briefly, 10 AA residues most relevant for peptide binding and/or TCR contact were selected, based on homology modeling and the available literature. Most of these AA residues are bimorphic (ie, only 2 different variants have been reported in the most frequent HLA-DPB1 alleles in Europeans),56 therefore only 1 variant with respect to WT HLA-DPB1*09:01 was analyzed. For two residues, namely at position 9 and 35, 3 different variants have been reported in the most frequent HLA-DPB1 alleles in Europeans,56 and both variants with respect to WT HLA-DPB1*09:01 were analyzed, for a total of 12 AA substitutions. FDAA scores for each of these 12 AAs were obtained as follows: the median RR of 17 clonal T-cell effectors allo-reactive to HLA-DPB1*09:01 as reference was experimentally determined and FDAA scores were then calculated as [1 – median RR]. The FDAllele score for individual HLA-DPB1 alleles, calculated as the sum of FDAA scores as shown in the figure, correlate well with TCE groups based on T-cell cross-reactivity patterns, and allow us to predict TCE group assignment for all known HLA-DPB1 alleles.32 (B) FDAllele scores of 19 HLA-DPB1 alleles occurring with a frequency of >0.5% in Europeans.56 TCE group assignment of the HLA-DPB1 alleles16,32,54 is shown on top.
HLA-DPB1 FD scores. (A) Schematic representation of the peptide antigen-binding groove encoded by HLA-DPB1*09:01 and definition of FDAA, FDAllele, and ΔFD scores. In the HLA-DPB1*09:01 molecule, the positions and side chains of 10 polymorphic AA residues used for the determination of FDAA scores are listed in boldface. The primary data for the development of FDAA and FDAllele scores were published previously.32 Briefly, 10 AA residues most relevant for peptide binding and/or TCR contact were selected, based on homology modeling and the available literature. Most of these AA residues are bimorphic (ie, only 2 different variants have been reported in the most frequent HLA-DPB1 alleles in Europeans),56 therefore only 1 variant with respect to WT HLA-DPB1*09:01 was analyzed. For two residues, namely at position 9 and 35, 3 different variants have been reported in the most frequent HLA-DPB1 alleles in Europeans,56 and both variants with respect to WT HLA-DPB1*09:01 were analyzed, for a total of 12 AA substitutions. FDAA scores for each of these 12 AAs were obtained as follows: the median RR of 17 clonal T-cell effectors allo-reactive to HLA-DPB1*09:01 as reference was experimentally determined and FDAA scores were then calculated as [1 – median RR]. The FDAllele score for individual HLA-DPB1 alleles, calculated as the sum of FDAA scores as shown in the figure, correlate well with TCE groups based on T-cell cross-reactivity patterns, and allow us to predict TCE group assignment for all known HLA-DPB1 alleles.32 (B) FDAllele scores of 19 HLA-DPB1 alleles occurring with a frequency of >0.5% in Europeans.56 TCE group assignment of the HLA-DPB1 alleles16,32,54 is shown on top.
FD at the AA level (FDAA) is a numerical score reporting the difference in the median relative response (RR) of 17 different allo-reactive T-cell effectors to WT HLA-DPB1*09:01 (arbitrarily set as 1) and to mutant HLA-DPB1*09:01 carrying 1 out of 12 naturally occurring AA substitutions at 10 different polymorphic positions, each as single-point mutation.32 FDAA scores thus represent the median functional impact of an individual AA substitution on T-cell allo-recognition of HLA-DPB1*09:01 (Figure 1A). The median FDAA score of the 12 AA substitutions analyzed is 0.71 (−0.12 to 0.96), with an FDAA score of 0.00 for AA encoded by WT HLA-DPB1*09:01 in each of the 10 relevant positions.
FD at the allele level (FDAllele) is a numerical score obtained as the sum of the FDAA scores of AAs encoded by any HLA-DPB1 allele in the 10 relevant positions32 (Figure 1A). Because all FDAA scores in WT HLA-DPB1*09:01 have the value 0.00, the FDAllele score of HLA-DPB1*09:01 is 0.00 also. AA substitutions different in position or type from those investigated for the 12 FDAA scores are not considered. The median FDAllele score of the most frequent HLA-DPB1 alleles in Europeans is 2.36 (−0.02 to 5.64). FDAllele scores of all HLA-DPB1 alleles known to date are presented in supplemental Table 1, available on the Blood Web site. FDAllele scores correlate well with the TCE group classification, with FDAllele score ranges of −0.16 to 0.59, 0.60 to 1.99, and 2.00 to 5.67 for TCE groups 1, 2, and 3, respectively16,32,54 (Figure 1B).
FD at the HCT recipient-donor pair level (ΔFD) is a numerical score obtained as the absolute difference (ie, no negative or positive sign and hence no graft-vs-host or a host-vs-graft vector) between the sum of the FDAllele scores of the 2 HLA-DPB1 alleles in the recipient and the sum of the FDAllele scores of the 2 HLA-DPB1 alleles in the donor (Figure 1A and supplemental Table 2). In case of HLA-DPB1 homozygosity, the FDAllele score of the relevant allele is counted twice. The ΔFD concept was coined in the present study and tested for its association with clinical outcome of unrelated HCT.
Patients and outcomes
A total of 416 adult patients who received a 10/10 HLA-matched unrelated donor HCT for the treatment of AML, ALL, or MDS at the Department of Bone Marrow Transplantation of the University Hospital Essen were included in the study. A total of 37 recipient-donor pairs were matched for both HLA-DPB1 alleles, whereas the remaining 379 pairs were HLA-DPB1 mismatched. The latter were used for evaluation of the association between HLA-DPB1 ΔFD scores and outcome, and their clinical and transplant characteristics are listed in Table 1.
With a median follow-up of 4 years for surviving patients, the estimates for different outcome end points in the entire cohort were as follows: OS 48%, EFS 42%, NRM 29%, relapse 29%, grades II-IV and III-IV acute GVHD 38% and 15%, respectively, and chronic GVHD 68%. No significant differences were found for any of these end points between HLA-DPB1 allele-matched or mismatched transplants (data not shown).
Impact of HLA-DPB1 ΔFD matching
The median ΔFD score of all 379 HLA-DPB1 mismatched recipient-donor pairs was 1.64 (0.01 to 7.46). ROC analysis indicated stratification into 2 subgroups with ΔFD scores ≤2.665 (n = 252 [67%]) and >2.665 (n = 127 [33%]) as the best predictor of OS, with a sensitivity of 39.6%, a specificity of 73.4%, and an area under the curve of .559 (P = .045). Similar data were found for EFS, whereas no significant ROC cutoff values were obtained for any of the other clinical end points. The 2 subgroups showed no significant differences for the distribution of major clinical variables including diagnosis, disease status at transplant, immune prophylaxis, and conditioning regimen (Table 1). However, the percentage of single HLA-DPB1 allele mismatches was significantly higher in the subgroup with ΔFD scores ≤2.665 (77% vs 46%; P < .0001). Moreover, the percentage of nonpermissive HLA-DPB1 TCE mismatches was significantly lower in the same subgroup (25% vs 88%; P < .0001) (Table 1). This shows a significant but not complete overlap between the HLA-DPB1 ΔFD and the TCE models, which were concordant in 301/379 (79.4%) pairs (ΔFD score ≤2.665 and TCE permissive [N = 189], or ΔFD scores >2.665 and TCE nonpermissive [N = 112]), and discordant in 78/379 (20.6%) pairs (ΔFD score ≤2.665 and TCE nonpermissive [N = 63], or ΔFD scores >2.665 and TCE permissive [N = 15]).
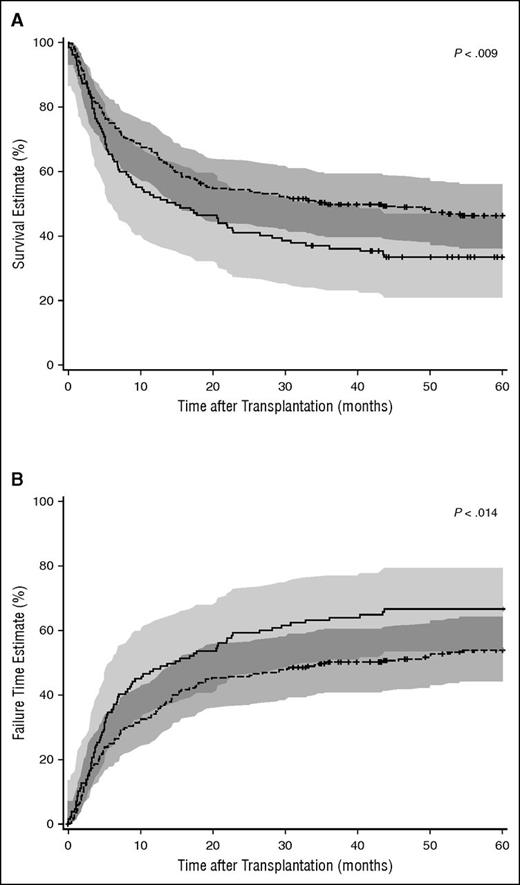
OS and EFS were significantly superior after HCT with ΔFD scores ≤2.665 compared with ΔFD scores >2.665, with respective Kaplan-Meier probabilities for the 2 subgroups of 52% vs 38% (P < .009; Figure 2A) for OS and 54% vs 66% (P < .014; Figure 2B) for treatment failure, the reverse of EFS. This was not due to the observed differences in single vs double HLA-DPB1 allele mismatches, because the Kaplan-Meier probabilities of OS and EFS were identical for these 2 groups (46% vs 47%, P = .93 for OS and 41% vs 44%, P = .72 for EFS), and also the multivariate HR of OS and EFS were not significantly different (Table 2). Likewise, it could not be entirely accounted for by the differences in percentages of nonpermissive TCE mismatches in the 2 subgroups, because Kaplan-Meier probabilities of OS were lower for nonpermissive compared with permissive TCE mismatches but this was not statistically significant (50% vs 44%; P = .31).
Kaplan–Meier OS and treatment failure estimates by HLA-DPB1 ΔFD. HLA-DPB1 ΔFD ≤2.665 and >2.665 are shown in dashed and solid lines, respectively, with 95% confidence limits in shades of gray above and below each curve. The significance of differences between strata was calculated by the log-rank test. (A) Kaplan-Meier estimates of OS. P < .009. (B) Kaplan-Meier estimates of treatment failure. P < .014.
Kaplan–Meier OS and treatment failure estimates by HLA-DPB1 ΔFD. HLA-DPB1 ΔFD ≤2.665 and >2.665 are shown in dashed and solid lines, respectively, with 95% confidence limits in shades of gray above and below each curve. The significance of differences between strata was calculated by the log-rank test. (A) Kaplan-Meier estimates of OS. P < .009. (B) Kaplan-Meier estimates of treatment failure. P < .014.
Multivariate analyses: effect of HLA-DPB1 ΔFD or TCE matching status and other non-HLA variables on clinical outcome
| Outcome and variable . | N* . | HR . | 95% CI . | P† . |
|---|---|---|---|---|
| OS | ||||
| HLA-DPB1 ΔFD | 132 (52) / 49 (39) | 1.40 | 1.05-1.87 | <.021 |
| HLA-DPB1 TCE | 103 (51) / 78 (44) | 1.15 | 0.86-1.54 | <.360 |
| Number of HLA-DPB1 allele mismatches | 120 (47) / 61 (48) | 0.97 | 0.71-1.32 | <.830 |
| Recipient age | 102 (54) / 79 (42) | 1.69 | 1.26-2.27 | <.0006 |
| Disease status at transplant | 96 (58) / 44 (49) / 41 (33) | 1.30 | 1.10-1.55 | <.003 |
| In vivo T-cell depletion (ATG) | 69 (37) / 112 (59) | 0.56 | 0.41-0.76 | <.0003 |
| EFS | ||||
| HLA-DPB1 ΔFD | 119 (47) / 43 (34) | 1.39 | 1.05-1.82 | <.021 |
| HLA-DPB1 TCE | 92 (45) / 70 (40) | 1.09 | 0.84-1.43 | <.510 |
| Number of HLA-DPB1 allele mismatches | 105 (42) / 57 (45) | 0.92 | 0.68-1.25 | <.588 |
| Recipient age | 93 (49) / 69 (36) | 1.59 | 1.20-2.10 | <.002 |
| Disease status at transplant | 84 (51) / 41 (46) / 37 (30) | 1.24 | 1.05-1.46 | <.010 |
| In vivo T-cell depletion (ATG) | 65 (35) / 97 (51) | 0.65 | 0.49-0.87 | <.004 |
| Relapse | ||||
| HLA-DPB1 ΔFD | 67 (27) / 41 (32) | 1.69 | 0.99-2.87 | <.055 |
| HLA-DPB1 TCE | 57 (28) / 51 (29) | 0.75 | 0.45-1.24 | <.261 |
| Number of HLA-DPB1 allele mismatches | 72 (28) / 36 (29) | 0.97 | 0.63-1.49 | <.901 |
| Recipient age | 54 (29) / 54 (28) | 1.33 | 0.88-2.01 | <.178 |
| Disease status at transplant | 49 (30) / 20 (22) / 39 (31) | 1.15 | 0.90-1.46 | <.255 |
| In vivo T-cell depletion (ATG) | 55 (29) / 53 (28) | 0.85 | 0.56-1.27 | <.422 |
| NRM | ||||
| HLA-DPB1 ΔFD | 67 (27) / 42 (33) | 1.48 | 0.88-2.51 | <.143 |
| HLA-DPB1 TCE | 54 (27) / 55 (31) | 0.98 | 0.59-1.61 | <.931 |
| Number of HLA-DPB1 allele mismatches | 76 (30) / 33 (26) | 0.81 | 0.53-1.26 | <.366 |
| Recipient age | 42 (22) / 67 (35) | 2.13 | 1.40-3.24 | <.0005 |
| Disease status at transplant | 34 (20) / 27 (30) / 48 (39) | 1.34 | 1.06-1.69 | <.016 |
| In vivo T-cell depletion (ATG) | 68 (36) / 41 (21) | 0.52 | 0.34-0.78 | <.002 |
| Acute GVHD grades II-IV | ||||
| HLA-DPB1 ΔFD | 96 (38) / 44 (35) | 0.99 | 0.63-1.57 | <.985 |
| HLA-DPB1 TCE | 80 (39) / 60 (34) | 0.82 | 0.53-1.27 | <.382 |
| Number of HLA-DPB1 allele mismatches | 93 (37) / 47 (37) | 1.08 | 0.74-1.58 | <.684 |
| Recipient age | 73 (39) / 67 (35) | 0.78 | 0.54-1.10 | <.158 |
| Disease status at transplant | 55 (33) / 32 (36) / 53 (43) | 1.10 | 0.90-1.36 | <.354 |
| In vivo T-cell depletion (ATG) | 74 (39) / 66 (34) | 0.91 | 0.63-1.33 | <.634 |
| Acute GVHD grades III-IV | ||||
| HLA-DPB1 ΔFD | 37 (15) / 18 (14) | 0.98 | 0.48-2.01 | <.967 |
| HLA-DPB1 TCE | 30 (15) / 25 (14) | 1.00 | 0.51-1.99 | <.993 |
| Number of HLA-DPB1 allele mismatches | 38 (15) / 17 (13) | 0.88 | 0.47-1.64 | <.691 |
| Recipient age | 28 (15) / 27 (14) | 1.02 | 0.58-1.79 | <.954 |
| Disease status at transplant | 21 (13) / 8 (9) / 26 (21) | 1.19 | 0.85-1.68 | <.304 |
| In vivo T-cell depletion (ATG) | 35 (19) / 20 (10) | 0.59 | 0.33-1.01 | <.090 |
| Chronic GVHD | ||||
| HLA-DPB1 ΔFD | 118 (47) / 67 (53) | 0.81 | 0.56-1.20 | <.282 |
| HLA-DPB1 TCE | 86 (42) / 99 (56) | 1.42 | 1.01-1.90 | <.016 |
| Number of HLA-DPB1 allele mismatches | 119 (47) / 66 (52) | 1.08 | 0.78-1.49 | <.656 |
| Recipient age | 96 (51) / 89 (47) | 0.86 | 0.63-1.18 | <.349 |
| Disease status at transplant | 82 (50) / 49 (55) / 54 (44) | 0.90 | 0.75-1.09 | <.278 |
| In vivo T-cell depletion (ATG) | 95 (51) / 90 (47) | 0.71 | 0.53-0.95 | <.021 |
| Outcome and variable . | N* . | HR . | 95% CI . | P† . |
|---|---|---|---|---|
| OS | ||||
| HLA-DPB1 ΔFD | 132 (52) / 49 (39) | 1.40 | 1.05-1.87 | <.021 |
| HLA-DPB1 TCE | 103 (51) / 78 (44) | 1.15 | 0.86-1.54 | <.360 |
| Number of HLA-DPB1 allele mismatches | 120 (47) / 61 (48) | 0.97 | 0.71-1.32 | <.830 |
| Recipient age | 102 (54) / 79 (42) | 1.69 | 1.26-2.27 | <.0006 |
| Disease status at transplant | 96 (58) / 44 (49) / 41 (33) | 1.30 | 1.10-1.55 | <.003 |
| In vivo T-cell depletion (ATG) | 69 (37) / 112 (59) | 0.56 | 0.41-0.76 | <.0003 |
| EFS | ||||
| HLA-DPB1 ΔFD | 119 (47) / 43 (34) | 1.39 | 1.05-1.82 | <.021 |
| HLA-DPB1 TCE | 92 (45) / 70 (40) | 1.09 | 0.84-1.43 | <.510 |
| Number of HLA-DPB1 allele mismatches | 105 (42) / 57 (45) | 0.92 | 0.68-1.25 | <.588 |
| Recipient age | 93 (49) / 69 (36) | 1.59 | 1.20-2.10 | <.002 |
| Disease status at transplant | 84 (51) / 41 (46) / 37 (30) | 1.24 | 1.05-1.46 | <.010 |
| In vivo T-cell depletion (ATG) | 65 (35) / 97 (51) | 0.65 | 0.49-0.87 | <.004 |
| Relapse | ||||
| HLA-DPB1 ΔFD | 67 (27) / 41 (32) | 1.69 | 0.99-2.87 | <.055 |
| HLA-DPB1 TCE | 57 (28) / 51 (29) | 0.75 | 0.45-1.24 | <.261 |
| Number of HLA-DPB1 allele mismatches | 72 (28) / 36 (29) | 0.97 | 0.63-1.49 | <.901 |
| Recipient age | 54 (29) / 54 (28) | 1.33 | 0.88-2.01 | <.178 |
| Disease status at transplant | 49 (30) / 20 (22) / 39 (31) | 1.15 | 0.90-1.46 | <.255 |
| In vivo T-cell depletion (ATG) | 55 (29) / 53 (28) | 0.85 | 0.56-1.27 | <.422 |
| NRM | ||||
| HLA-DPB1 ΔFD | 67 (27) / 42 (33) | 1.48 | 0.88-2.51 | <.143 |
| HLA-DPB1 TCE | 54 (27) / 55 (31) | 0.98 | 0.59-1.61 | <.931 |
| Number of HLA-DPB1 allele mismatches | 76 (30) / 33 (26) | 0.81 | 0.53-1.26 | <.366 |
| Recipient age | 42 (22) / 67 (35) | 2.13 | 1.40-3.24 | <.0005 |
| Disease status at transplant | 34 (20) / 27 (30) / 48 (39) | 1.34 | 1.06-1.69 | <.016 |
| In vivo T-cell depletion (ATG) | 68 (36) / 41 (21) | 0.52 | 0.34-0.78 | <.002 |
| Acute GVHD grades II-IV | ||||
| HLA-DPB1 ΔFD | 96 (38) / 44 (35) | 0.99 | 0.63-1.57 | <.985 |
| HLA-DPB1 TCE | 80 (39) / 60 (34) | 0.82 | 0.53-1.27 | <.382 |
| Number of HLA-DPB1 allele mismatches | 93 (37) / 47 (37) | 1.08 | 0.74-1.58 | <.684 |
| Recipient age | 73 (39) / 67 (35) | 0.78 | 0.54-1.10 | <.158 |
| Disease status at transplant | 55 (33) / 32 (36) / 53 (43) | 1.10 | 0.90-1.36 | <.354 |
| In vivo T-cell depletion (ATG) | 74 (39) / 66 (34) | 0.91 | 0.63-1.33 | <.634 |
| Acute GVHD grades III-IV | ||||
| HLA-DPB1 ΔFD | 37 (15) / 18 (14) | 0.98 | 0.48-2.01 | <.967 |
| HLA-DPB1 TCE | 30 (15) / 25 (14) | 1.00 | 0.51-1.99 | <.993 |
| Number of HLA-DPB1 allele mismatches | 38 (15) / 17 (13) | 0.88 | 0.47-1.64 | <.691 |
| Recipient age | 28 (15) / 27 (14) | 1.02 | 0.58-1.79 | <.954 |
| Disease status at transplant | 21 (13) / 8 (9) / 26 (21) | 1.19 | 0.85-1.68 | <.304 |
| In vivo T-cell depletion (ATG) | 35 (19) / 20 (10) | 0.59 | 0.33-1.01 | <.090 |
| Chronic GVHD | ||||
| HLA-DPB1 ΔFD | 118 (47) / 67 (53) | 0.81 | 0.56-1.20 | <.282 |
| HLA-DPB1 TCE | 86 (42) / 99 (56) | 1.42 | 1.01-1.90 | <.016 |
| Number of HLA-DPB1 allele mismatches | 119 (47) / 66 (52) | 1.08 | 0.78-1.49 | <.656 |
| Recipient age | 96 (51) / 89 (47) | 0.86 | 0.63-1.18 | <.349 |
| Disease status at transplant | 82 (50) / 49 (55) / 54 (44) | 0.90 | 0.75-1.09 | <.278 |
| In vivo T-cell depletion (ATG) | 95 (51) / 90 (47) | 0.71 | 0.53-0.95 | <.021 |
ΔFD, δ-functional distance.
Numbers (N) refer to the number of censored patients (% total) for OS and EFS, and the number of events (% total) for the other end points, in the reference group before the slash vs the group of interest after the slash, as follows: HLA-DPB1 ΔFD ≤2.665 / >2.665; HLA-DPB1 TCE permissive/nonpermissive; number of HLA-DPB1 allele mismatches one/two; recipient age < median / > median (53 y); disease status at transplant early/intermediate/advanced; in vivo T-cell depletion (ATG) No/Yes. The total number of patients in each reference group before the slash vs the group of interest after the slash were as follows: HLA-DPB1 ΔFD 252/127; HLA-DPB1 TCE 203/176; number of HLA-DPB1 allele mismatches 253/126; recipient age 189/190; disease status at transplant 166/89/124; in vivo T-cell depletion (ATG) 188/191.
Proportional hazards P value.
In multivariate analysis, the stratified ΔFD value was an independent predictor of OS (HR, 1.40; 95% confidence interval [CI], 1.05-1.87; P < .021), along with other pretransplant variables known to impact on this end point (ie, patient age, disease status at transplant, and the use of ATG) (Table 2). No significant difference was observed in OS between transplants with ΔFD scores ≤2.665 and HLA-DPB1 allele-matched transplants (HR, 1.17; 95% CI, 0.66-2.06; P = .59). For nonpermissive HLA-DPB1 TCE mismatches, the multivariate hazards of OS (HR, 1.15; 95% CI, 0.86-1.54; P < .360) were similar to those reported previously4,9 (Table 2), but this was not significant in the present cohort. This finding is consistent with some previous reports,31,55 suggesting that statistical power is an important parameter for the ability to appreciate the association between nonpermissive TCE mismatches and OS.
The stratified ΔFD value was also an independent predictor of EFS (HR, 1.39; 95% CI, 1.05-1.82; P < .021) but not of acute or chronic GVHD (Table 2). Although the hazards of relapse and NRM were higher for ΔFD scores >2.665 compared with ΔFD scores ≤2.665, this was not statistically significant (HR 1.69; 95% CI, 0.99-2.87; P < .055 for relapse; and HR, 1.48; 95% CI, 0.88-2.51; P < .143 for NRM). For nonpermissive TCE mismatches, a significant multivariate association was observed only with chronic GVHD in this cohort (HR, 1.42; 95% CI, 1.01-1.90; P < .016), possibly reflecting the use of the updated version 2.0 of the online “DPB1 T-Cell Epitope Algorithm”32,33 (Table 2).
Relative relevance of AA substitutions in HLA-DPB1
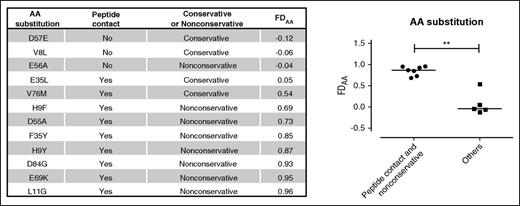
In order to investigate the mechanism underlying the observed clinical associations between patient-donor ΔFD scores and HCT outcome, we correlated the biochemical and structural characteristics of polymorphic AA in HLA-DPB1 with their FDAA scores (Figure 3). The 12 AA substitutions at 10 different positions in HLA-DPB1*09:01 studied for FDAA scores32 were divided into 2 groups. Group 1 consisted of 7 AA substitutions that were both nonconservative (ie, different in biochemical characteristics) with respect to the AA present in WT HLA-DPB1*09:01 and peptide contact residues, according to homology modeling predictions. Group 2 consisted of the 5 remaining AA substitutions that were either conservative (ie, similar biochemical characteristics) or not predicted to bind peptide, or both. The median FDAA scores varied significantly between the 2 groups, from 0.87 (0.69 to 0.96) for group 1 and −0.04 (−0.12 to 0.54) for group 2 (P = .0025) (Figure 3). No apparent correlations were found between FDAA scores and the predicted ability to make contact with the TCR (data not shown).
FDAA scores of 12 AA substitutions in HLA-DPB1. Shown are 12 AA substitutions in ten polymorphic residues of HLA-DPB1, along with their FDAA scores as determined previously.32 Statistical comparison between the FDAA scores of 7 nonconservative AA substitutions with peptide-binding characteristics (filled circles), and 5 other AA substitutions (filled squares) was performed by the nonparametric Mann-Whitney U test. **P = .0025.
FDAA scores of 12 AA substitutions in HLA-DPB1. Shown are 12 AA substitutions in ten polymorphic residues of HLA-DPB1, along with their FDAA scores as determined previously.32 Statistical comparison between the FDAA scores of 7 nonconservative AA substitutions with peptide-binding characteristics (filled circles), and 5 other AA substitutions (filled squares) was performed by the nonparametric Mann-Whitney U test. **P = .0025.
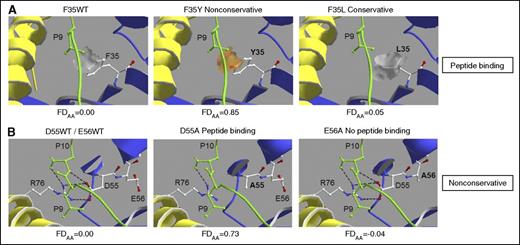
To further corroborate these results, we used homology modeling of HLA-DPB1*09:01 for pairwise comparison of AA substitutions with opposing FDAA scores correlated with the presence or absence of only one of the two criteria (Figure 4). The nonconservative F35Y but not the conservative F35L substitution is predicted to introduce a marked change in the overall charge of the peptide-binding pocket interacting with the P9 residue side chain of the bound peptide, resulting in a potentially significant interference with the hydrophobic side chain of this residue in the MP-10R13 peptide (Figure 4A). Similarly, the nonconservative D55A substitution but not the equally nonconservative E56A substitution is predicted to disrupt a hydrogen bond network formed between the side chain of peptide residue 9, the conserved R76 residue of the HLA-DP α chain, and the charged D55 residue of the HLA-DP β chain in WT HLA-DPB1*09:01, because position 55 but not position 56 is predicted to interact with bound peptide (Figure 4B).
Homology modeling of AA substitutions with opposing FDAA scores. Homology modeling was performed using the HLA-DPB1*09:01-HLA-DPA1*02:01 heterodimer with the bound 10-mer peptide MP-10R13 derived from the streptococcal M12 protein.47 HLA-DP α and β chains are shown in yellow and blue, respectively, and peptide is shown in green. (A) Comparative analysis of 2 AA substitutions at the peptide-binding residue 35, either nonconservative (F35Y; FDAA 0.85) or conservative (F35L; FDAA 0.05). Orange or gray patches indicate hydrophilic or hydrophobic areas at the molecular surface introduced by the side chains of polymorphic residue 35. (B) Comparative analysis of 2 nonconservative AA substitutions at neighboring residues with (D55A; FDAA 0.73) or without (E56A, with FDAA −0.04) peptide-binding characteristics. Electrostatic interactions between peptide residue P9, conserved R76 of the HLA-DP α chain, and polymorphic D55 of the HLA-DPB1*09:01 β chain are shown as black dotted lines.
Homology modeling of AA substitutions with opposing FDAA scores. Homology modeling was performed using the HLA-DPB1*09:01-HLA-DPA1*02:01 heterodimer with the bound 10-mer peptide MP-10R13 derived from the streptococcal M12 protein.47 HLA-DP α and β chains are shown in yellow and blue, respectively, and peptide is shown in green. (A) Comparative analysis of 2 AA substitutions at the peptide-binding residue 35, either nonconservative (F35Y; FDAA 0.85) or conservative (F35L; FDAA 0.05). Orange or gray patches indicate hydrophilic or hydrophobic areas at the molecular surface introduced by the side chains of polymorphic residue 35. (B) Comparative analysis of 2 nonconservative AA substitutions at neighboring residues with (D55A; FDAA 0.73) or without (E56A, with FDAA −0.04) peptide-binding characteristics. Electrostatic interactions between peptide residue P9, conserved R76 of the HLA-DP α chain, and polymorphic D55 of the HLA-DPB1*09:01 β chain are shown as black dotted lines.
Frequency and association of high impact AA substitutions in HLA-DPB1
A total of 19/480 HLA-DPB1 alleles encoding different proteins33 occur with an allelic frequency of >0.5% and cover >99% of the genetic variability in Europeans56 (Table 3). The number of AAs with high impact FDAA scores ≥0.69 in these alleles varies from 0 to 5 and shows an almost linear correlation with FDAllele scores (R2 = 0.96), as well as a good correlation with TCE groups32 (Table 3). As already observed for TCE groups,57 FDAllele scores also correlated well with a rs9277534 G/A SNP in the HLA-DPB1 3′ untranslated region, which was recently reported to modulate HLA-DP protein expression and to associate with GVHD after unrelated HCT.58,59 With a median of 2.36 for the 19 FDAllele scores overall, 6 HLA-DPB1 alleles associated with the low expression rs9277534 A-variant had a significantly higher median FDAllele score compared with 13 HLA-DPB1 alleles associated with the high expression rs9277534 G-variant (3.89 vs 2.29; P < .02).
HLA-DPB1 allele frequencies, FDAA scores, FDAllele scores, TCE groups, and rs9277534 SNP
| HLA-DPB1* . | Allele frequency (%)† . | N. AA with FDAA score ≥0.69 . | FDAllele score‡ . | TCE group . | Linkage rs9277534 SNP§ . |
|---|---|---|---|---|---|
| 10:01 | 1.66 | 0 | − 0.12 | 1 | G |
| 09:01 | 0.72 | 0 | 0 | 1 | G |
| 17:01 | 1.5 | 0 | 0.54 | 1 | A |
| 14:01 | 1.17 | 1 | 0.95 | 2 | G |
| 06:01 | 1.83 | 1 | 1.41 | 2 | G |
| 19:01 | 0.72 | 2 | 1.43 | 2 | G |
| 03:01 | 10.11 | 2 | 1.82 | 2 | G |
| 16:01 | 0.52 | 2 | 2.01 | 3 | G |
| 20:01 | 0.57 | 2 | 2.36 | 3 | G |
| 13:01 | 1.57 | 3 | 2.29 | 3 | G |
| 11:01 | 2.29 | 3 | 2.83 | 3 | G |
| 02:01 | 12.74 | 3 | 2.94 | 3 | A |
| 02:02 | 0.67 | 3 | 2.95 | 3 | A |
| 05:01 | 2.05 | 3 | 2.97 | 3 | G |
| 15:01 | 0.71 | 4 | 3.79 | 3 | G |
| 04:02 | 11.52 | 4 | 3.89 | 3 | A |
| 01:01 | 5.06 | 5 | 4.2 | 3 | G |
| 04:01 | 43.85 | 5 | 4.58 | 3 | A |
| 23:01 | 0.54 | 5 | 4.58 | 3 | A |
| HLA-DPB1* . | Allele frequency (%)† . | N. AA with FDAA score ≥0.69 . | FDAllele score‡ . | TCE group . | Linkage rs9277534 SNP§ . |
|---|---|---|---|---|---|
| 10:01 | 1.66 | 0 | − 0.12 | 1 | G |
| 09:01 | 0.72 | 0 | 0 | 1 | G |
| 17:01 | 1.5 | 0 | 0.54 | 1 | A |
| 14:01 | 1.17 | 1 | 0.95 | 2 | G |
| 06:01 | 1.83 | 1 | 1.41 | 2 | G |
| 19:01 | 0.72 | 2 | 1.43 | 2 | G |
| 03:01 | 10.11 | 2 | 1.82 | 2 | G |
| 16:01 | 0.52 | 2 | 2.01 | 3 | G |
| 20:01 | 0.57 | 2 | 2.36 | 3 | G |
| 13:01 | 1.57 | 3 | 2.29 | 3 | G |
| 11:01 | 2.29 | 3 | 2.83 | 3 | G |
| 02:01 | 12.74 | 3 | 2.94 | 3 | A |
| 02:02 | 0.67 | 3 | 2.95 | 3 | A |
| 05:01 | 2.05 | 3 | 2.97 | 3 | G |
| 15:01 | 0.71 | 4 | 3.79 | 3 | G |
| 04:02 | 11.52 | 4 | 3.89 | 3 | A |
| 01:01 | 5.06 | 5 | 4.2 | 3 | G |
| 04:01 | 43.85 | 5 | 4.58 | 3 | A |
| 23:01 | 0.54 | 5 | 4.58 | 3 | A |
SNP, single nucleotide polymorphism.
*HLA-DPB1 alleles are listed in the order of increasing number of AAs with FDAA scores ≥0.69 (see text).
See as reported in Hollenbach et al.56 The combined frequency of HLA-DPB1 alleles with 3 or more AAs with FDAA scores ≥0.69 is 81% compared with 18.8% for HLA-DPB1 alleles with 2 or less such AAs.
The median FDAllele score in the 19 HLA-DPB1 alleles listed is 2.36.
See as reported in Petersdorf et al.59
Discussion
In this study, we present clinical evidence for the innovative concept of ΔFD between mismatched HLA-DPB1 alleles in recipient and donor for risk prediction after unrelated HCT. ΔFD reflects the combined impact of AA polymorphism in HLA-DPB1 on T-cell allo-reactivity, and shows a significant but not complete 79.4% overlap with the TCE group model of nonpermissive HLA-DPB1 mismatches, which we and others have shown previously to correlate with mortality after unrelated HCT.4,9,17 We found that ΔFD is a significant independent predictor of OS and EFS but not of acute or chronic GVHD, although for grade III-IV acute GVHD, the number of events was limited and therefore the results should be interpreted with caution. High ΔFD scores were associated with markedly albeit not significantly increased hazards of relapse and NRM, which might contribute cumulatively to the observed survival associations, as indicated by the significant association with treatment failure. HLA-DPB1 ΔFD scores do not have a graft-vs-host or a host-vs-graft vector, because they represent the absolute difference between the FDAllele scores of recipient and donor. Association between high ΔFD scores and increased rather than reduced relapse risk without concomitant significant association with GVHD, might therefore reflect indirect mechanisms such as modulation of donor immune reconstitution after transplantation. These observations suggest that in contrast to the commonly accepted genetic distance between polymorphic HLA alleles, the novel concept of FD might allow an improved dissection of GVL from GVHD, although further studies are needed to verify this important point.
Stratification of ΔFD scores used the cutoff value 2.665, determined by ROC analysis concordantly for the end points OS and EFS, whereas no significant ROC cutoffs were obtained for any of the other clinical end points. Independent studies will be needed to establish if the ΔFD cutoff 2.665 is the most appropriate to discriminate between low and high risk. An elegant approach to solve this issue would be to adapt ΔFD scores as a continuous variable. This type of analysis however, requires high statistical power and is warranted in subsequent studies of sufficient size.
The multivariate associations between ΔFD scores and survival were superior to those of TCE matching in this study, suggesting that ΔFD matching might eventually be recommended as a refinement of TCE matching for clinical donor selection. Moreover, given the large degree of overlap between the 2 models, ΔFD scores could be used to investigate structural correlates of nonpermissive HLA-DPB1 TCE mismatches. The association between AA polymorphism in mismatched HLA and HCT outcome is the subject of intensive debate. Most of these studies investigated 1 AA substitution at a time,11,18-20 whereas ours is, to our best knowledge, the first to analyze the combined role of individual AA substitutions. A limit of our approach is that we considered only a selection of polymorphic residues for the calculation of the FDAA scores, and that we assumed an additive rather than a compensatory effect of combined AA substitutions. However, the strong correlation between HLA-DPB1 FDAllele scores and TCE groups,32 and the significant clinical risk associations of ΔFD scores shown in this study, support this approach despite these limitations. Our results show that different from current dogma, the functional impact of a given AA substitution is determined not only by its position but also by its biochemical type, and that the clinical relevance of HLA-DPB1 disparity reflects the combined rather than the individualistic effect of polymorphic high-impact residues. A dominant association between AA substitutions at peptide-binding positions and HCT outcome has been suggested previously by others for HLA class I,12,20,21,25 but this is the first study to provide similar evidence also for mismatches encoded by HLA class II. Moreover, we show that 2 different AA substitutions at the same peptide-binding position 35 in HLA-DPB1*09:01 had opposing functional effects, which were suggested by homology modeling to be related to biochemical changes introduced by the nonconservative high-impact F35Y but not by the conservative low-impact F35L substitution. In line with this notion, the nonconservative peptide-binding L156D difference between HLA-B*44:02 and HLA-B*44:03 was shown to be involved in rejection and GVHD after unrelated HCT.60,61 It is tempting to speculate that the subsequent failure to confirm a predominant role of AA substitutions at position 156 for the outcome of unrelated HCT18,20 might be related to the confounding effect of conservative substitutions at this position. For HLA-DPB1, AA substitutions at several of the positions covered by our FDAA scores have been shown to be important for in vitro T-cell allo-recognition and HCT outcome.19,62,63 The ΔFD concept reconciles these data by supporting a model in which the combined structural dissimilarity of AA polymorphism encoded by mismatched HLA-DPB1 alleles results in functional dissimilarity, which in turn is associated with clinical outcome of HCT.
The risk of acute GVHD after unrelated HCT has recently been shown to be associated with the presence of an HLA-DPB1 mismatch with high expression levels determined by the rs9277534 SNP G-variant in the HLA-DPB1 3′ untranslated region, compared with the A-variant of the same SNP.59 Association between these two SNP variants and HLA-DPB1 alleles revealed a strong correlation with TCE groups57 and with FDAllele scores in the present study. Low- and high-FDAllele scores were predominantly associated with the high-expression rs9277534 SNP G-variant and the low-expression A-variant, respectively. This might suggest evolutionary pressure onto HLA-DP antigens with similar structure-function characteristics to be presented at higher or lower density on the cell surface. The observed associations lead to a higher probability for unrelated HCT recipient-donor pairs with high ΔFD scores to be mismatched for the rs9277534 SNP, and for recipient-donor pairs with low ΔFD scores to be matched for the rs9277534 SNP. ΔFD scores and rs9277534 SNP variant matching could thus potentiate each other, or alternatively, be surrogates for each other. Further experimental and clinical studies are needed to clarify this point.
In conclusion, the concept of HLA-DPB1 ΔFD-matching presented in this study sheds new light onto the mechanisms governing the associations between AA polymorphism and the clinical outcome of HLA-DPB1 mismatched unrelated HCT. These novel findings might prove useful for developing models of clinical risk associations also at other HLA loci, which are urgently warranted.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Monika Lindemann and Falko M. Heinemann for help with HLA typing and useful discussions, as well as Sabine Riebschläger for performing donor searches. Luca Vago and Esteban Arrieta-Bolaños are gratefully acknowledged for critically reading the manuscript.
This work was supported by grants from the Deutsche José Carreras Leukämie Stiftung (DJCLS R 15/02) (K.F., D.W.B., and P.A.H.); European Commission Transcan JTC2012 (Cancer12-045-HLALOSS) (K.F. and D.W.B.), the Joseph Senker Stiftung (K.F.), and from the Deutsche Gesellschaft für Immungenetik e.V. (A.H. and P.A.H.).
Authorship
Contribution: P.C. designed the experimental part, performed the ΔFD classifications and HLA-DP homology modeling, and wrote the manuscript; A.H. participated in the project design and performed HLA typing; V.R. performed the ROC analyses; H.D.O. participated in the clinical part and supervised unrelated donor selection; P.A.H. participated in the project design and supervised HLA typing; D.W.B. directed the clinical part and performed the statistical analyses; and K.F. designed and supervised the study, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Katharina Fleischhauer, University Hospital Essen, Institute for Experimental Cellular Therapy, Institutsgruppe 1 (IG1), 11th Floor, Hufelandstrasse 55, D-45122 Essen, Germany; e-mail: katharina.fleischhauer@uk-essen.de.
References
Author notes
D.W.B. and K.F. contributed equally to this study.

![Figure 1. HLA-DPB1 FD scores. (A) Schematic representation of the peptide antigen-binding groove encoded by HLA-DPB1*09:01 and definition of FDAA, FDAllele, and ΔFD scores. In the HLA-DPB1*09:01 molecule, the positions and side chains of 10 polymorphic AA residues used for the determination of FDAA scores are listed in boldface. The primary data for the development of FDAA and FDAllele scores were published previously.32 Briefly, 10 AA residues most relevant for peptide binding and/or TCR contact were selected, based on homology modeling and the available literature. Most of these AA residues are bimorphic (ie, only 2 different variants have been reported in the most frequent HLA-DPB1 alleles in Europeans),56 therefore only 1 variant with respect to WT HLA-DPB1*09:01 was analyzed. For two residues, namely at position 9 and 35, 3 different variants have been reported in the most frequent HLA-DPB1 alleles in Europeans,56 and both variants with respect to WT HLA-DPB1*09:01 were analyzed, for a total of 12 AA substitutions. FDAA scores for each of these 12 AAs were obtained as follows: the median RR of 17 clonal T-cell effectors allo-reactive to HLA-DPB1*09:01 as reference was experimentally determined and FDAA scores were then calculated as [1 – median RR]. The FDAllele score for individual HLA-DPB1 alleles, calculated as the sum of FDAA scores as shown in the figure, correlate well with TCE groups based on T-cell cross-reactivity patterns, and allow us to predict TCE group assignment for all known HLA-DPB1 alleles.32 (B) FDAllele scores of 19 HLA-DPB1 alleles occurring with a frequency of >0.5% in Europeans.56 TCE group assignment of the HLA-DPB1 alleles16,32,54 is shown on top.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/128/1/10.1182_blood-2015-12-686238/4/m_120f1.jpeg?Expires=1769244021&Signature=UD1jYC6CrIKdYNzaXyBdqhogkDOPmfV-oRP2CSiYqgbCARPg0HcY3mYCPIwmHpktmukNhMiqntLSmnxsUCL5c7qqkxTO5iK8Gz3IoqqrCcs-S2R7iGrZglsDuIZRfryo6WF3iPgU6Pw9aJ0IbHJRchvhFrybdUI5kNbnx4CxJUEOws7NPIw2MCuaVwZIU2rSfKmqiKAPPY5mJGFgwYA7IbwbYLrZnr4UtzR-MF4gn5AV3ga9K1J0us0vw6NtcewofVFAaopYl6wXYbB3ur-Vgq3Rk~o6pwpM1hQJwBqK777adgSbgZwL3sq2VWjd2~59ytZfEHiuxvC2I~DQxSCfNQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal