Key Points
Whole-exome sequencing of CLL patients who relapsed after FCR treatment revealed frequent mutations in RPS15.
RPS15 mutations are likely to be early clonal events and confer poor prognosis.
Abstract
Fludarabine, cyclophosphamide, and rituximab (FCR) is first-line treatment of medically fit chronic lymphocytic leukemia (CLL) patients; however, despite good response rates, many patients eventually relapse. Although recent high-throughput studies have identified novel recurrent genetic lesions in adverse prognostic CLL, the mechanisms leading to relapse after FCR therapy are not completely understood. To gain insight into this issue, we performed whole-exome sequencing of sequential samples from 41 CLL patients who were uniformly treated with FCR but relapsed after a median of 2 years. In addition to mutations with known adverse-prognostic impact (TP53, NOTCH1, ATM, SF3B1, NFKBIE, and BIRC3), a large proportion of cases (19.5%) harbored mutations in RPS15, a gene encoding a component of the 40S ribosomal subunit. Extended screening, totaling 1119 patients, supported a role for RPS15 mutations in aggressive CLL, with one-third of RPS15-mutant cases also carrying TP53 aberrations. In most cases, selection of dominant, relapse-specific subclones was observed over time. However, RPS15 mutations were clonal before treatment and remained stable at relapse. Notably, all RPS15 mutations represented somatic missense variants and resided within a 7 amino-acid, evolutionarily conserved region. We confirmed the recently postulated direct interaction between RPS15 and MDM2/MDMX and transient expression of mutant RPS15 revealed defective regulation of endogenous p53 compared with wild-type RPS15. In summary, we provide novel insights into the heterogeneous genetic landscape of CLL relapsing after FCR treatment and highlight a novel mechanism underlying clinical aggressiveness involving a mutated ribosomal protein, potentially representing an early genetic lesion in CLL pathobiology.
Introduction
The clinical course of patients with chronic lymphocytic leukemia (CLL) is highly variable and ranges from rapid disease progression requiring early treatment to survival for decades without any need for therapy. Today, the gold standard first-line regimen in young, medically fit CLL patients is chemoimmunotherapy (ie, the combination of fludarabine, cyclophosphamide, and an anti-CD20-antibody, rituximab [FCR]).1,2 Although this therapy is initially effective, achieving a 90% overall response rate, most patients will relapse, often with a more aggressive disease and in rapid need of secondary treatment, especially if relapse occurs within a short time (<2-3 years) after receiving FCR.1,2
With the advent of next-generation sequencing, new insights into the molecular landscape of CLL have been achieved.3-8 In addition to the well-documented TP53 aberrations, recurrent somatic mutations were recently discovered within genes involved in key cellular processes (eg, NOTCH signaling, RNA splicing, nuclear factor κB signaling). Such mutations tend to be enriched in high-risk CLL patients and have been associated with inferior outcome and even chemo-refractory disease.4-13 New technologies have also facilitated the exploration into the clonal architecture of CLL, not only at a single time point, but also throughout the disease course, by analyzing longitudinal samples.14,15 From these pioneering studies, it became evident that subclonal mutations (ie, variants detected in only a fraction of the tumor population) could be present at low frequencies, even remaining undetected, at early stages of the disease; however, they can be positively selected as the disease progresses, particularly after several lines of therapy.14,15 The detrimental effect of low-frequency variants in CLL was recently illustrated whereby microclones carrying TP53 mutations could be detected at diagnosis using ultra-deep sequencing; these microclones appeared to expand over time and were found to have a clinical impact similar to clonal TP53 mutations.16,17 At present, definitive conclusions cannot be drawn regarding the dynamics of clonal evolution in relation to specific treatment regimens because relevant studies have investigated relatively small cohorts of, more importantly, heterogeneously treated patients.
To overcome these limitations, we applied whole-exome sequencing (WES) of longitudinal samples collected from 41 patients with CLL who were homogeneously treated with FCR and exhibited a good initial response but relapsed within a median of 2 years. In addition to gene mutations previously associated with poor outcome, we identified recurrent mutations in RPS15, a gene encoding a ribosomal protein, in almost 20% of FCR-relapsing patients. Screening in extended cohorts confirmed an increased frequency of RPS15 mutations in adverse prognostic CLL. In addition to the established role of RPS15 in protein translation, we verified the recently reported interaction between RPS15 and MDM2/MDMX18 and showed reduced stabilization and increased p53 degradation in RPS15 mutants compared with wild-type (wt) RPS15, indicating a novel molecular mechanism involved in the pathobiology of CLL.
Methods
Patient material
Forty-one CLL patients from 7 collaborating institutions in Sweden, Greece, Italy, France, the Czech Republic, the United Kingdom, and Germany were included for mutational screening using WES. All cases were diagnosed according to the International Workshop on Chronic Lymphocytic Leukemia guidelines and displayed a typical CLL phenotype.19 All patients had received FCR treatment and had either obtained a complete remission (CR; n = 32) or partial remission (n = 9); at least 4 rounds of treatment were required for patients exhibiting partial remission, whereas patients with CR were included in the study irrespective of the number of treatment cycles. Response and relapse criteria were adopted according to the International Workshop on Chronic Lymphocytic Leukemia guidelines.19 Patients relapsed at a median of 2.17 years (range, 1-11 years) after the first round of treatment; the main characteristics of the analyzed cases are summarized in supplemental Table 1, available on the Blood Web site. Peripheral blood samples were collected before the start of FCR treatment (median, 0.1 years; range, 0-2.85 years) and after treatment relapse (median, 0.51 years; range, 0-3.46 years). Constitutional DNA from sorted T cells or buccal swabs was available for 28 patients. Our extended cohort for targeted resequencing of exon 4 in RPS15 comprised an additional 790 CLL samples (605 adverse prognostic and 185 favorable prognostic patients) from the previously mentioned institutions and from additional institutions in Greece, The Netherlands, and the United States as well as 30 cases with Richter transformation. In a second validation cohort, samples from 329 untreated patients enrolled in the UK Leukaemia Research Fund Chronic Lymphocytic Leukaemia Trial 4 (CLL4) clinical trial were investigated.20 The main clinicobiological characteristics of the analyzed cases are summarized in supplemental Tables 2 and 3. Informed consent was collected according to the Helsinki Declaration and approval was granted by local ethical review committees.
WES and data analysis
Before WES, all CLL samples were fluorescence-activated cell–sorted or underwent negative selection to enrich the tumor cell population (>95%), with the exception of 6 samples for which viable frozen cells were not available (tumor percent within these 6 samples was >60%). High-quality genomic DNA was obtained using Qiagen extraction kits. Libraries for WES were constructed using the TruSeq Exome Enrichment Kit (Illumina) and sequenced on an Illumina HiSeq 2000. The raw sequencing reads were processed using the bcbio-nextgen framework. Reads were aligned to the Hg19 reference genome using BWA-mem, version 0.7.10,21 realigned using GATK, version 3.2,22 and polymerase chain reaction duplicates were marked using Sambamba, version 0.4.7. Single nucleotide variants (SNV) and indels were detected using VarScan2, version 2.3.6.23 Samples with matched germline DNA were analyzed using the somatic mode and the remaining samples using the mpileup2cns mode, both with a 10% variant allele frequency cutoff. Samples with matched constitutional DNA were analyzed in all targeted regions as specified by the manufacturer. From this screening, genes mutated in ≥2 samples at relapse were screened in samples without matched normal DNA. Somatic copy-number aberrations were derived from WES data using EXCAVATOR24 and the output was segmented using the DNAcopy R package. A high concordance was observed between WES somatic copy-number aberrations and aberrations detected by fluorescence in situ hybridization (supplemental Table 4).
Sanger sequencing
Bidirectional Sanger sequencing of selected variants and the hotspot region of RPS15 (exon 4) was performed using the BigDye Terminator, v3.1, Cycle Sequencing Kit and an ABI 3730 DNA Analyzer (Life Technologies). Primers are available upon request.
Analysis of clonal composition
SciClone25 enabled clustering analysis of variant allele frequencies of both the pretreatment and relapse mutations. All somatic variants, including synonymous variants and variants in noncoding regions, with coverage ≥20× were included in the analysis. The algorithm excludes regions affected by copy-number gain or losses; these data were obtained from EXCAVATOR.24 This analysis focuses on detecting mutation clusters and delineating their intrinsic relationship rather than assigning the exact cell fraction affected by the mutation. For analysis of mutations specific to the pretreatment or relapse sample, the position was examined in the reference-called sample to ensure sufficient read depth. Assuming that the variant allele frequency of the largest detectable cluster corresponds to the total tumor population, the tumor fraction of each cluster was calculated in relation to the largest cluster at each time point.
Cell culture
Because of the inherent difficulties in transfecting existing CLL cell lines, a well-characterized TP53wt colorectal cancer cell line HCT116 (ATCC) was selected; the cell line was maintained in McCoy’s 5A medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin (all from Thermo Fisher Scientific Inc) and cultured in a humidified atmosphere of 5% CO2 and 37°C.
Transient transfections
Myc-DDK tagged RPS15 (RC210640), turbo green fluorescent protein (tGFP) tagged MDM2 (RG219518), and tGFP tagged MDMX constructs (RG209620) were purchased from Origene. To generate RPS15 expression vectors with p.P131S or p.G132A mutations, site-directed mutagenesis was performed using the QuikChange II kit (Agilent Technologies) with the following primers: GGCCCCGATGGGGACCGGCC (P131S fwd), GGCCGGTCCCCCATCGGGGCC (P131S rev), GGCCCCGATGGCGGGCCGGCC (G132A fwd), and GGCCGGCCCGCCATCGGGGCC (G132A rev). The presence of mutations was confirmed by direct Sanger sequencing. Transfection of HCT116 colorectal cancer cells was performed using lipofectamine 2000 (Thermo Fisher Scientific Inc) according to the manufacturer’s instructions.
Western blot
Cells were lysed in NP40 buffer supplemented with phosphatase/protease inhibitors (Roche); protein levels were quantified by colorimetric assay (Thermo Fisher Scientific Inc). An equal amount of total protein was run on sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel (NuPAGE Novex 4% to 12% Bis-Tris Gels; Thermo Fisher Scientific Inc) and transferred to nitrocellulose membranes using iBlot (Thermo Fisher Scientific Inc). Following incubation with primary antibodies, the membranes were washed in phosphate-buffered saline and incubated with secondary horseradish peroxidase–coupled goat anti-mouse and horse anti-rabbit antibodies (#7074 Sigma, 1:10 000; #7076 Sigma, 1:5000). Immunoreactive proteins were visualized using ECL western blotting detection reagent (GE Healthcare) on the ImageQuant LAS 4000 imaging system (GE Healthcare).
Coimmunoprecipitation
HCT116 cells were cotransfected, as described previously, using an equal amount of plasmids. For immunoprecipitation, cells were harvested 24 hours’ posttransfection and 500 μg of the total cell lysates were precleared using Protein A/G Plus Agarose beads (20423, Thermo Scientific) followed by incubation with 2 μg of either anti-DDK (TA50011-100, Origene) or anti-tGFP (TA150041, Origene) antibody overnight at +4°C. Proteins were captured on preblocked Protein A/G Plus agarose over 3 hours and immunocomplexes were washed with 1× phosphate-buffered saline and 0.1% NP40. Bound proteins were detected by western blot using anti-DDK (dilution 1:1000) and anti-tGFP (dilution 1:1000) antibodies.
Cycloheximide-chase assay
The stability of p53 in the presence of RPS15 was detected by blocking protein synthesis using cycloheximide. Briefly, 24 hours after transfection, HCT116 cells were treated with 100 μg/mL cycloheximide and harvested at indicated time points. Cell lysates (40 μg) were analyzed by western blot (detailed previously) using anti-p53 (DO-1, Santa Cruz Biotechnology), anti-b-actin (#A5316 Cell Signaling, dilution 1:2000), and anti-c-DDK (TA100023 Origene, dilution 1:2000) antibodies. Protein bands were quantified using Image J software (http://imagej.nih.gov/ij).
Ubiquitination assay
For p53 ubiquitination assay, 24 hours after transfection with indicated plasmids, cells were treated with 30 μM MG132 (Selleckchem, S2619) for 4 hours and lysed in NP40 lysis buffer supplemented with 10 mM N-Ethylmaleimide (Sigma-Aldrich). Cell lysates were processed as described previously and an equal amount of total protein was resolved on 4% to 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Immunoblotting was performed with anti-p53 (DO-1, Santa Cruz Biotechnology) and anti-DDK antibodies.
Statistical analysis
Differences in frequencies were evaluated using descriptive statistics. Associations between categorical variables were assessed using the χ2 test for independence. Overall survival (OS) was calculated from the date of sampling until the date of last follow-up or death. Survival curves were constructed using the Kaplan-Meier method, and the log-rank test was used to determine differences between survival proportions. For all comparisons, P values were 2-sided and a significance level of P < .05 was set. All statistical analyses were performed using Statistica Software 10.0 (Stat Soft Inc).
Results
Mutational landscape in FCR-relapsing CLL
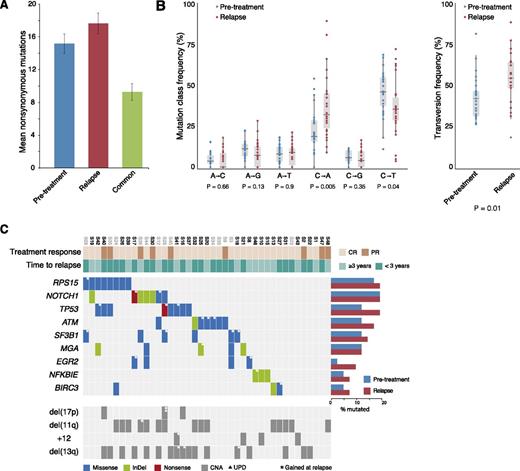
WES of 110 samples collected from 41 CLL patients who relapsed after receiving FCR treatment reached a mean effective coverage of ∼60× (supplemental Table 5). For the 28 patients with matched germline DNA, 1191 somatic variants were detected in the pretreatment samples and 1334 in the relapse samples. A total of 430 variants in the pretreatment samples and 499 in the relapse samples concerned nonsynonymous mutations, rendering an average of 15.2 (range, 3-24) and 17.6 (range, 2-32) nonsilent mutations per case, respectively (Figure 1A). When comparing the 6 classes of transition and transversion mutations in the pretreatment sample vs the relapse-specific mutations, we observed a significant increase in C•G→A•T transversions at relapse (P = .005, Figure 1B).
Somatic mutation frequencies in CLL relapsing after FCR treatment. (A) Average number of nonsynonymous mutations in both the pretreatment and relapse samples and the number of shared mutations in the 28 samples with matched constitutional DNA. (B) Frequency of the 6 mutation classes for pretreatment and relapse-specific mutations in the 28 samples with matched constitutional DNA. Statistical significance was assessed using 796 pretreatment mutations and 425 relapse-specific mutations. (C) Recurrently mutated genes. Columns represent patients (n = 41) and rows genes or genetic lesions. Color-coding indicates the type of mutation or genomic alteration. Case names in gray were analyzed without matched normal DNA. The majority of cases with TP53 aberrations harbored a mutation without coexisting del(17p); this is explained by the fact that the TP53 mutation status was not known in most cases before the start of the FCR regime in contrast to fluorescence in situ hybridization detection of del(17p), which had been performed in all cases. All TP53 mutations were deemed damaging and have been reported previously. CNA, copy-number aberration; PR, partial relapse; UPD, uniparental disomy.
Somatic mutation frequencies in CLL relapsing after FCR treatment. (A) Average number of nonsynonymous mutations in both the pretreatment and relapse samples and the number of shared mutations in the 28 samples with matched constitutional DNA. (B) Frequency of the 6 mutation classes for pretreatment and relapse-specific mutations in the 28 samples with matched constitutional DNA. Statistical significance was assessed using 796 pretreatment mutations and 425 relapse-specific mutations. (C) Recurrently mutated genes. Columns represent patients (n = 41) and rows genes or genetic lesions. Color-coding indicates the type of mutation or genomic alteration. Case names in gray were analyzed without matched normal DNA. The majority of cases with TP53 aberrations harbored a mutation without coexisting del(17p); this is explained by the fact that the TP53 mutation status was not known in most cases before the start of the FCR regime in contrast to fluorescence in situ hybridization detection of del(17p), which had been performed in all cases. All TP53 mutations were deemed damaging and have been reported previously. CNA, copy-number aberration; PR, partial relapse; UPD, uniparental disomy.
Mutational screening revealed high frequencies of mutations linked to poor outcome in both pretreated and relapse samples, including NOTCH1, TP53, ATM, SF3B1, MGA, and BIRC3; similar frequencies were detected in genes recently found to be enriched in patients with aggressive disease (ie, NFKBIE and EGR2)6-13,26-28 (supplemental Table 6; a complete list of mutated genes is provided in supplemental Tables 7 and 8). Twenty-six (63.4%) of 41 patients carried mutations in at least 1 of the previously mentioned genes at pretreatment rising to 33/41 (80.5%) at relapse (Figure 1C), whereas 12 (29.2%) and 15 cases (36.6%) harbored 2 or more coexisting mutations before the start of treatment and at relapse, respectively.
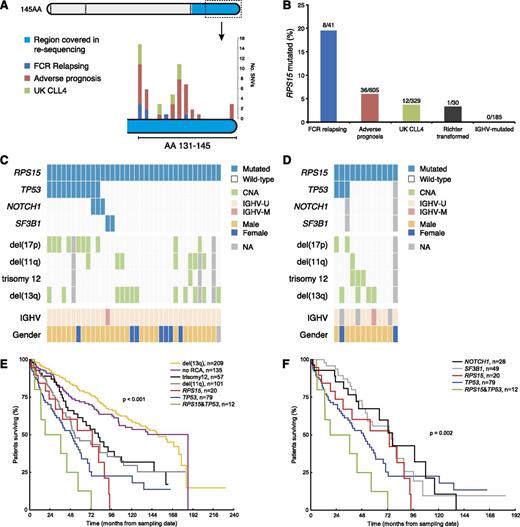
Intriguingly, a high frequency of mutations was detected in RPS15 (pretreatment 7/41 cases, 17.1%; at relapse 8/41 cases, 19.5%), which encodes the ribosomal protein S15, a component of the 40S ribosomal subunit; thus far, very few CLL cases carrying mutations within this gene have been reported.9,28 All mutations found within RPS15 were missense SNVs (n = 7) or multiple nucleotide variants (n = 1) and, with the exception of 1 SNV, clustered to a 7 amino-acid evolutionarily conserved region within exon 4 (Figure 2A; supplemental Figure 1). Of note, 3 RPS15-wt cases carried mutations within 2 other ribosomal genes (ie, RPSA and RPS20) (supplemental Table 7). All RPS15 mutations were confirmed by Sanger sequencing and their somatic origin was verified in all cases with available matched germline DNA (supplemental Table 9). All 8 RPS15-mutant patients carried unmutated immunoglobulin heavy variable (IGHV) genes (U-CLL); 3 had TP53 aberrations and another 3 had 11q deletion (Figure 1C). None of the RPS15 mutated cases showed any indication of copy number aberrations involving the RPS15 gene (19p13.3), based on copy number WES data.24 Six of the 8 RPS15-mutant cases achieved CR and 5/8 relapsed within 3 years (Figure 1C). However, no difference in time to relapse (median 22 months vs 26 months, P = .78) or OS (median 114 months vs not reached, P = .89) was seen between RPS15-mutated vs wt patients.
RPS15 mutations in CLL. (A) Localization of SNVs detected by either WES or with targeted resequencing of exon 4 in the extension cohorts (data for each cohort are provided in supplemental Table 10). With the exception of a single mutation in amino acid 33 (not shown in figure), all mutations clustered to the C-terminal of the RPS15 protein. Exons are marked with dashed lines and the region covered in the targeted resequencing is color coded. (B) Frequency of RPS15 mutations in the FCR relapse cohort (n = 41), the extended screening cohort (n = 605), the CLL4 trial cohort (n = 329), in RS cases (n = 30) and IGHV-mutated/stage A patients (n = 185). (C-D) Concurrent mutations and genetic lesions in RPS15-mutated cases in the extended screening cohort and the UK CLL4 cohort. (E-F) Overall survival for subgroups carrying recurrent cytogenetic and molecular aberrations. Pairwise log-rank test: RPS15mut vs del(13q), P < .001; RPS15mut vs TP53abn, P = .42; RPS15mut/TP53abn vs TP53abn, P = .15; RPS15mut vs RPS15mut/TP53abn, P = .12. Ten-year survival rate for RPS15mut/TP53wt was similar to RPS15mut/TP53abn and RPS15wt/TP53abn patients (0%, 0%, and 22%, respectively), but lower than the remaining RPS15wt/TP53wt patients (59%, see supplemental Figure 7). NA, not available.
RPS15 mutations in CLL. (A) Localization of SNVs detected by either WES or with targeted resequencing of exon 4 in the extension cohorts (data for each cohort are provided in supplemental Table 10). With the exception of a single mutation in amino acid 33 (not shown in figure), all mutations clustered to the C-terminal of the RPS15 protein. Exons are marked with dashed lines and the region covered in the targeted resequencing is color coded. (B) Frequency of RPS15 mutations in the FCR relapse cohort (n = 41), the extended screening cohort (n = 605), the CLL4 trial cohort (n = 329), in RS cases (n = 30) and IGHV-mutated/stage A patients (n = 185). (C-D) Concurrent mutations and genetic lesions in RPS15-mutated cases in the extended screening cohort and the UK CLL4 cohort. (E-F) Overall survival for subgroups carrying recurrent cytogenetic and molecular aberrations. Pairwise log-rank test: RPS15mut vs del(13q), P < .001; RPS15mut vs TP53abn, P = .42; RPS15mut/TP53abn vs TP53abn, P = .15; RPS15mut vs RPS15mut/TP53abn, P = .12. Ten-year survival rate for RPS15mut/TP53wt was similar to RPS15mut/TP53abn and RPS15wt/TP53abn patients (0%, 0%, and 22%, respectively), but lower than the remaining RPS15wt/TP53wt patients (59%, see supplemental Figure 7). NA, not available.
RPS15 mutations are enriched within aggressive CLL
Prompted by our finding of RPS15 mutations, we next performed targeted resequencing of the RPS15 hotspot (exon 4) in an extended CLL series (n = 790), intentionally enriched for cases displaying adverse prognostic profiles (n = 605; supplemental Table 2), and found an additional 36 mutations in RPS15 (36/605, 6%, Figure 2B-C). In contrast, none of the 185 more favorable prognostic, IGHV-mutated CLL patients carried RPS15 mutations (supplemental Table 2). Enrichment of TP53 aberrations (ie, TP53 mutations and/or del(17p)) was identified in RPS15-mutant versus wt RPS15 cases (36% vs 18%, P < .01) and a paucity of trisomy 12 (3% vs 13%, P < .05) and SF3B1 mutations (6% vs 20%, P < .05), similar to the discovery cohort.
Survival analysis (n = 613) showed an equally poor OS of RPS15-mutant cases with coexisting TP53 aberrations (noteworthy is that these cases generally carried biallelic TP53 mutation/deletion) as cases harboring TP53 aberrations only (Figure 2E); a nonsignificant trend (P = .15) of worse OS for RPS15mut/TP53abn cases was observed. RPS15-mutant patients without concomitant TP53 aberrations had an OS similar to other aggressive CLL subgroups (patients with del(11q), NOTCH1, or SF3B1 mutations vs RPS15-mutated; P = .30, Figure 2E-F), whereas the 10-year survival was lower for this subgroup compared with the remaining RPS15wt/TP53wt patients (0% vs 59%, P = .031; supplemental Figure 2A), pointing to a dismal prognosis for RPS15-mutated CLL. Similar results were obtained in the UK CLL4 trial cohort,29 which included CLL cases requiring treatment and randomized to 1 of chlorambucil, fludarabine, or fludarabine and cyclophosphamide. Overall, 12/329 (3.6%) cases within this cohort carried RPS15 mutations (Figure 2D), and RPS15mut/TP53wt patients displayed an OS similar to other adverse prognostic groups (supplemental Figure 2B, supplemental Table 3B). Finally, we analyzed 30 cases with Richter syndrome (RS; ie, CLL transformed into diffuse large B-cell lymphoma) and only a single case was found to carry an RPS15 mutation (Figure 2B); this mutation was also observed in the preceding CLL phase.
RPS15 mutations as potential early initiating events
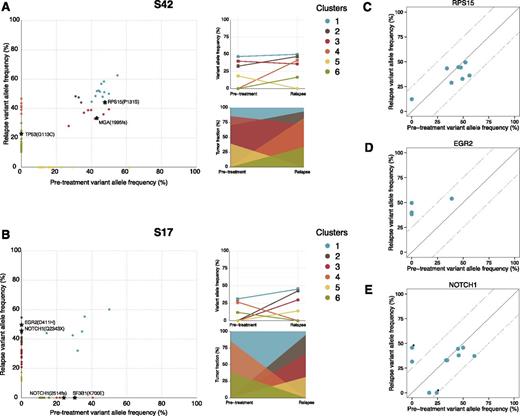
Temporal dynamics of subclonal composition were investigated for 25/28 samples with matched germline DNA using SciClone.25 This analysis revealed clusters of mutations that potentially correspond to individual (sub)clones. Overall, 2 general patterns were observed; the vast majority of cases exhibited either relapse-specific mutations/clusters that had expanded to become dominant or clusters that expanded more than 20% in (median) allele frequency (n = 23), whereas the remaining 2 cases demonstrated a more stable intraclonal composition over time (supplemental Figures 3 and 4, supplemental Table 11). Pretreatment and relapse-specific mutations were observed in all cases, indicating a branched, rather than linear, evolution pattern:30 2 characteristic examples are shown in Figure 3. Although the majority of EGR2 and BIRC3 mutations expanded upon relapse, NOTCH1 and TP53 mutations remained stable or expanded and mixed patterns were observed for ATM and SF3B1 mutations (Figure 3C-E; supplemental Figure 5). Notably, in all but 1 case with RPS15 mutations, the variants were detected at high allelic frequencies at both time points (median pretreatment 47.7%, median at relapse 40.2%, range 29% to 56%), implying that RPS15 mutations may represent early initiating events in pathogenesis of CLL. RNA sequencing data were available for 4 RPS15-mutated cases and revealed similar variant allele frequency as WES data (supplemental Table 12). Of the available cases from both the screening and validation cohorts with variant allele frequency data for both RPS15 and TP53 (n = 7), 3 showed clonal RPS15 and subclonal TP53 mutations (<25%), whereas the remaining cases showed both clonal RPS15 and TP53 mutations (supplemental Table 13). The timing of the analysis in relation to diagnosis, treatment initiation, and relapse for RPS15 mutant cases is depicted graphically in supplemental Figure 6.
Temporal dynamics of somatic mutations. SciClone analysis of variant allele frequencies for case S42 revealed 6 predicted subclones (A). Although cluster 1 (harboring a mutation in RPS15), clusters 2 and 3 (carrying a frame-shift deletion in MGA, a gene previously found disrupted in high-risk CLL)26,27 remained stable over time; clusters 4 and 6 represent relapse-specific (sub)clones, with the latter harboring a TP53 mutation that affects 25% of tumor cells, whereas cluster 5 was eliminated. In case S17, a single stable population was detected; however, a drastic shift in (sub)clonal populations was also observed (B); clusters 4 and 6 were successfully eliminated by treatment. This is noteworthy because the former harbored the classical 2-bp deletion of NOTCH1 and the recurrent p.K700E SNV in SF3B1. On the other hand, clusters 2, 3, and 5 represent relapse-specific subclones with cluster 2, which was the dominant clone at relapse, harboring a stop-gain mutation in NOTCH1 and an SNV in EGR2. Results for the remaining samples are provided in supplemental Figure 4. (A-B) The mean variant allele frequency of each cluster at both time points (top right) and the fraction of each cluster in comparison with the major cluster (cluster 1; bottom right). (C-E) Variant allele frequencies for FCR relapsing cases with RPS15, EGR2, and NOTCH1 mutations before treatment and at relapse. *Denote 2 unique mutations in the same case (S17).
Temporal dynamics of somatic mutations. SciClone analysis of variant allele frequencies for case S42 revealed 6 predicted subclones (A). Although cluster 1 (harboring a mutation in RPS15), clusters 2 and 3 (carrying a frame-shift deletion in MGA, a gene previously found disrupted in high-risk CLL)26,27 remained stable over time; clusters 4 and 6 represent relapse-specific (sub)clones, with the latter harboring a TP53 mutation that affects 25% of tumor cells, whereas cluster 5 was eliminated. In case S17, a single stable population was detected; however, a drastic shift in (sub)clonal populations was also observed (B); clusters 4 and 6 were successfully eliminated by treatment. This is noteworthy because the former harbored the classical 2-bp deletion of NOTCH1 and the recurrent p.K700E SNV in SF3B1. On the other hand, clusters 2, 3, and 5 represent relapse-specific subclones with cluster 2, which was the dominant clone at relapse, harboring a stop-gain mutation in NOTCH1 and an SNV in EGR2. Results for the remaining samples are provided in supplemental Figure 4. (A-B) The mean variant allele frequency of each cluster at both time points (top right) and the fraction of each cluster in comparison with the major cluster (cluster 1; bottom right). (C-E) Variant allele frequencies for FCR relapsing cases with RPS15, EGR2, and NOTCH1 mutations before treatment and at relapse. *Denote 2 unique mutations in the same case (S17).
Mutated RPS15 leads to impaired p53 stability
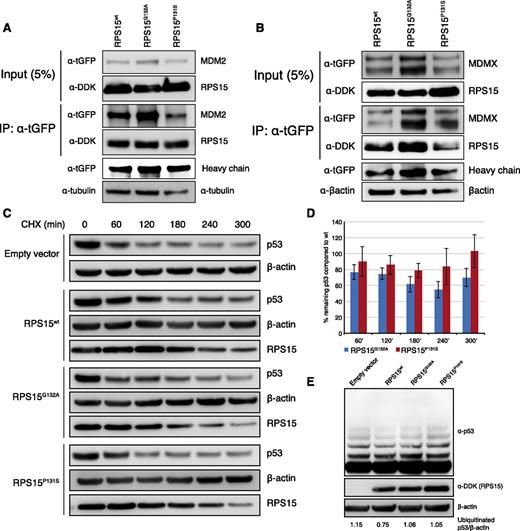
In addition to its role in protein translation, free RPS15 protein has been shown to stabilize p53 by interfering with the MDM2-p53-MDMX network by directly binding to MDM2 and inhibiting p53 degradation.18 We thus sought to investigate the potential effect of RPS15 mutations on p53 regulation. Because most (if not all) established CLL cell lines are Epstein-Barr virus—transformed, and 1 of the most used cell lines (MEC-1) carries a TP53 defect, we chose to use a well-characterized TP53wt cell line (HCT116) for the transient expression of 2 recurrent RPS15 mutations (p.P131S and p.G132A; Figure 2A) and wt RPS15. First, we performed coimmunoprecipitation and confirmed the physical interaction both between RPS15 and MDM2 and RPS15 and MDMX (Figure 4A-B). A higher binding efficiency to MDM2 was noted for RPS15P131S compared with RPS15G132A and wt. Second, we studied the impact of these 2 mutants on p53 stabilization and found that the RPS15G132A mutant consistently displayed an impaired ability to regulate endogenous p53, showing a distinct reduction in p53 stabilization over time compared with wt RPS15. The analysis for the RPS15P131S mutant disclosed a minor reduction (3 independent experiments are shown in Figure 4C-D and supplemental Figure 7). Finally, to test whether reduced p53 is a consequence of ubiquitin-mediated degradation, we performed p53 ubiquitination experiments that revealed a higher level of ubiquitination in both mutants compared with wt RPS15 (Figure 4E).
In vitro characterization of RPS15 mutations. (A-B) RPS15G132A and RPS15P131S interact with MDM2 and MDMX. Immunoblot analysis of whole-cell lysates (input) and immunoprecipitates (IP) of HCT116 cells cotransfected with 1 of the Myc-DDK-RPS15 vectors (wt, G132A or P131S) and either tGFP-MDM2 (A) or tGFP-MDMX (B). Twenty-four hours posttransfection 500 μg of cell lysates were subjected to immunoprecipitation using anti-tGFP antibody followed by immunoblotting with indicated antibodies. Immunoglobulin G heavy chain was used as loading control for IP fraction. (C-D) Transient expression of RPS15wt, RPS15P131S, and RPS15G132A in HCT116 cells revealed an impaired ability to stabilize endogenous p53, in particular for the RPS15G132A mutant. Western blot images from 1 representative experiment and quantification results from 3 independent experiments are shown. Images from 2 additional experiments are presented in supplemental Figure 7. (E) Ubiquitination experiments revealed increased ubiquitin-mediated p53 degradation in both RPS15 mutants (40.1% and 40.6% increase for RPS15P131S and RPS15G132A, respectively, measuring the intensity for the upper 4 bands) compared with wild-type RPS15.
In vitro characterization of RPS15 mutations. (A-B) RPS15G132A and RPS15P131S interact with MDM2 and MDMX. Immunoblot analysis of whole-cell lysates (input) and immunoprecipitates (IP) of HCT116 cells cotransfected with 1 of the Myc-DDK-RPS15 vectors (wt, G132A or P131S) and either tGFP-MDM2 (A) or tGFP-MDMX (B). Twenty-four hours posttransfection 500 μg of cell lysates were subjected to immunoprecipitation using anti-tGFP antibody followed by immunoblotting with indicated antibodies. Immunoglobulin G heavy chain was used as loading control for IP fraction. (C-D) Transient expression of RPS15wt, RPS15P131S, and RPS15G132A in HCT116 cells revealed an impaired ability to stabilize endogenous p53, in particular for the RPS15G132A mutant. Western blot images from 1 representative experiment and quantification results from 3 independent experiments are shown. Images from 2 additional experiments are presented in supplemental Figure 7. (E) Ubiquitination experiments revealed increased ubiquitin-mediated p53 degradation in both RPS15 mutants (40.1% and 40.6% increase for RPS15P131S and RPS15G132A, respectively, measuring the intensity for the upper 4 bands) compared with wild-type RPS15.
Discussion
Previous large-scale screening efforts in CLL have mainly focused on treatment-naïve patients or included samples at various time points from mixed patient groups receiving different treatment regimens.4-7,14 By applying WES to samples collected longitudinally from CLL patients relapsing after FCR therapy, we aimed to describe the genomic landscape and the temporal dynamics of genetic lesions in a homogeneously treated patient group. Our prime finding concerns mutations within RPS15, a gene encoding a component of the ribosomal S40 subunit, which were present in almost 20% of relapsing CLL patients. In extended analyses, we noted that RPS15 mutations were primarily found in aggressive CLL and were clonal in the pretreatment sample in the majority of cases. Finally, our pilot functional analysis indicates that RPS15 mutations may lead to defective p53 stability and increased degradation compared with wt RPS15, representing a potential novel mechanism in CLL pathobiology.
In line with previous sequencing studies of CLL patients treated with chemoimmunotherapy, we observed that the average frequency of somatic mutations remained largely unchanged at relapse (15.2 and 17.6 mutations per case before therapy and at relapse, respectively), indicating that FCR treatment per se is not intrinsically mutagenic.6,14 Nevertheless, a significant increase in C•G→A•T transversions at relapse was noted when comparing pretreatment and relapse-specific mutations. This is in line with previous observations in relapsing acute myeloid leukemia31 ; however, the mechanism responsible for this enrichment of transversions at relapse remains elusive.
In a substantial proportion of cases, mutations within genes recently suggested to impact outcome or even treatment response were identified (eg, NOTCH1, TP53, ATM, SF3B1, MGA, BIRC3, EGR2, NFKBIE),6-13,26,27 and the frequency of mutations within at least 1 of these genes increased from 63.5% before treatment to 80.5% at relapse (Figure 1C), indicating significant enrichment over time at least for certain genes. In agreement with a very recent study,32 we also noted that mutations within known driver genes frequently occurred simultaneously (Figure 1C), hence highlighting the need to introduce targeted next-generation sequencing within the diagnostic setting,13 because single-gene analysis will not suffice for correct risk stratification and outcome prediction.
In addition to the previously mentioned genes, we detected a surprisingly high number of mutations within RPS15 (8/41 patients, 19.5%), a gene recently found mutated at only a low frequency in CLL14,28 and rarely described in other cancer types. Intriguingly, with the exception of a single mutation, all mutations detected within RPS15 resided within a 7 amino-acid evolutionary conserved region; this “hotspot” of clustered mutations points to an oncogenic rather than tumor-suppressor role of the gene. Mutations within ribosomal genes have been identified in various syndromes or inherited conditions, mainly affecting the erythroid lineage (eg, Diamond-Blackfan anemia); however, mutations within RPL5, RPL10, and RPL22 were recently reported in T-cell acute lymphoblastic leukemia.33,34 Prompted by our findings, we screened a larger number of cases to not only obtain deeper insights into the frequency of RPS15 in CLL, but also, for the first time, to investigate their association with clinicobiological features and their impact on disease outcome. In the adverse prognostic patient group (mainly comprising U-CLL), we found that 6% of patients carried RPS15 mutations, of which one-third (11/36) also harbored TP53 aberrations; on the other hand, no RPS15 mutations were detected in the more favorable prognostic, IGHV-mutated CLL. Overall, CLL patients with RPS15 mutations (without coexisting TP53 aberration) had a shorter 10-year survival compared with wt CLL patients, and an OS similar to patients carrying other adverse-prognostic markers (ie, 11q deletion, SF3B1/NOTCH1 mutations). Cases with concomitant RPS15 and TP53 aberrations tended to have a worse OS than TP53 aberrant/RPS15 wt cases; however, the significance of this finding, if any, has to be further studied. We next screened CLL patients included in the UK CLL4 trial29 and detected 12/329 patients (3.6%) carrying RPS15 mutations. Akin to our first validation cohort, RPS15mut/TP53wt patients had a prognosis similar to other (non-TP53) adverse-prognostic patient groups. Finally, we analyzed 30 cases with RS and found only 1 case carrying an RPS15 mutation (also present in the CLL phase). Hence, although RPS15 mutations appear to be linked to clinical aggressiveness in CLL, they do not represent a mechanism underlying the histological transformation of CLL into RS.
In addition to a role in protein translation, free RPS15 has more recently been implicated in the regulation of the MDM2-p53-MDMX-axis because it binds to MDM2 and inhibits MDM2-mediated p53 degradation.18 Using a well-characterized TP53wt cell line, we transiently expressed RPS15 mutants, representing 2 common mutations (p.P131S and p.G132A), along with wt MDM2, and corroborated the recently reported physical interaction between RPS15 and MDM2/MDMX.18 Although a higher binding efficiency was seen for the RPS15P131S mutant compared with the other mutant and wt, the relevance of this finding is currently unclear. Next, we investigated the impact of RPS15 mutants on endogenous p53 regulation and, based on 3 independent experiments, observed decreased p53 stabilization compared with wt, in particular for the RPS15G132A mutant. In parallel, an increase of ubiquitin-mediated p53 degradation was observed in both RPS15 mutants compared with wt RPS15, pointing to reduced p53 stabilization as a consequence of higher p53 degradation. Although we cannot exclude the possibility that RPS15 mutants may exert their activity via non-p53–mediated pathways (eg, ribosomal regulation or other unknown functions), these data suggest a functional impairment by mutant RPS15 in regulating p53 stability compared with the wt RPS15 protein. This argument is also supported by the extreme clustering of RPS15 mutations within an evolutionarily conserved region, alluding to altered binding properties to MDM2 for mutant RPS15. However, our observations should be considered as preliminary, in particular considering that the function of RPS15 has yet not been fully elucidated, and further studies are warranted for a more in-depth analysis of the functional role of recurrent RPS15 mutations.
Recent seminal studies have revealed that subclones containing driver mutation(s) may confer adverse outcome in CLL and that treatment may speed up the clonal selection and outgrowth of an evolutionarily fit subclone.14,16 From these studies, genetic lesions were defined to be either clonal (ie, affecting virtually all tumor cells) or subclonal (ie, where a proportion of the tumor cells carry a particular lesion). Although variant allele frequencies compensated for copy-number alterations can be exploited to determine if a somatic mutation is clonal or subclonal, groups of mutations with similarly compensated variant allele frequencies can be assigned to clusters potentially representing clones and subclones. Following this approach, we analyzed all mutations present at both pretreatment and relapse and assigned them to separate clusters using the SciClone25 clustering tool. Indeed, in most cases (23/25), relapse-specific subclones and/or clusters had expanded significantly to become the dominant clone at relapse. Contrary to this, RPS15 mutations were detected at clonal levels before treatment in all but 1 case and remained stable at relapse. Hence, in contrast to cases carrying subclonal lesions, which are enriched over time or after treatment, our data imply that RPS15 mutations represent early initiating events and additional subclonal genetic events, such as TP53 aberrations, may occur as the disease progresses.
In summary, we provide novel insights into the heterogeneous genomic landscape of relapsing CLL and report recurrent mutations in a “hotspot” region of RPS15. Validation in a larger cohort revealed enrichment of RPS15 mutations among cases with adverse prognosis, and in vitro characterization of 2 recurrent RPS15 mutations indicated reduced p53 stability and increased p53 degradation. Because most RPS15 mutations were found at clonal levels even before treatment, these aberrations may represent an early genetic event in CLL pathobiology and be linked to a more aggressive disease. Hence, we propose recurring mutations within a ribosomal protein as a novel mechanism underlying clinical aggressiveness in CLL, potentially linked to p53 deregulation. These findings have implications for the optimal management of patients with CLL, raising the possibility that RPS15-mutant cases should be treated with alternative regimens that act independently of the p53 pathway and that mutations might be worth screening at the time of treatment decision.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Swedish Cancer Society; the Swedish Research Council, Uppsala University, Uppsala University Hospital; Lion’s Cancer Research Foundation; and Selander’s Foundation, Uppsala, Sweden; research project CZ.1.05/1.1.00/02.0068 of the Czech Ministry of Education, Youth and Sports, project NT13493-4/2012 of the Internal Grant Agency of the Czech Ministry of Health, and project AZV 15-31834A of the Czech Ministry of Health; the Kay Kendall Leukemia Fund, Leukemia and Lymphoma Research [the Leukaemia Research Fund Chronic Lymphocytic Leukemia 4 trial was funded by a core grant from Leukemia and Lymphoma Research], Cancer Research UK, the Bournemouth Leukemia Fund, Wessex Medical Research; Associazione Italiana per la Ricerca sul Cancro (AIRC) investigator grant (P.G., L.T.) and Special Program Molecular Clinical Oncology (5 per mille #9965 and #10007), Milano, Italy; Ricerca Finalizzata 2010 (#2318823) (P.G.) and Progetto Giovani Ricercatori (#GR-2010-2317594), Ministero della Salute, Rome, Italy; PRIN 2010-2011 (L.T.), Ministero dell’Istruzione, dell’Università e della Ricerca, Rome, Italy; Progetto di Ateneo, Università di Padova, Italy; and H2020 “AEGLE, An analytics framework for integrated and personalized healthcare services in Europe,” by the European Commission. A.A. is a fellow of AIRC (triennial fellowship “Guglielmina Lucatello é Gino Mazzega”). Sequencing was performed by the SNP&SEQ Technology Platform, SciLifeLab at Uppsala University, a national infrastructure supported by the Swedish Research Council (Council for Research Infrastructures [VRRFI]) and the Knut and Alice Wallenberg Foundation. The computations were performed on resources provided by Swedish National Infrastructure for Computing through Uppsala Multidisciplinary Center for Advanced Computational Science under Project b2013136. Fluorescence-activated cell sorting was performed at the BioVis imaging facility, SciLifeLab, Uppsala.
Authorship
Contribution: V.L. performed research, analyzed data, and wrote the paper. D.C., E.Y., T.P., L.M., P.B., R.C., L.-A.S., and S.J.B. performed research and analyzed data. K.P., S.N., N.S., A.A., D.R., M.H., J.K., G.J., C.B., N.C., P.P., A.W.L., K.E.S., D.O., G.G., A.S., F.D., C.P., J.C.S., L.T., S.P., and P.G. contributed samples/patient data and interpreted data. K.S., T.S., and R.R. designed the study, supervised research, and wrote the paper. All authors contributed to the preparation of the manuscript and approved the submission in its current form.
Conflict-of-interest disclosure: The authors declare no competing financial interests
Correspondence: Richard Rosenquist, Department of Immunology, Genetics and Pathology, Rudbeck Laboratory, Uppsala University, SE-751-85 Uppsala, Sweden; e-mail: richard.rosenquist@igp.uu.se
References
Author notes
V.L. and D.C. contributed equally to this study.
T.S. and R.R. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal