In this issue of Blood, Ljungström and colleagues1 expand the scope of knowledge regarding genetic drivers of chronic lymphocytic leukemia by describing and characterizing new recurrent mutations in the ribosomal protein S15.
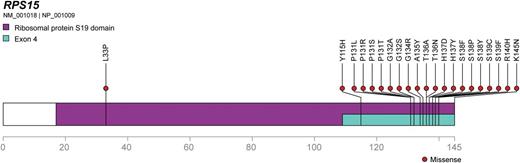
RPS15 protein is depicted as a linear peptide sequence, with the S19 ribosomal protein domain and exon 4, the mutational hotspot, highlighted. Missense mutations found by Ljungström et al are indicated, demonstrating an obvious hotspot.
RPS15 protein is depicted as a linear peptide sequence, with the S19 ribosomal protein domain and exon 4, the mutational hotspot, highlighted. Missense mutations found by Ljungström et al are indicated, demonstrating an obvious hotspot.
Chronic lymphocytic leukemia (CLL) has been the beneficiary of considerable investigation into genetic and biologic mechanisms of disease development and progression. Easy access to tumor specimens plus an often prolonged disease course, lasting up to decades, enables serial sampling and has afforded investigators the further opportunity to study clonal dynamics of putative genetic and epigenetic drivers of CLL, an area that is under active investigation.2-6
While some previous efforts have focused on well-characterized, high-prevalence recurrent mutations like TP53 using targeted sequencing panels to achieve ultra-deep coverage and ferret out small subclones of known CLL drivers,7 Ljungström and colleagues instead took the costlier and more time-consuming but unbiased approach of whole exome sequencing (WES) in 41 paired pretreatment-relapse samples. Importantly, they enriched for an aggressive disease phenotype by case selection: all but 4 of the 41 had IGHV unmutated disease and the median time to relapse was about 2 years after treatment with fludarabine, cyclophosphamide, and rituximab. WES followed up with large-scale validation (n = 1119) by Sanger sequencing confirmed the presence of recurrent mutations clustering in the C-terminal region of RPS15 (see figure), the gene encoding ribosomal protein S15 and the human ortholog of rig, which was originally isolated from a rat insulinoma.8 The gene is highly conserved in evolution, and according to COSMIC9 rarely implicated in human cancers in terms of mutation, although its overexpression may influence cancer growth and development (predictably, in insulinoma, but also in esophageal cancer and colon cancer).10
RPS15 bears many similarities to a housekeeping gene, including orthologous ultraconservation (indeed, the C-terminal 15 amino acid stretch where most of the mutations are located has nearly 100% identity in vertebrates as well as the lower eukaryote Caenorhabditis elegans), fundamental cellular function, relatively ubiquitous expression, numerous pseudogene paralogues scattered throughout the genome, few previously described mutations, and high, consistent transcript expression. That this protein could be an important driver in cancer is surprising, and is therefore notable that Ljungström et al found such an enrichment of RPS15 mutations in high-risk CLL patients.
Among 41 patients in the discovery cohort, 8 (19.5%) had RPS15 mutations. Interestingly, of 28 cases with matched germline DNA, 4 (14%) had mutations in other ribosomal protein genes including 40S protein encoding genes RPSA and RPS20, and the putative tumor suppressor kinase RPS6KA2. Mutations in these genes, as in RPS15, appear rare among all cancers. The variant allele frequency (VAF) of RPS15 mutations in the 8 serially-analyzed cases (depicted in supplemental Figure 6 in the article by Ljungström et al) remained relatively static even among patients who achieve complete response, with only one case gaining a mutation not detected at diagnosis, whereas mutations in ATM, BIRC3, NFKBIE, and TP53 predominantly appeared or increased in VAF at relapse, providing support for the authors’ hypothesis that RPS15 mutations may be an early-acquired lesion and clonal driver in high-risk disease. Consistent with this, the log-rank test indicates that RPS15 mutation is similar to abnormalities of TP53 in terms of overall survival (OS). Although the difference in OS between the combination of the two versus TP53 abnormalities alone was not statistically significant, there was a trend toward worsened OS when both lesions were present. Finally, the authors complement their clinical data by performing pilot experiments that demonstrate reduced p53 stability in the presence of RPS15 mutants.
Mutations in RPS15 have also been reported recently by Landau et al in a comprehensive exome-wide survey of mutations in CLL.6 In that study, RPS15 is described as a putative clonal driver whose mutation had an adverse impact on progression-free survival. Contemporaneously Ljungström et al confirm and expand on these findings by demonstrating an OS detriment and providing a preliminary basis of understanding the mechanisms of these mutations; one hopes that these mechanistic studies will be expanded and that the pathogenic basis of other mutations enriched in aggressive disease will be similarly explored.
The discovery of recurrent RPS15 mutation in CLL and support for its status as a driver in this disease raises questions and provides opportunities for further investigation. Are the deleterious consequences of mutated RPS15 limited to its role in p53 stabilization, or do they also have a role in the dynamics of other proteins as might be suggested by the discovery of additional mutations in other 40S ribosomal proteins? It is worthwhile to note that RPS15 is not included on common academic and commercial sequencing panels currently used to screen patients in the clinic or en masse. Do we then systematically underestimate the number of other diseases with RPS15 mutation? More generally, are there other cancers with subgroups enriched for other benign-appearing genes? Revelation of recurrent mutations in a surprising place and the preliminary functional investigations undertaken in this issue of Blood provide both a template for discovery in other diseases and a starting point to better define and understand the pathogenesis of CLL.
Conflict-of-interest disclosure: The author declares no competing financial interests.