Key Points
EVI-1–positive AML cases are sensitive to ATRA.
Abstract
Enhanced expression of ecotropic viral integration site 1 (EVI-1) occurs in ∼10% of acute myeloid leukemia (AML) patients and is associated with a very poor disease outcome. Patients with EVI-1–positive AML have poor initial responses to chemotherapy and high relapse rates, indicating an urgent need for alternative treatment strategies improving clinical outcome for these patients. Because treatment of acute promyelocytic patients with all-trans retinoic acid (ATRA) has improved the survival of these patients substantially, we investigated whether ATRA might also be effective for the subgroup of AML patients with EVI-1 overexpression. Here, we show that a substantial part of the EVI-1–positive AML cases respond to ATRA by induction of differentiation and decreased clonogenic capacity of myeloid blasts. Most importantly, we demonstrate that in vivo treatment of primary EVI-1–positive AML with ATRA leads to a significant reduction in leukemic engraftment. Altogether, our results show that a considerable part of the EVI-1–positive primary AML cases are sensitive to ATRA, suggesting that combining ATRA with the currently used conventional chemotherapy might be a promising treatment strategy decreasing relapse rates and enhancing complete remissions in this poor prognostic subgroup of AML patients.
Introduction
Acute myeloid leukemia (AML) is a heterogeneous disease that can be classified based on morphology, immunophenotypic features, and, more importantly, cytogenetic aberrations, molecular abnormalities, gene expression, and methylation signatures.1-3 Aberrant expression of the transcriptional regulator ecotropic viral integration site 1 (EVI-1) occurs in ∼10% of human adult AML patients and is associated with particularly aggressive disease and a very poor outcome. Up to 95% of EVI-1–positive patients have an overall survival of <1 year,4-6 and novel treatment strategies to improve the survival of AML patients with aberrant expression of EVI-1 are urgently needed. For more than 40 years, most AML patients, including the subgroup with EVI-1 overexpression, have been treated with a combination chemotherapy consisting of cytarabine-arabinoside and an antracycline like daunorubicin.7,8 Importantly, EVI-1-positive AML cases have an extremely poor initial response to the currently used combination chemotherapy; 39% of EVI-1–positive patients do not achieve complete remission after induction therapy, which is in sharp contrast to the 18% of patients in the other AML subgroups.5
To date, none of other tested alternative regimens have proved to be more effective in AML treatment than the combination of cytarabine and an anthracycline. The exception to this is the treatment of acute promyelocytic (APL) patients with all-trans retinoic acid (ATRA), which increased their survival chances significantly and has turned APL from a poor prognostic leukemia to one that can be cured.9,10
Because treatment of APL patients with ATRA is very successful, we hypothesize that ATRA might also be effective in treatment of AML subtypes other than APL cases. Several clinical trials have already evaluated the combination of chemotherapy with ATRA in non-APL AML patient subgroups.11-15 In general, these clinical studies showed disappointing results, apart from a trial for older (>65 years) AML patients with an NPM1 mutation who showed a better outcome after treatment with the combination of chemotherapy and ATRA as compared with chemotherapy alone.15 This clinical study probably already indicated the sensitivity of NPM1-mutated AML cells for ATRA because it was recently reported that NPM1-mutated AML is vulnerable to the combination of ATRA and arsenic trioxide.16,17 Better recognition and selection of ATRA-responsive AML patients might therefore be the key to more successful treatment yielding better clinical outcomes for particular AML subgroups.
Herein, we investigated the response of primary AML cases with overexpression of EVI-1 to ATRA. We report that cells from EVI-1–positive AML respond to ATRA by induction of differentiation and decreased clonogenic capacity of AML blasts, whereas patients from other AML subgroups lack an ATRA response. Moreover, in vivo treatment of primary EVI-1–positive AML with ATRA showed a significant reduction in leukemic engraftment.
Study design
Patient samples
At the time of diagnosis, bone marrow (BM) or peripheral blood from AML patients hospitalized at the VU University Medical Center was collected after informed consent according to a protocol approved by the medical ethical committee of the VU University Medical Center. All the studies were conducted in accordance with the Declaration of Helsinki. Mononuclear cells were isolated using Ficoll-Paque Plus (Amersham Biosciences, Uppsala, Sweden) to assess molecular aberrancies and were also viably frozen for research purposes.
Molecular diagnostics
DNA and/or RNA from the mononuclear cells was studied for the presence of t(9;22), t(8;21), t(15;17), and core binding factor and mixed lineage leukemia translocations; CEBPα, FLT3-ITD, NPM1, IDH1/2, c-Kit, and Jak2 mutations; and the overexpression of EVI-1 following standard procedures (http://www.modhem.nl).
Cytogenetics were determined according to standard techniques, and karyotypes were described according to the International System for Human Cytogenetic Nomenclature.
In vitro culturing of primary AML cells
Cryopreserved AML samples were rapidly thawed at 37°C and washed with phosphate-buffered saline containing 0.1% human serum albumin. Cells were incubated for 30 minutes with 100 μg/mL DNAse I (Roche, Basel, Switzerland) and 10 mM magnesium chloride (Sigma Aldrich, St. Louis, MO) and cultured in CellGro-SCGM medium (Cellgenix, Vancouver, BC, Canada) supplemented with penicillin/streptomycin (Gibco Life Technologies, Carlsbad, CA), FLT3 ligand (50 ng/mL), interleukin 3 (20 ng/mL), and stem cell factor (100 ng/mL) (Peprotech, Rocky Hill, NJ). Cells were seeded, and ATRA (Sigma Aldrich) or ATRA and doxorubicin (Pharmachemie, Haarlem, The Netherlands) were added for the indicated times/concentrations. Concentrations and incubation times are indicated in the particular assays as described in “Study design.”
Colony-forming unit assay
After incubation with ATRA or vehicle, AML cells were transferred to methocult (Stemcell Technologies, Grenoble, France) and seeded into 6-well plates. Colonies were quantified after 7 to 14 days of incubation by using an Axiovert 25 bright field microscope (Zeiss, Jena, Germany).
Immunostaining and flow cytometry analysis
After incubation with drugs or vehicle, cells were harvested and washed in phosphate-buffered saline containing 0.1% human serum albumin and stained with CD45 Krome Orange (Clone J.33, IgG1; Beckman Coulter), CD34 phycoerythrin cyanin 7 (Clone 581, IgG1; Beckman Coulter), CD11b phycoerythrin (D12, IgG2a), CD33-allophycocyanin (Clone P67.6, IgG1; BD), and CD3-V450 (clone UCHT1; BD). Flow count beads (Beckman Coulter, Pasadena, CA) were added according to the manufacturer’s instructions. Flow cytometry was performed with a FACS-CANTO flow cytometer (Becton Dickinson, Franklin Lakes, NJ), and analysis was done using FACS-Diva software (Becton Dickinson). To detect human myeloid cells in the mouse BM, the hCD45 V500-C (clone 2D1) antibody from Becton Dickinson was used. For the detection of apoptosis, cells were treated with 0.1 µM or 1 µM ATRA for 7 days and subsequently stained with annexin-V fluorescein isothiocyanate (Tau Technologies, Albuquerque, NM) and 7-amino-actinomycin D (7-AAD) (BD Technologies) according to the manufacturer’s instructions.
Xenograft mouse model
NOD/SCID/IL2r gamma (null) mice (females and males) (NSG) were purchased from Charles River Laboratory (Wilmington, MA). The described research was approved by the Animal Experimental Committee. Mice were bred and maintained in filter-top cages under specific opportunistic pathogen-free conditions and received sterile water and radiation-sterilized Teklad food pellets (Harlan Laboratories, Indianapolis, IN) ab libitum. The weight of the female mice was between 18 and 22 g, and the weight of the male mice was between 26 and 30 g at the start of the experiment. One day prior to injection of primary AML cells, NSG mice were irradiated with 200 cGy. To deplete T cells, primary AML cells were thawed and incubated with anti-CD3 microbeads (Miltenyi, Leiden, the Netherlands) according to manufacturer’s instructions, and subsequently, MACS columns (Miltenyi, Bergisch Gladbach, Germany) were used to deplete the CD3-expressing cells.
T-cell–depleted AML cells were intravenously injected (1-2 × 106 cells) in NSG mice 7 to 10 weeks of age. Fourteen days after injection, a 21-day-release 10-mg ATRA pellet (Innovative Research of America, Sarasota, FL) was subcutaneously implanted in the middle of the neck close to the posterior triangle, whereas the control group received a placebo pellet. This procedure was done under anesthesia (induction with 3% isoflurane and maintenance with 2% isoflurane, maximum 3 minutes) on a heating pad (30°C) to maintain normothermia. Because ATRA influences the appetite of the mice, soft Teklad food (Harlan Laboratories) was provided during the 21 days of the treatment. Mice were euthanized when disease features such as weight loss were observed or at the end point (16 weeks) of the experiment. BM (right and left femur pooled) and spleen were analyzed for the presence of human AML cells (human CD45+CD33+) using flow cytometry.
May-Grünwald Giemsa staining
Primary AML cells (50 000-100 000), incubated with or without ATRA for 4 days, were seeded on an object glass, using a Shandon Cytospin centrifuge (Thermo Scientific, Waltham, MA). Cells were dried for at least 30 minutes at room temperature and stained using a Mirastainer (VWR international, Radnor, PA) with Giemsa and May-Grünwald staining (Merck, Kenilworth, NJ).
Statistical analysis
Statistical analysis was performed using GraphPad Prism software and calculated using an unpaired 2-tailed Student t test. Statistical analysis is presented as follows: not significant (NS), P > .05; *P ≤ .05; **P ≤ .01;***P ≤ .001.
Results and discussion
AML cases with aberrant EVI-1 expression respond poorly to the currently used chemotherapy regimens, and thus a demand for novel therapeutic strategies for this subgroup of patients is needed. Here we investigated the response to ATRA of EVI-1–positive AML (n = 14), and we compared this with the response of EVI-1–negative AML cases. We studied the effect of ATRA on myeloid blast differentiation, leukemic clonogenic capacity, and the survival of leukemic cells in vitro and in vivo.
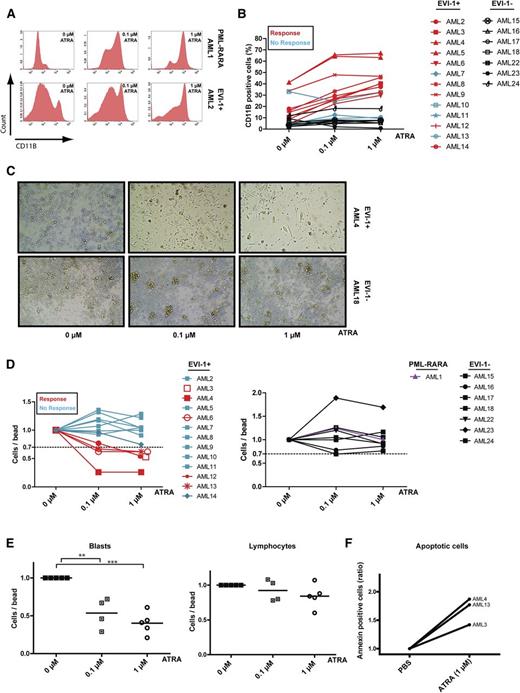
As treatment of APL cells with ATRA leads to differentiation of leukemic blast cells toward the neutrophil lineage and induced expression of CD11b,18 we examined the induction of differentiation by measuring cell surface CD11b expression on EVI-1–positive AML cells after treatment with ATRA. Like induction of CD11b expression on APL cells (Figure 1A, upper panels), we observed increased CD11b expression (example in Figure 1A, lower panels) on the cell membrane of the majority of EVI-1–positive AML cases (9/13) after 7 days of incubation with ATRA (Figure 1B, red lines). We compared these results with those in 7 AML cases without aberrant EVI-1 expression (1 AML with a single CEBPα mutation, 3 with an FLT3-ITD and an NPM1 mutation, 1 with an FLT3-ITD, 1 with an inv16, and 1 with a t[8;21]). In contrast to EVI-1–positive AML cases, the EVI-1–negative cases had no induction of differentiation after ATRA treatment (Figure 1B, black lines).
ATRA induces differentiation in EVI-1–positive AML. (A) Flow cytometric analysis of CD11b expression in PML-RARA mutated (upper) and EVI-1–positive (lower) AML cases incubated with or without ATRA for 7 days. (B) Quantification of CD11b expression in 13 EVI-1–positive AML cases (red lines, responders; blue lines, nonresponders) and 7 EVI-1–negative AML cases (black lines). (C) Morphologic examination (×10 0.25 objective) of cells from an EVI-1–positive AML case (upper) and an EVI-1–negative AML case (lower) before and after treatment with ATRA for 14 days. (D) Quantification (by flow cytometry) of cell viability (number of cells per number of beads) in AML cases incubated with or without ATRA for 7 days. Relative number of cells in EVI-1–positive AML cases (left: red lines, responders; blue lines, nonresponders), in EVI-1–negative cases (right, black lines) and in a PML-RARA–mutated AML case (right, purple line). (E) Number of myeloid blasts (CD45dim/CD33+) and lymphocytes (CD45high/SSClow) relative to the number of measured beads in EVI-1–positive AML cases that showed a reduced number of total cells upon ATRA incubation (responders, D, left, 5/13 cases). Incubation of AML3 with 0.1 µM ATRA was not done in this experiment. (F) Flow cytometric analysis of AML cells treated with ATRA for 7 days and labeled with annexin-V and 7-AAD. Shown is the number of annexin-V–positive cells relative to the number of beads.
ATRA induces differentiation in EVI-1–positive AML. (A) Flow cytometric analysis of CD11b expression in PML-RARA mutated (upper) and EVI-1–positive (lower) AML cases incubated with or without ATRA for 7 days. (B) Quantification of CD11b expression in 13 EVI-1–positive AML cases (red lines, responders; blue lines, nonresponders) and 7 EVI-1–negative AML cases (black lines). (C) Morphologic examination (×10 0.25 objective) of cells from an EVI-1–positive AML case (upper) and an EVI-1–negative AML case (lower) before and after treatment with ATRA for 14 days. (D) Quantification (by flow cytometry) of cell viability (number of cells per number of beads) in AML cases incubated with or without ATRA for 7 days. Relative number of cells in EVI-1–positive AML cases (left: red lines, responders; blue lines, nonresponders), in EVI-1–negative cases (right, black lines) and in a PML-RARA–mutated AML case (right, purple line). (E) Number of myeloid blasts (CD45dim/CD33+) and lymphocytes (CD45high/SSClow) relative to the number of measured beads in EVI-1–positive AML cases that showed a reduced number of total cells upon ATRA incubation (responders, D, left, 5/13 cases). Incubation of AML3 with 0.1 µM ATRA was not done in this experiment. (F) Flow cytometric analysis of AML cells treated with ATRA for 7 days and labeled with annexin-V and 7-AAD. Shown is the number of annexin-V–positive cells relative to the number of beads.
Morphologic examination of EVI-1–positive AML cells (Figure 1C, upper panels) supports our results obtained by flow cytometric analyses (Figure 1B) and showed that incubation with ATRA leads to a differentiated phenotype (flat cells) of EVI-1–positive cells in contrast to cells without aberrant EVI-1 expression (Figure 1C, lower panels; supplemental Figure 1A, available on the Blood Web site). Quantification revealed a significant increase of the differentiated flat cells after ATRA incubation (supplemental Figure 1B). Moreover, staining with May-Grünwald Giemsa shows that EVI-1–positive AML cells treated with ATRA exhibit increased differentiated features such as a larger cytoplasm and more vacuoles (supplemental Figure 1C).
Next, we studied the effect of ATRA on AML cell survival and observed that particular EVI-1–positive AML cases (Figure 1D, left panel) showed decreased total cell numbers after ATRA treatment (5 of the 13 cases, red lines), whereas the 7 EVI-1–negative cases (Figure 1D, right panel, black lines) and the APL (Figure 1D, right panel, purple line) did not show an effect on the number of leukemic cells. Moreover, in the EVI-1–positive cases that showed decreased cell numbers, ATRA treatment resulted in a significant reduction of CD45dim blasts, whereas the lymphocytes did not show this reduction (Figure 1E), suggesting a direct effect of ATRA on survival of AML blasts in several EVI-1–positive cases. This reduction in cell numbers is because of the induction of apoptosis in the 3 tested AML patient samples (Figure 1F; supplemental Figure 2). Collectively, these data suggest that ATRA treatment of EVI-1–positive AML cases can induce differentiation and, in some cases, apoptosis.
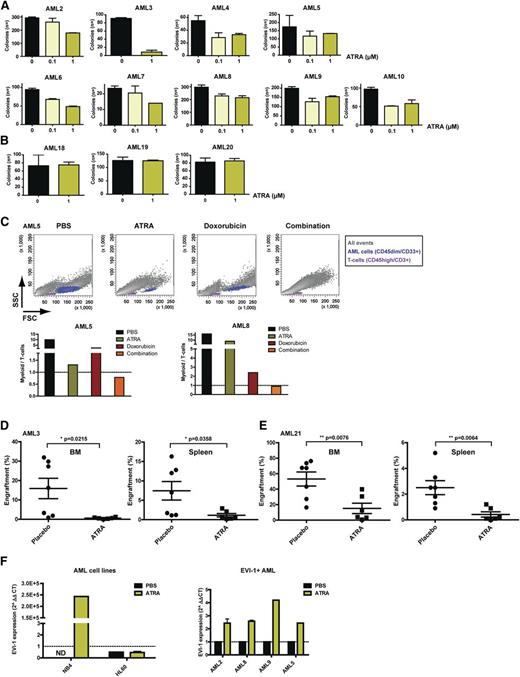
Besides low complete remission rates after chemotherapy treatment, the extreme poor prognosis of EVI-1–positive patients is because of high relapse rates. This relapse is thought to be caused by a subpopulation of AML cells with stem cell–like features named leukemic progenitors or leukemic stem cells (LSCs).19-21 Eradication or induction of differentiation of these leukemic progenitors/LSCs is crucial to improve AML outcome. Because we observed that treatment of EVI-1–positive AML with ATRA results in differentiation of AML blasts (Figure 1), we sought to determine whether ATRA also leads to decreased progenitor/LSC survival and/or impairs LSC and/or leukemic progenitor clonogenic capacity. We performed a clonogenic assay after ATRA incubation of 13 EVI-1–positive AML cases. Four EVI-1–positive AML cases did not form colonies and could therefore not be quantified (AML11-14). For the other EVI-1–positive AML cases, we observed a significant reduction in the colony-forming capacity of leukemic progenitors/LSCs after treatment with ATRA (Figure 2A). In contrast, AML cells lacking aberrant expression of EVI-1 did not show a change in colony-forming capacity after ATRA treatment (Figure 2B).
ATRA reduces colony-forming capacity and in vivo leukemic engraftment of EVI-1–positive cells. Number of colonies generated from EVI-1–positive (A) and EVI-1–negative (B) AML cells after 7 days of treatment with the indicated concentrations of ATRA. Bars represent the average numbers of colonies from a duplicate, and error bars indicate the standard deviation. AML11 to 14 did not show colonies. (C) Flow cytometric analysis (upper panels) of cells treated with 1 µM ATRA (4 days), 0.5 µM doxorubicin (3 days), or the combination (pretreatment with ATRA) and the quantification (lower panels) of myeloid (CD33+/CD45dim/CD3−) (blue) and lymphoid (CD33−/CD45high/CD3+) (purple) cells, which were negative for 7-AAD. (D-E) Quantification of human myeloid cells (hCD45dim/hCD33+) from BM and spleen of NSG mice injected with primary AML cells and treated with ATRA or placebo. (F) Quantitative reverse-transcription polymerase chain reaction analysis of EVI-1 expression upon 5 days of ATRA treatment of the AML cell lines NB4 and HL60 (left) and upon 7 days of ATRA treatment of EVI-1–positive AML cases (right).
ATRA reduces colony-forming capacity and in vivo leukemic engraftment of EVI-1–positive cells. Number of colonies generated from EVI-1–positive (A) and EVI-1–negative (B) AML cells after 7 days of treatment with the indicated concentrations of ATRA. Bars represent the average numbers of colonies from a duplicate, and error bars indicate the standard deviation. AML11 to 14 did not show colonies. (C) Flow cytometric analysis (upper panels) of cells treated with 1 µM ATRA (4 days), 0.5 µM doxorubicin (3 days), or the combination (pretreatment with ATRA) and the quantification (lower panels) of myeloid (CD33+/CD45dim/CD3−) (blue) and lymphoid (CD33−/CD45high/CD3+) (purple) cells, which were negative for 7-AAD. (D-E) Quantification of human myeloid cells (hCD45dim/hCD33+) from BM and spleen of NSG mice injected with primary AML cells and treated with ATRA or placebo. (F) Quantitative reverse-transcription polymerase chain reaction analysis of EVI-1 expression upon 5 days of ATRA treatment of the AML cell lines NB4 and HL60 (left) and upon 7 days of ATRA treatment of EVI-1–positive AML cases (right).
To investigate whether ATRA can enhance the response to chemotherapy, we studied the survival of AML cells after treatment with doxorubicin with or without pretreatment with ATRA in 2 AML cases (AML5 and AML8; Figure 2C). In both tested cases, the preincubation with ATRA leads to decreased AML cell survival after doxorubicin incubation as compared with treatment with doxorubicin alone (Figure 2C, AML5, upper and lower panels, and AML8, lower panel).
To evaluate the effect of ATRA in vivo, we intravenously injected leukemic cells from 3 primary EVI-1–positive AML cases into NOD SCID-IL2g knockout (NSG) mice and treated the mice for 21 days with ATRA or placebo (Figure 2D-E; supplemental Figure 3). Analysis of both the BM and the spleen of the mice showed a significant decrease in myeloid engraftment after treatment with ATRA in 2 of the 3 injected EVI-1–positive AML cases (AML3 and AML21; Figure 2D-E and supplemental Figure 3). Two of the 3 in vivo tested cases were also investigated in vitro (Figure 1) and showed that the in vitro and in vivo response to ATRA corresponds. AML3 showed an effect in vivo (Figure 2D) and showed induced differentiation and decreased cell viability in vitro (Figure 1). AML11 showed no effect in vivo (supplemental Figure 3D) and has no induction of differentiation and decreased cell viability after ATRA treatment in vitro (Figure 1).
Because ATRA can induce differentiation in EVI-1–positive AML cases, we hypothesize that EVI-1 expression levels might be important for the response to ATRA. For this reason, we determined the EVI-1 expression levels before and after ATRA treatment of the ATRA-sensitive APL cell line NB4, the ATRA nonresponsive cell line HL60, 1 EVI-1–negative AML case, and several ATRA-responsive EVI-1–positive AML cases. We observed that NB4 cells and EVI-1–positive primary AML cells show, in contrast to the EVI-1–negative AML case (not shown) and HL60 cells, enhanced EVI-1 expression levels upon treatment with ATRA (Figure 2F), suggesting that EVI-1 might be functionally involved in ATRA-induced AML differentiation.
In conclusion, our results show that ATRA leads to an induction of differentiation and reduction of leukemic clonogenic capacity for most EVI-1–positive AML cases. Moreover, we observed that ATRA leads to a significant reduction in leukemic cell survival of EVI-1–positive AML in vitro and in in vivo. Altogether, our results suggest that addition of ATRA to the currently used chemotherapy might increase complete remission rates or prevent relapses in EVI-1–positive AML patients. Steinmetz et al have already shown that enhanced expression of EVI-1 in HL60 cells increased the response to ATRA22 and that the protein GDF15 is part of the ATRA-induced cell cycle block, supporting our results showing that part of the EVI-1–positive AML cases are sensitive to ATRA by inducing differentiation, cell death, and decreased leukemic engraftment.
To search for aberrancies that might be responsible for the lack of response to ATRA in particular EVI-1–positive AML cases, we compared the molecular and cytogenetic aberrancies present in the nonresponsive patients with those in the responsive ones (supplemental Table 1). Although the patient groups are small, we observed that unresponsiveness to ATRA in EVI-1–positive cases is not because of the presence of particular additional mutations, the presence of a translocation involving the EVI-1 locus, an MLL translocation, or the level of EVI-1 overexpression at diagnosis (supplemental Table 1 and data not shown).
Because AML cases that do not respond to ATRA are thought to have a block in the ATRA-induced differentiation pathway, which can in some cases be activated by inhibition of the demethylase LSD1,23 it might well be that EVI-1 expression activates the ATRA-driven differentiation pathway by induction of a particular epigenetic state because of inhibition of a demethylase. This hypothesis is strengthened by the fact that the subgroup of EVI-1–positive AML cases has a distinct methylation profile24 and that downregulation of EVI-1 results in epigenetic alterations.25 Several EVI-1–positive AML cases (3/14 AML cases tested in vitro and 1/3 AML cases tested in vivo) are insensitive to ATRA, which might be because of their distinct epigenetic state as a result of the presence of an additional unknown aberrancy.
As in the majority of tested cases addition of ATRA results in induction of differentiation without inhibition of cell survival, combining ATRA with chemotherapy is likely necessary to induce the eradication of EVI-1–positive leukemic (stem) cells, analogs to the crucial combination of ATRA and arsenic trioxide in depletion of APL cells. Altogether, our results are an impetus to test the clinical implications of the treatment of EVI-1–positive AML patients with ATRA.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Stan Heukelom and Dr Phil W. Koken for radiation of the mice, and the members from the Amsterdam Animal Research Center, in particular Jerry Middelberg and Aafje Vossenaar, for their assistance with the animal experiments. The authors also thank the staff of the Hematology Department from the VU University Medical Center for the generation of a biobank of AML samples, and Guus Westra for support with the May-Grünwald Giemsa staining.
This work was supported by a grant from the Cancer Center Amsterdam, VU University Medical Center.
Authorship
Contribution: H.J.M.P.V., M.A.S., and L.S. designed the study; H.J.M.P.V., M.A.S., A.R., F.D., P.J.P., and P.A.M. performed experiments; G.J.O. collected patient material and provided clinical data; H.J.M.P.V., M.A.S., and L.S. analyzed results; H.J.M.P.V., M.A.S., and L.S. wrote the manuscript; and G.J.O. suggested improvements to the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Linda Smit, Department of Hematology, Cancer Center Amsterdam, VU University Medical Center, Boelelaan 1117, 1081 HV Amsterdam, The Netherlands; e-mail: li.smit@vumc.nl.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal