Key Points
BCR stimulation promotes mRNA translation in CLL cells, including of the oncoprotein, MYC, and is inhibited by ibrutinib or tamatinib.
Differences in mechanisms of regulation of mRNA translation in CLL and normal blood B cells may highlight potential targets for therapy.
Abstract
Antigenic stimulation via the B-cell receptor (BCR) is a major driver of the proliferation and survival of chronic lymphocytic leukemia (CLL) cells. However, the precise mechanisms by which BCR stimulation leads to accumulation of malignant cells remain incompletely understood. Here, we investigated the ability of BCR stimulation to increase messenger RNA (mRNA) translation, which can promote carcinogenesis by effects on both global mRNA translation and upregulated expression of specific oncoproteins. Re-analysis of gene expression profiles revealed striking upregulation of pathways linked to mRNA translation both in CLL cells derived from lymph nodes, the major site of antigen stimulation in vivo, and after BCR stimulation in vitro. Anti-IgM significantly increased mRNA translation in primary CLL cells, measured using bulk metabolic labeling and a novel flow cytometry assay to quantify responses at a single-cell level. These translational responses were suppressed by inhibitors of BTK (ibrutinib) and SYK (tamatinib). Anti–IgM-induced mRNA translation was associated with increased expression of translation initiation factors eIF4A and eIF4GI, and reduced expression of the eIF4A inhibitor, PDCD4. Anti-IgM also increased mRNA translation in normal blood B cells, but without clear modulatory effects on these factors. In addition, anti-IgM increased translation of mRNA-encoding MYC, a major driver of disease progression. mRNA translation is likely to be an important mediator of the growth-promoting effects of antigen stimulation acting, at least in part, via translational induction of MYC. Differences in mechanisms of translational regulation in CLL and normal B cells may provide opportunities for selective therapeutic attack.
Introduction
B-cell receptor (BCR) signaling is a major driver of malignant behavior in B-cell cancers. BCR signaling has been relatively well studied in chronic lymphocytic leukemia (CLL), and the balance between antigen-induced anergy and positive (growth-promoting) signaling is a key determinant of variable clinical behavior.1 CLL cell anergy is associated with reduced surface (s)IgM expression and signaling capacity.2 It is observed at variable levels in all CLL samples, but is most prominent in samples expressing mutated IGHV genes (M-CLL) and is associated with indolent disease. By contrast, samples expressing unmutated IGHV genes (U-CLL) tend to retain sIgM signaling capacity, and positive signaling is associated with more progressive disease. These findings and others provided a strong impetus for clinical testing of new inhibitors of BCR-associated signaling kinases. These have shown impressive responses and the BTK inhibitor, ibrutinib, is now approved for treatment of CLL and mantle cell lymphoma.3-5
Despite intense interest in the biology and drug targeting of BCR pathways in B-cell malignancies, understanding the mechanisms that drive accumulation of malignant cells remains incomplete. One important response may be increased messenger RNA (mRNA) translation. mRNA translation is subject to tight control, and recent studies performed using mouse fibroblasts indicated that mRNA translation regulation may play a similar role to transcription regulation in determining variation in protein expression.6,7
Although it is relatively well studied in solid tumors, comparatively little is known about the control of mRNA translation in B-cell malignancies, especially in primary malignant cells. Two recent studies identified increased expression/phosphorylation of specific mRNA translation initiation factors (eIFs), eIF4B and eIF4E, in diffuse large B-cell lymphoma.8,9 In CLL, stimulation of malignant cells by CD40L-expressing stromal cells increased global mRNA translation and formation of the eIF4F complex, which binds the 5′CAP of mRNAs and enhances mRNA translation.10 Previous comparisons of gene-expression profiling (GEP) and proteomic analysis revealed differences in the transcriptomic and proteomic response after sIgM stimulation of CLL cells, pointing to posttranscriptional regulation.11,12 However, whether mRNA translation is modulated by BCR signaling in CLL, where it is such a critical determinant of tumor behavior and target for therapy, is unknown.
Increased mRNA translation plays important roles in carcinogenesis, via both global effects to support increased cell growth and upregulation of specific oncoproteins.13,14 MYC RNA has a highly structured 5′-untranslated region and is one of a family of malignancy-associated mRNAs for which translation is tightly dependent on eIF activity.15,16 In our previous studies, we demonstrated that MYC protein was induced after stimulation of sIgM of CLL cells in vitro and that MYC is highly expressed in proliferation centers in CLL lymph nodes (LNs),17 which are presumed to be the major sites of antigen-induced cell division in vivo.18 Therefore, we hypothesized that MYC may be an important target for translational regulation in CLL.
In this work, we investigated the effects of BCR stimulation on mRNA translation in vitro and in vivo. Bio-informatic analysis of GEP revealed that antigen stimulation of CLL cells in LNs in vivo was associated with strong induction of multiple components of the mRNA translation machinery. Anti-IgM stimulation of primary CLL cells in vitro resulted in increased mRNA translation, measured using bulk metabolic labeling and at a single-cell level using a novel flow cytometry–based assay. Anti-IgM also resulted in increased translation of MYC mRNA and was associated with increased expression of the translation initiation factors eIF4GI and eIF4A. Furthermore, ibrutinib and the SYK inhibitor, tamatinib, inhibited these responses. mRNA translation is likely to be an important mediator of growth-promoting effects of antigen stimulation acting, at least in part, via translational induction of MYC.
Materials and methods
Patients and cells for studies in vitro
Patients provided written informed consent in accordance with Ethics Committee approvals and the Declaration of Helsinki. Heparinized peripheral blood mononuclear cells (PBMCs) were obtained from patients attending clinics at the Southampton General Hospital or the Royal Berkshire Hospital (both UK) (supplemental Table 1, available on the Blood Web site). Diagnosis of CLL was made according to the IWCLL-NCI 2008 criteria,19 and the monoclonal B-lymphocyte population in the peripheral blood had a typical IgM+IgD+ CLL phenotype in all circumstances.20 The vast majority of samples were obtained before treatment. Where treatment of CLL had taken place, this was at least 6 months before sample collection. IGHV usage and homology to germline and expression of cell surface CD5, CD19, CD23, and CD38, and ZAP70 were determined as previously described.2,21 sIgM signaling capacity was determined by measuring the percentage of cells with increased intracellular Ca2+ after stimulation with soluble goat F(ab′)2 anti-IgM and using a cutoff value of ≥5% responding cells to define samples as sIgM-responsive as previously described.2 PBMC samples from healthy donors were processed as previously described2 and cryopreserved. B cells were isolated by negative selection using the human MACS B cell isolation kit II, according to the manufacturer’s protocol (Miltenyi Biotec, Surrey, UK).
CLL cells were either used directly (“fresh”) or after cryopreservation. When cryopreserved cells were used, recovered cells were rested for 1 hour at 37°C before use. CLL cell viability determined by trypan blue exclusion was ≥90%, and the median proportion of CD5+CD19+ CLL cells was 95% (range, 62%-99%). For sIg stimulation, samples were incubated with soluble or bead-bound goat F(ab′)2 anti-human IgM, anti-human IgD, or control antibodies.22 CpG-ODN 2006 was obtained from Source Bioscience (Nottingham, UK) and was used at 7.5 μg/mL. Ibrutinib and tamatinib were obtained from SelleckChem (Suffolk, UK) and were used at 10 μM. Cells were pretreated with these compounds for 1 hour before stimulation. Cycloheximide (Sigma Chemicals, Poole, UK) was used as a positive control for inhibition of mRNA translation and was used at 10 μg/mL in the final 5 minutes of incubation. For incubations >6 hours, the caspase inhibitor Q-VD-OPh (5 μM; Sigma) was added to minimize secondary events caused by apoptosis.
Protein synthesis assays
Metabolic labeling was performed using 2 × 106 cells per assay. Tran35S-Label (MP Biomedicals, Illkirch, France) was added to the culture medium (0.37 MBq/mL) for the final 4 hours of culture. Cells were collected by centrifugation and washed in ice-cold phosphate-buffered saline (PBS). Cells were resuspended in water containing 10 mg/mL l-cysteine and 10 mg/mL l-methionine, and subjected to 2 freeze-thaw cycles. Lysates were then incubated at 37°C for 15 minutes and clarified by centrifugation. Lysates were applied to Whatmann filter discs (GE Healthcare, Amersham, UK) and allowed to air dry. Bound proteins were precipitated by the addition of 10% (wt/vol) trichloroacetic acid (TCA; Sigma Chemicals). Boiling 5% (wt/vol) TCA was added to the filter discs before washing in 100% (vol/vol) ethanol and 100% (vol/vol) acetone. The filter discs were air-dried before scintillation counting using OptiScint “HiSafe” scintillation fluid (PerkinElmer, Cambridgeshire, UK) and a WALLAC 1409 liquid scintillation counter (PerkinElmer). All assays were performed in duplicate. As a control, cells were treated with cycloheximide and counts from cycloheximide-treated samples were subtracted from experimental values.
Click-iT assays were performed using 1 × 106 cells per assay. O-propargyl-puromycin (OPP; 20 μM) (Life Technologies) was added to the cells and incubated for 30 minutes. Cells were washed in ice-cold PBS and then fixed and permeabilized using the Cytofix/Cytoperm Fixation Permeabilization Kit (BD Biosciences). Alexa-Fluor-647 (Life Technologies) was conjugated to OPP as described in the manufacturer’s instructions and cells were stained with anti-CD5-PerCyP5.5 and anti-CD19-Pacific Blue antibodies (BD Biosciences) for 15 minutes on ice. Cells were washed in Cytoperm buffer (BD Biosciences) and resuspended in fluorescence-activated cell sorting buffer before data acquisition using a FACS CantoII flow cytometer (BD Biosciences). Data analysis was performed using FlowJo v9.7.6 (FlowJo, Ashland, OR). As a control, a proportion of cells were treated with cycloheximide for 5 minutes before OPP addition and fluorescence of cycloheximide-treated cells was subtracted from all experimental values.
Statistics
Statistical comparisons were performed using Student t tests (Prism 6 software, GraphPad Software, La Jolla, CA). Experimental details for analysis of GEP data, immunoblotting and polysome profiling are provided in the supplemental material, found on the Blood Web site.
Results
Gene expression network analysis reveals increased mRNA translation as a prominent response to antigen engagement in vivo
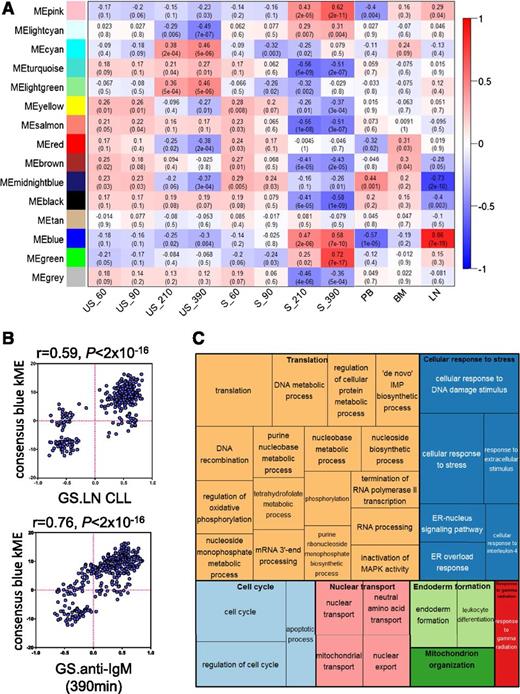
We used unsupervised consensus-weighted gene coexpression network analysis (WGCNA)23 of 2 published GEP data sets to identify growth-promoting pathways activated after antigen stimulation of CLL cells in vivo. The data sets were derived from the study of Herishanu et al, which compared gene expression of matched samples of CLL cells derived from LN, blood and bone marrow (BM),18 and the study of Vallat et al, which investigated effects of anti-IgM on CLL cell gene expression in vitro.11 For further details of WGCNA, please see the supplemental material.
WGCNA identified 14 modules of genes that had similar patterns of expression across the 11 conditions comprising the 2 data sets (supplemental Figure 1A-B). The blue module was particularly interesting because expression of this module’s eigengene (the gene considered to be the first principal component)23 correlated very strongly with both anti-IgM stimulation (especially at 210 and 390 minutes) and LN derivation (Figure 1A). There was also a very strong positive correlation between the kME (a connectivity measure that reflects the “centrality” of each gene to the module) of individual genes comprising the blue module and variation in gene expression between LN and other tissues, and after anti-IgM stimulation in vitro (Figure 1B). Overall, WGCNA identified a module of genes (the blue module) with very strong coordinate regulation in vitro after sIgM stimulation, and in LN samples compared with other tissues, indicating that this module comprises genes most strongly induced after antigen engagement in vivo.
Analysis of gene-expression networks using WGCNA. (A) Correlations between consensus modules identified by WGCNA and specific experimental conditions. The y-axis shows the 14 consensus modules that were randomly assigned different colors for identification, and the x-axis shows the 11 experimental conditions defined within the 2 GEP studies.11,18 The heat map shows the correlation between the expression of each module’s eigengene and experimental condition; cells show correlation coefficient (top) and corresponding P value (bottom). Cells are color-coded using correlation values according to scale on the right. For the Vallat et al study: S, anti-IgM stimulated; US, no stimulation. Time of stimulation (60, 90, 210, and 390 minutes) is indicated. For the Herishanu et al study, the source of the sample is shown. BM, bone marrow; LN, lymph node; PB, peripheral blood. (B) Correlations between kME and fold difference in expression between LN compared with other sites (top) and after anti-IgM stimulation in vitro (bottom) for the 344 genes comprising the blue module. (C) “TreeMap” view of over-represented GO terms in the blue module, generated using REVIGO. Each rectangle represents a cluster of related GO terms. The representatives are joined into “superclusters” of loosely related terms, visualized with different colors. Size of the rectangles reflects Benjamini-Hochberg adjusted P values.
Analysis of gene-expression networks using WGCNA. (A) Correlations between consensus modules identified by WGCNA and specific experimental conditions. The y-axis shows the 14 consensus modules that were randomly assigned different colors for identification, and the x-axis shows the 11 experimental conditions defined within the 2 GEP studies.11,18 The heat map shows the correlation between the expression of each module’s eigengene and experimental condition; cells show correlation coefficient (top) and corresponding P value (bottom). Cells are color-coded using correlation values according to scale on the right. For the Vallat et al study: S, anti-IgM stimulated; US, no stimulation. Time of stimulation (60, 90, 210, and 390 minutes) is indicated. For the Herishanu et al study, the source of the sample is shown. BM, bone marrow; LN, lymph node; PB, peripheral blood. (B) Correlations between kME and fold difference in expression between LN compared with other sites (top) and after anti-IgM stimulation in vitro (bottom) for the 344 genes comprising the blue module. (C) “TreeMap” view of over-represented GO terms in the blue module, generated using REVIGO. Each rectangle represents a cluster of related GO terms. The representatives are joined into “superclusters” of loosely related terms, visualized with different colors. Size of the rectangles reflects Benjamini-Hochberg adjusted P values.
To probe the biological pathways represented by the blue module, we identified enriched gene ontology terms and organized these into a “TreeMap” using REVIGO (Figure 1C).24 Enriched terms included “regulation of cell cycle” consistent with the idea that the LN is the major site for CLL cell proliferation in vivo. However, the most commonly over-represented ontologies were associated with stress responses, including the endoplasmic reticulum–located unfolded protein response (UPR), metabolism, and mRNA translation.
We performed a similar analysis using Ingenuity Pathway Analysis (IPA) (supplemental Figure 2). This revealed that the blue module comprised networks linked to protein synthesis, energy production, and amino-acid/nucleic acid metabolism. For example, network 3 comprised multiple eIFs (eg, eIF3B, eIF4A1, eIF4E) and factors involved in ribosome biogenesis (eg, NOLC1, NOP10). IPA “upstream analysis” identified MYC as the strongest driver of the blue module (P = 1.91 × 10−30), and the highest scoring network (network 1) was a MYC-centered network with linkage to metabolism (eg, LDHA, ENO1, MCT1, IDH3A) and mRNA translation (eg, ABCE1, NPM1), including DDX21, recently identified as central coordinator of ribosome biogenesis.25
Finally, we examined the functions of individual genes comprising the blue module (supplemental Table 2). Nearly half of the 50 highest kME genes with known function had established or potential links to protein synthesis, including factors involved in amino-acid transport (SLC3A2, SLC7A5), ribosome biosynthesis (BYSL, RRP12, RPF2) and translation initiation/termination (eIF3B, ABCE1, BZW2).
Overall, network analysis revealed prominent regulation of biosynthetic and stress response pathways after BCR engagement in vivo. Regulation of the UPR and MYC is consistent with our previous reports demonstrating induction of a partial UPR and MYC in CLL cells after sIgM stimulation in vitro, and in CLL cells in LN.17,26 However, an important new finding was the particularly strong regulation of genes associated with mRNA translation. Because network analysis suggested a prominent role for increased mRNA translation after engagement of the BCR of CLL cells in vivo, we next investigated its potential regulation by anti-IgM in vitro.
sIgM stimulation increases global mRNA translation in signal-responsive CLL samples in vitro
We used metabolic labeling to quantify effects of sIgM stimulation on mRNA translation in CLL samples. sIgM signaling capacity is variable in CLL, and we therefore selected a cohort of samples (n = 21), all classified as sIgM signal–competent based on Ca2+ response after soluble anti-IgM stimulation.2 This cohort comprised U-CLL samples and representatives of the smaller proportion of signal-responsive M-CLL. Effects on mRNA translation were analyzed using a bead-bound form of anti-IgM, which induces relatively strong signaling responses compared with soluble antibody,22,26 and with “fresh” (n = 11) and cryopreserved (n = 10) samples to control for potential effects of storage. CLL cells undergo variable levels of spontaneous apoptosis in vitro, so cells were additionally treated with a caspase inhibitor (Q-VD-OPh) to minimize potentially confounding effects of apoptosis.
Metabolic labeling demonstrated that anti-IgM beads increased mRNA translation in all samples (Figure 2A and supplemental Figure 3A). As expected for this cohort selected on the basis of retained signal-responsiveness, the fold increase in anti–IgM-induced metabolic labeling was not significantly different between U-CLL and M-CLL (supplemental Figure 3B). Anti–IgM-responses were also not significantly influenced by cryopreservation (supplemental Figure 3C). Therefore, all subsequent experiments were performed using frozen samples.
Effect of anti-IgM on mRNA translation in CLL cells. (A) Signaling-responsive CLL samples (n = 21) were incubated with anti-IgM beads or control beads, or left untreated as a control for 24 hours. mRNA translation was analyzed by quantifying metabolic labeling. (B) As in (A), except cells were pretreated for 1 hour with ibrutinib (n = 5) or tamatinib (n = 4), or dimethyl sulfoxide (DMSO) as a control, before addition of anti-IgM or control beads. The graphs show fold increase in metabolic labeling (means ± standard error of the mean [SEM]) compared with untreated cells (set to 1.0). (C-D) Immunoblot analysis of phosphorylated/total p70S6K and ERK1/2 expression at 30 minutes poststimulation with anti-IgM beads (ibrutinib, n = 6; tamatinib, n = 5). (C) Representative immunoblots. (D) Quantitation of multiple experiments after 30 minutes of stimulation; graphs show normalized p70S6K/ERK1/2 phosphorylation (means ± SEM) as a percentage of control (DMSO) anti–IgM-treated cells. Statistical comparisons between groups are shown (Student t test).
Effect of anti-IgM on mRNA translation in CLL cells. (A) Signaling-responsive CLL samples (n = 21) were incubated with anti-IgM beads or control beads, or left untreated as a control for 24 hours. mRNA translation was analyzed by quantifying metabolic labeling. (B) As in (A), except cells were pretreated for 1 hour with ibrutinib (n = 5) or tamatinib (n = 4), or dimethyl sulfoxide (DMSO) as a control, before addition of anti-IgM or control beads. The graphs show fold increase in metabolic labeling (means ± standard error of the mean [SEM]) compared with untreated cells (set to 1.0). (C-D) Immunoblot analysis of phosphorylated/total p70S6K and ERK1/2 expression at 30 minutes poststimulation with anti-IgM beads (ibrutinib, n = 6; tamatinib, n = 5). (C) Representative immunoblots. (D) Quantitation of multiple experiments after 30 minutes of stimulation; graphs show normalized p70S6K/ERK1/2 phosphorylation (means ± SEM) as a percentage of control (DMSO) anti–IgM-treated cells. Statistical comparisons between groups are shown (Student t test).
We also investigated anti–IgM-induced mRNA translation in 4 additional M-CLL samples that were considered as sIgM-nonresponsive based on Ca2+ mobilization analysis.2 Anti-IgM beads increased metabolic labeling in these samples but the overall response (∼twofold mean induction) was clearly lower than for signaling responsive samples (∼sixfold) (supplemental Figure 3D).
Finally, we compared response to bead-bound anti-IgM and anti-IgD in 17 of the 21 samples initially analyzed for anti-IgM responses. Anti-IgD enhanced mRNA translation; however, stimulatory effects of anti-IgD (∼twofold) were significantly reduced compared with anti-IgM (∼sixfold; supplemental Figure 4).
Effect of tamatinib and ibrutinib on anti–IgM-induced mRNA translation
We used tamatinib and ibrutinib to investigate potential roles of SYK and BTK in anti–IgM-induced mRNA translation. SYK is one of the first kinases activated after BCR stimulation and coordinates activation of downstream signaling within the signalosome, including BTK. Tamatinib and ibrutinib were used at 10 μM, based on previous publications and our own pilot studies.27 Inhibitory effects of ibrutinib were partial, decreasing anti–IgM-induced metabolic labeling by ∼60%, whereas tamatinib completely blocked the response (Figure 2B). Tamatinib also modestly reduced “basal” protein synthesis in cells treated with control beads. Analysis of cell death demonstrated that tamatinib and ibrutinib did not significantly reduce cell viability under our experimental conditions where Q-VD-OPh was added to decrease caspase activation (supplemental Figure 5).
Parallel analysis of signaling confirmed inhibition of anti–IgM-induced ERK1/2 phosphorylation in these experiments (Figure 2C-D). In addition, ibrutinib or tamatinib effectively inhibited anti–IgM-induced phosphorylation of p70S6K, a positive regulator of mRNA translation, which is phosphorylated by mTORC1 downstream of the PI3K→AKT pathway (Figure 2C-D).28 Consistent with the longer duration of signaling induced by bead-bound anti-IgM, increased phosphorylation of ERK1/2 and p70S6K was maintained at 6 hours post-stimulation (data not shown).
Single-cell analysis of mRNA translation
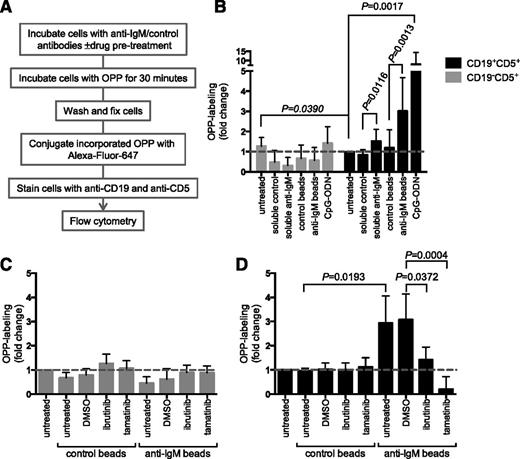
We extended our analysis using flow cytometry to quantify mRNA translation on a single-cell basis.29 In this assay, cells were incubated with a puromycin analog (OPP), which is incorporated into nascent polypeptide chains and then fluorescently labeled via “Click Chemistry” (Figure 3A). We gated on viable cells and OPP labeling was combined with staining with anti-CD19 and anti-CD5 antibodies to enable separate quantification of protein synthesis in CLL cells and nonmalignant T cells, which are also present at variable levels in PBMC samples from CLL patients (supplemental Figure 6).
Analysis of mRNA translation using OPP labeling. (A) Overview of experimental procedure for OPP labeling. (B) CLL samples (n = 13) were treated with soluble anti-IgM or anti-IgM beads, control antibodies, or CpG-ODN2006 or left untreated for 24 hours before OPP labeling. Graphs show fold increase in OPP labeling (means ± SEM) in CLL (CD19+CD5+) and T cells (CD19–CD5+), with values for untreated CLL cells set to 1.0. Statistical comparisons between untreated CLL and T cells, and between control and anti–IgM-treated CLL cells are shown (Student t test). (C-D) As in (B), except cells were pretreated for 1 hour with ibrutinib (n = 5) or tamatinib (n = 5), or DMSO was used as a control, before the addition of anti-IgM/control antibodies. The graphs show fold increase in OPP labeling (means ± SEM) for T cells (C) and CLL cells (D), with values for untreated cells set to 1.0. Statistical comparisons between groups are shown (Student t test).
Analysis of mRNA translation using OPP labeling. (A) Overview of experimental procedure for OPP labeling. (B) CLL samples (n = 13) were treated with soluble anti-IgM or anti-IgM beads, control antibodies, or CpG-ODN2006 or left untreated for 24 hours before OPP labeling. Graphs show fold increase in OPP labeling (means ± SEM) in CLL (CD19+CD5+) and T cells (CD19–CD5+), with values for untreated CLL cells set to 1.0. Statistical comparisons between untreated CLL and T cells, and between control and anti–IgM-treated CLL cells are shown (Student t test). (C-D) As in (B), except cells were pretreated for 1 hour with ibrutinib (n = 5) or tamatinib (n = 5), or DMSO was used as a control, before the addition of anti-IgM/control antibodies. The graphs show fold increase in OPP labeling (means ± SEM) for T cells (C) and CLL cells (D), with values for untreated cells set to 1.0. Statistical comparisons between groups are shown (Student t test).
In CLL cells, anti-IgM beads significantly increased OPP labeling by ∼threefold on average (Figure 3B). We also tested responses to CpG-ODN, which is a relatively strong stimulating agent for CLL cells.30 CpG-ODN increased OPP labeling to a greater extent than anti-IgM (∼eightfold on average). We used a “B-cell–specific” CpG-ODN (2006) in these experiments and, as expected, neither CpG-ODN nor anti-IgM increased OPP labeling in T cells (Figure 3B), demonstrating specificity of the response.31
Similar to metabolic labeling, OPP labeling in CLL cells was significantly reduced by ibrutinib and tamatinib (Figure 3D). Tamatinib reduced anti–IgM-induced OPP labeling below the level of unstimulated cells. Effects of ibrutinib were partial (∼50% reduction). The drugs had no significant effect on OPP labeling in T cells (Figure 3C). Analysis using OPP labeling confirms that sIgM stimulation of CLL cells increases mRNA translation and demonstrates that this occurs within the malignant clone.
We also used the OPP labeling assay to investigate the effects of varying BCR signaling strength by comparing responses to anti-IgM beads and soluble anti-IgM. In CLL cells, responses to soluble anti-IgM are generally weak and short-lived, even in responsive samples. Soluble anti-IgM significantly increased mRNA translation, but to a lower extent than bead-bound antibody (Figure 3B). Therefore, increased mRNA translation in CLL cells is sensitive to signal strength, and efficient activation requires protracted/stronger sIgM-induced signaling.
Regulation of eIFs and PDCD4 in CLL and normal B cells
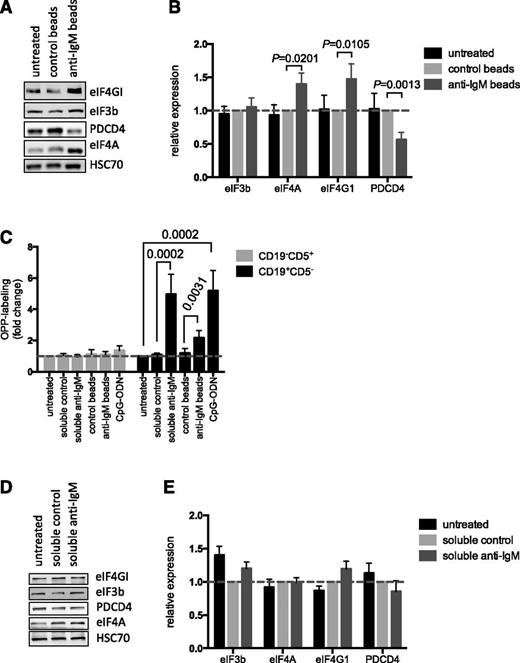
We investigated the mechanisms that mediated anti–IgM-induced mRNA translation in CLL cells by analyzing expression of key mRNA translation initiation factors. Direct analysis of the Vallat data set11 confirmed that anti-IgM increased expression of eIF4A1, eIF4G1, and eIF3b mRNAs (supplemental Figure 7), as initially revealed by network analysis (supplemental Figure 2). Immunoblot analysis demonstrated that bead-bound anti-IgM increased expression of eIF4A and eIF4GI at the protein level. However, we did not detect any changes in eIF3b protein expression. Anti-IgM also significantly downregulated expression of PDCD4, a negative regulator of eIF4A (Figure 4A-B),32 although there was no change in PDCD4 mRNA expression after anti-IgM treatment (supplemental Figure 7).
Effect of anti-IgM on eIF expression and comparison with normal B cells. (A-B) CLL samples (n = 12) were stimulated with anti-IgM beads or control beads, or were left untreated. After 24 hours, expression of eIF4A, eIF4GI, eIF3B, PDCD4, and HSC70 (loading control) was analyzed by immunoblotting. (A) Representative immunoblot. (B) Quantitation of multiple experiments; the graph shows normalized expression (means ± SEM) relative to control beads. Statistical comparisons between groups are shown (Student t test). (C) PBMCs from healthy donors (n = 7) were treated with soluble anti-IgM or anti-IgM beads, control antibodies, CpG-ODN2006 or left untreated for 24 hours before OPP labeling. The graphs show fold increase in OPP labeling (means ± SEM) in B (CD19+CD5–) and T cells (CD19–CD5+), with values for untreated B cells set to 1.0 for each donor. Statistical comparisons between groups are shown (Student t test). (D-E) Normal B cells (n = 5) were isolated from PBMCs by negative selection and stimulated with soluble anti-IgM or control antibody, or were left untreated as a control, for 24 hours. Expression of eIF4A, eIF4GI, eIF3B, PDCD4, and HSC70 were analyzed by immunoblotting. (D) Representative immunoblot. (E) Quantification of multiple experiments; the graph shows normalized expression (means ± SEM) relative to control beads. Statistical comparisons between groups are shown (Student t test).
Effect of anti-IgM on eIF expression and comparison with normal B cells. (A-B) CLL samples (n = 12) were stimulated with anti-IgM beads or control beads, or were left untreated. After 24 hours, expression of eIF4A, eIF4GI, eIF3B, PDCD4, and HSC70 (loading control) was analyzed by immunoblotting. (A) Representative immunoblot. (B) Quantitation of multiple experiments; the graph shows normalized expression (means ± SEM) relative to control beads. Statistical comparisons between groups are shown (Student t test). (C) PBMCs from healthy donors (n = 7) were treated with soluble anti-IgM or anti-IgM beads, control antibodies, CpG-ODN2006 or left untreated for 24 hours before OPP labeling. The graphs show fold increase in OPP labeling (means ± SEM) in B (CD19+CD5–) and T cells (CD19–CD5+), with values for untreated B cells set to 1.0 for each donor. Statistical comparisons between groups are shown (Student t test). (D-E) Normal B cells (n = 5) were isolated from PBMCs by negative selection and stimulated with soluble anti-IgM or control antibody, or were left untreated as a control, for 24 hours. Expression of eIF4A, eIF4GI, eIF3B, PDCD4, and HSC70 were analyzed by immunoblotting. (D) Representative immunoblot. (E) Quantification of multiple experiments; the graph shows normalized expression (means ± SEM) relative to control beads. Statistical comparisons between groups are shown (Student t test).
We also investigated mRNA translation and protein modulation in normal cells using peripheral B cells from independent donors as a direct comparator for CLL blood samples. All donors had a high proportion of IgM-expressing CD19+ cells (mean, 84%; supplemental Figure 8A). Anti-IgM increased OPP labeling of normal (CD19+CD5–) B cells in all samples, with no effect on CD19–CD5+ T cells (Figure 4C). In contrast to CLL cells, soluble anti-IgM was an effective inducer of mRNA translation in normal B cells. In fact, response to soluble anti-IgM appeared to be greater than bead-bound anti-IgM in these cells.
To investigate protein expression, we isolated CD19+ B cells (>99% purity) and stimulated these cells with soluble anti-IgM. Soluble anti-IgM was used because it appeared to exert the strongest effect on OPP-labeling in normal B cells (Figure 4C). There was a trend toward increased eIF4GI and decreased PDCD4 expression in stimulated cells (Figure 4D-E). However, eIF4A was clearly unchanged and differences in expression of eIF4GI and PDCD4 were not significant. Analysis of phospho-ERK1/2 and phospho-p70S6K confirmed activation of signaling pathways (supplemental Figure 8B). Thus, sIgM stimulation increases mRNA translation in both normal B cells and CLL cells. However, responses in CLL cells may involve a broader reprogramming of the translation machinery compared with normal B cells.
Increased mRNA translation contributes to increased MYC expression after sIgM stimulation
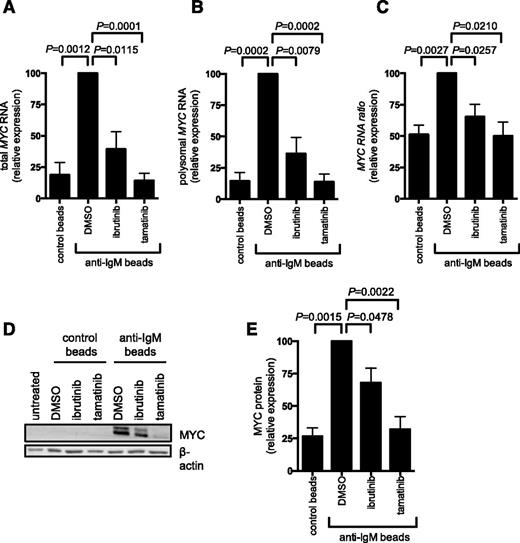
We investigated the effect of bead-bound anti-IgM on translation of mRNA-encoding MYC. Analysis of MYC mRNA translation was based on polysome profiling. Analysis of sucrose gradients revealed that the overall abundance of polysome-associated mRNAs was very low in unstimulated CLL cells (supplemental Figure 9). Consistent with metabolic labeling and OPP assays, there was an increase in the abundance of polysome-associated mRNAs after sIgM stimulation, although this was modest and not clearly observed in all samples.
To analyze MYC expression, we quantified MYC mRNA in fractions from the polysome profiles using quantitative polymerase chain reaction. Consistent with previous studies demonstrating transcriptional increases,11,17 anti-IgM increased the total amount of MYC mRNA detected in all fractions (compare first 2 bars in Figure 5A). There was also a clear increase in the amount of MYC mRNA present in polysome-associated fractions, demonstrating that anti-IgM also increased MYC mRNA translation. This was observed regardless of whether we determined the absolute amount of MYC mRNA in polysome fractions (Figure 5B) or the proportion of polysome-associated MYC mRNA (Figure 5C), which measures translation changes, independent of changes in the overall levels of MYC mRNA. Consistent with increased MYC translation and transcription, anti-IgM significantly increased MYC protein expression (Figure 5D-E). Ibrutinib and tamatinib significantly reduced anti–IgM-induced MYC mRNA translation (and transcription) (Figure 5A-C). Similar to effects on overall mRNA translation, effects of ibrutinib were partial, whereas tamatinib had more pronounced effects. The compounds also significantly inhibited anti–IgM-induced MYC protein expression (Figure 5D-E).
Anti-IgM regulation of MYC mRNA transcription and translation. CLL samples were pretreated for 1 hour with ibrutinib or tamatinib, or DMSO as a control, and then incubated with anti-IgM or control beads for 24 hours. (A-C) Monosome- and polysome-associated MYC mRNA was quantified using quantitative polymerase chain reaction; the graphs show total MYC mRNA (monosomal plus polysomal) (A), polysome-associated MYC mRNA (B), and polysome/monosome ratio for MYC mRNA (C) for ibrutinb (n = 5) and tamatinib (n = 6). (D-E) MYC and β-actin (loading control) protein analysis by immunoblotting. (D) Representative immunoblots and (E) quantitation of multiple experiments for 4 samples. The graphs show mean fold increases (± SEM), with values for anti–IgM/DMSO-treated cells set at 100%. Statistical comparisons between anti–IgM/DMSO-treated cells are shown (Student t test).
Anti-IgM regulation of MYC mRNA transcription and translation. CLL samples were pretreated for 1 hour with ibrutinib or tamatinib, or DMSO as a control, and then incubated with anti-IgM or control beads for 24 hours. (A-C) Monosome- and polysome-associated MYC mRNA was quantified using quantitative polymerase chain reaction; the graphs show total MYC mRNA (monosomal plus polysomal) (A), polysome-associated MYC mRNA (B), and polysome/monosome ratio for MYC mRNA (C) for ibrutinb (n = 5) and tamatinib (n = 6). (D-E) MYC and β-actin (loading control) protein analysis by immunoblotting. (D) Representative immunoblots and (E) quantitation of multiple experiments for 4 samples. The graphs show mean fold increases (± SEM), with values for anti–IgM/DMSO-treated cells set at 100%. Statistical comparisons between anti–IgM/DMSO-treated cells are shown (Student t test).
Discussion
mRNA translation is a key cancer-associated pathway but has not been well studied in B-cell malignancies. Here we show for the first time that stimulation of the BCR of CLL cells promotes mRNA translation in vitro and in vivo. Increased mRNA translation is likely to contribute to antigen-induced CLL cell accumulation via effects on both global mRNA translation and expression of specific oncoproteins, including MYC. This study therefore reveals an important new promalignancy pathway operating downstream of the BCR in CLL cells with clear relevance for drug targeting.
We used an integrated bioinformatical approach to probe transcriptional signatures induced by antigen engagement of CLL cells in LN in vivo. Previous analysis of cells isolated from different compartments highlighted BCR and NF-κB signaling, and modulation of cell cycle, as key pathways activated in LN.18 As expected, there was overlap between the genes and pathways identified in our analysis and this previous work, including identification of MYC as a key driver of transcriptional responses. However, the important new finding from our analysis was the extent to which the response in vivo was dominated by genes linked to mRNA translation.
Our studies in vitro focused on signal-responsive samples, demonstrating that anti-IgM increased mRNA translation in samples from both the U-CLL and M-CLL subsets. Although analysis was restricted to a few samples, anti–IgM-induced mRNA translation was substantially lower in M-CLL samples that were considered nonresponsive on the basis of calcium mobilization. Similar to other signaling readouts,26 mRNA translation in CLL cells was also lower in cells treated with soluble anti-IgM. Moreover, responses to bead-bound anti-IgD were clearly reduced compared with anti-IgM, consistent with our previous study demonstrating that although most CLL samples retain initial sIgD signaling responsiveness, anti-IgD fails to effectively induce MYC expression.17 Further studies are required to probe the relationship between increased mRNA translation and other sIgM signaling responses, but increased mRNA translation is likely to be part of a constellation of responses coregulated downstream of sIgM in signal-responsive samples and sensitive to signal strength.1
In addition to analyzing global mRNA translation, we also showed for the first time that MYC is a target for translational regulation in CLL after sIgM stimulation. We confirmed induction of MYC mRNA after sIgM stimulation11,17 but also showed that increased mRNA translation contributed to induction of MYC protein. Interestingly, in addition to being translationally regulated, MYC may also play a role in upregulation of translation as part of a positive feedback loop.16 For example, MYC is a major regulator of ribosome biosynthesis33 and can also induce eIF expression.34,35 MYC-mediated effects could also involve translational modulation via miRNAs; BCR stimulation modulates miRNA expression in CLL cells and MYC is a master regulator of miRNA networks.36,37
Results with inhibitors are consistent with a pivotal role for SYK in mediating translational induction downstream of the BCR because tamatinib very effectively blocked anti–IgM-induced mRNA translation, including of MYC mRNA. Responses to ibrutinib were partial, perhaps indicating bifurcation of translation-promoting pathways between SYK and BTK. However, these results should be interpreted with caution and further work is required to “map” translational regulation signaling pathways. For example, tamatinib and ibrutinib may have “off-target” effects.38,39 Although in line with other studies, the drug concentrations used for our studies exceed plasma concentrations after administration to patients.3,40 Thus, further work is also required to determine whether ibrutinib, for example, results in decreased mRNA translation in patients.
It was particularly interesting to note that tamatinib was a more effective inhibitor of mRNA translational responses, whereas ibrutinib appears to yield superior clinical responses in patients. There are, of course, many potential explanations for the apparent inconsistency. One possibility is that on-target inhibition of SYK in other cell types, or off-target effects, limits clinical effectiveness of fostamatinib (the prodrug for tamatinib). Therefore, concentrations for effective inhibition of BCR signaling in CLL cells may not be achieved or sustained in patients. Alternately, the more dramatic clinical responses to ibrutinib may be driven by its effects on other pathways independent of the BCR, on other cell types that express BTK, or may be BTK-independent.38,39,41-43 Definitive answers will require clinical evaluation of additional (especially more selective) inhibitors to determine whether this is a class effect, and parallel pharmacokinetic/pharmacodynamic analysis to define the relationship between molecular responses in vitro and clinical outcome.
We investigated the mechanisms by which sIgM stimulation caused increased mRNA translation in CLL and normal B cells. A recent study demonstrated that anti–IgM-induced translation in normal human splenic B cells was associated with decreased expression of the eIF4A inhibitor PDCD4, and increased eIF4A and eIF4E 5′CAP complex formation, but without changes in the overall expression of these eIFs.44 In our study using normal blood B cells, increased mRNA translation occurred without clear changes in expression of eIF4A, PDCD4, or eIF4GI. By contrast, in CLL cells, anti-IgM resulted in clear increases in expression of eIF4A and eIF4GI, as well as decreased PDCD4 (confirming previous observations in CLL cells by Perrot et al).12 Thus, although BCR stimulation increased mRNA translation in both CLL and normal B cells, mechanisms of regulation appear to differ between these cell types, with CLL responses associated with broader modulation of the translation machinery.
In conclusion, BCR stimulation of CLL cells triggers a profound increase in mRNA translation in vitro and in vivo. This response is likely to be important for cell accumulation (via effects on both global mRNA translation and expression of oncoproteins), and its inhibition might contribute to clinical effects of kinase inhibitors, such as ibrutinib. Direct inhibition of mRNA translation is an exciting area for therapy for hematologic malignancies with compounds such ribavirin, homoharringtonine, and silvestrol undergoing assessment in multiple clinical trials. Activity has focused on myeloid leukemia, whereas our study suggests that such agents could also be useful in B-cell lymphoma. In fact, various mRNA translation inhibitors accelerate apoptosis of CLL cells in vitro.10,45,46 Recent studies in colorectal cancer demonstrate that targeted inhibition of mRNA translation is an effective strategy to counter MYC-driven tumorigenesis in vivo, whereas inhibition of upstream signaling was confounded by complex compensatory crosstalk.47 Thus, targeted inhibition of mRNA translation in CLL may also be an effective strategy to counter MYC function. Importantly, the different mechanisms of translational regulation downstream of the BCR in CLL and normal B cells may provide opportunities for therapeutic attack.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the patients involved in this study for their kind gift of samples; Professor Simon Morley (University of Sussex, UK) for the kind gift of anti-eIF4GI and anti-eIF4A antibodies; and Kathy Potter, Isla Henderson, Ian Tracy, Joanne Cowan, and Valentina Iadevaia for their support.
This study was supported by the Gerald Kerkut Charitable Trust, Leukemia and Lymphoma Research, Cancer Research UK, Worldwide Cancer Research, CLL Global Research Foundation, the Experimental Cancer Medicine Centre, and the South Coast Tissue Bank.
Authorship
Contribution: A.Y., S.M.T., B.V.-A., A.L., S.K., M.S.H., E.L., and M.I. performed research and analyzed data; A.Y., S.M.T., S.D.W., A.E.W., A.J.S., F.K.S., F.F., M.J.C., and G.P. designed the research and analyzed data; F.F. provided patient samples and analyzed clinical data; A.Y. and G.P. wrote the initial draft of the manuscript; and all authors contributed to the modification of the draft and approved the final submission.
Conflict-of-interest disclosure: M.S.H. received financial sponsorship from the Jose Castillejo National Programme from the Spanish Ministry of Education. The remaining authors declare no competing financial interests.
The current affiliation for M.S.H. is Department of Pharmacology, Faculty of Pharmacy, University of Seville, Seville, Spain.
The current affiliation for E.L. is XPE Pharma & Science, Wavre, Belgium.
The current affiliation for S.K. is Bart’s Cancer Institute, Queen Mary, University of London, London, United Kingdom.
Correspondence: Alison Yeomans, Somers Cancer Science Building, Southampton General Hospital, University of Southampton, Southampton SO16 6YD, United Kingdom; e-mail: a.m.yeomans@soton.ac.uk.

![Figure 2. Effect of anti-IgM on mRNA translation in CLL cells. (A) Signaling-responsive CLL samples (n = 21) were incubated with anti-IgM beads or control beads, or left untreated as a control for 24 hours. mRNA translation was analyzed by quantifying metabolic labeling. (B) As in (A), except cells were pretreated for 1 hour with ibrutinib (n = 5) or tamatinib (n = 4), or dimethyl sulfoxide (DMSO) as a control, before addition of anti-IgM or control beads. The graphs show fold increase in metabolic labeling (means ± standard error of the mean [SEM]) compared with untreated cells (set to 1.0). (C-D) Immunoblot analysis of phosphorylated/total p70S6K and ERK1/2 expression at 30 minutes poststimulation with anti-IgM beads (ibrutinib, n = 6; tamatinib, n = 5). (C) Representative immunoblots. (D) Quantitation of multiple experiments after 30 minutes of stimulation; graphs show normalized p70S6K/ERK1/2 phosphorylation (means ± SEM) as a percentage of control (DMSO) anti–IgM-treated cells. Statistical comparisons between groups are shown (Student t test).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/127/4/10.1182_blood-2015-07-660969/4/m_449f2.jpeg?Expires=1767697335&Signature=DzKbaByDN5zJJ-eeyprsmLPLZsM8nwr0fn0mQi4msvSubi18FfGMmmz5zK1mwUDA84v5Y62XWJRtNE2oj4L1WesBKyh264uXrR~~WO5d13mEBldYFqGzkeeUma7~v94hH8Qm2iiw20Qb8cL0ddylKdlnWsSwBNXzEYvbFQiGcsCV2Nlh6pxbDU~KXuWEmlJZQekrq0KHfxfBH4P-yg8Z-fY54R8Sf7aC0h4J98EvKGlzNCr~5Q9N4Fnu1mBILShXD1HzloLMLU7tb3GLc7v-0vBTUnlzcOtwMzSg-ne1R~wKt5v8~UWUEy~ocpmXe1IVUrkJC9H~TMoefP9JuhsfQA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal