Key Points
Activation of innate immune receptors induces an antiapoptotic signal and proliferation in ZAP-70–positive CLL dependent on Syk activation.
TLR9 activation autonomously induces BCR signaling in ZAP-70–positive CLL based on an auto/paracrine feedback loop involving immunoglobulin M.
Abstract
The crucial dependence of chronic lymphocytic leukemia (CLL) cells on signals derived from the B cell receptor (BCR) has encouraged the development of new inhibitors, which interfere with BCR signaling and demonstrate clinical benefits in nearly all patients. In addition, signaling through Toll-like receptor (TLR) 9 of the innate immune system has been shown to further contribute to the activation of CLL cells. However, responses to TLR9 engagement are not uniform, but diametrically opposed with cell death in some patients and cell proliferation in others. We now provide evidence that heterogeneous responses to TLR agonists are related to differences in the ability of CLL cells to activate the BCR-associated kinase Syk. Notably, expression of ZAP-70 appears to be of crucial importance for TLR9-mediated activation of Syk. We show that the activation of Syk provides an antiapoptotic signal, which is independent of Mcl-1, Bcl-2, and Bcl-XL, but related to the degradation of the proapoptotic Bim. Mechanistically, TLR9-mediated antiapoptotic signals in ZAP-70–positive CLL trigger secretion of immunoglobulin M, which then serves as (auto-) antigen for a prosurvival BCR signal. Thus, our data show that single activation of the innate immune receptor TLR9 is sufficient to fully engage BCR signaling in ZAP-70–positive CLL, protecting malignant cells from apoptosis. We conclude that the integration of TLR signaling into an adaptive immune response can further promote survival of CLL cells and may contribute to the unfavorable prognosis of ZAP-70–positive CLL.
Introduction
The heterogeneity of chronic lymphocytic leukemia (CLL), ranging from a smoldering, benign variant to an aggressive, fatal disease has clinically been recognized for many decades. More recently, the distinction of CLL cases carrying unmutated immunoglobulin variable region genes (IGVH) from those with mutated IGVH genes has provided a genetic correlation of this observation and now allows us to prospectively predict disease outcome.1,2 In an attempt to understand the pathological implications of these genetic disease variants, transcriptome analyses identified that the ζ-associated protein (ZAP) 70 is aberrantly and predominantly expressed in unmutated CLL.3,4 ZAP-70 is a Syk-related tyrosine kinase whose expression in T and natural killer cells is required for proximal T-cell receptor signaling and signaling through natural killer cell receptors such as CD16, respectively. Experimental evidence indicates that its role in CLL is to enhance signaling through the B cell receptor (BCR) even though this function is not dependent on its kinase activity.5,6
Importantly, it appears that cell survival of both subgroups, unmutated and mutated CLL, relies on continuous activation of this pathway, although their responsiveness to BCR activation is not uniform. Besides differences in the expression levels of ZAP-70, mutated CLL cells express lower levels of soluble immunoglobulin M (sIgM) and are less responsive to BCR engagement.7 In addition, differences in glycosylation of the μ-chain, indicative of antigen exposure in vivo, suggest continued activation of this pathway particularly in unmutated CLL.8
Although BCR signals are of critical importance for leukemic B-cell survival, additional factors provided by their surrounding microenvironment also contribute. Specifically, triggering of innate Toll-like receptor 9 (TLR9) through unmethylated cytosine guanine dinucleotide (CpG) oligodeoxynucleotides (ODNs) has been demonstrated to activate CLL cells.9 Intriguingly, the response of CLL cells to TLR9 agonists is dramatically different between subgroups.10,11 TLR9 agonists protect unmutated CLL from spontaneous apoptosis and drive proliferation, whereas cell death is induced in CLL cells with mutated IGVH genes.12-14 Because differences in the expression levels of TLR9 receptors are not accountable for these opposing effects, the mechanisms underlying these diametrically different responses remain elusive.
Here we report a mechanism explaining this dichotomy based on the activation of Syk by CpGs in Zap-70–positive CLL.
Materials and methods
Patients and CLL cell preparations
After informed patients’ consent and in accordance with the Helsinki declaration, peripheral blood cells were obtained from untreated patients with a diagnosis of CLL by Ficoll-Paque density gradient centrifugation and purified (>96%) by magnetic depletion of CD2/CD14-positive cells (Dynabeads; Life Technologies, Carlsbad, CA). ZAP-70 expression was quantified by flow cytometry with phycoerythrin-labeled Ab clone 1E7.2 (Molecular Probes, Life Technologies) using a cutoff of ≤20% for the ZAP-70–negative subset. These studies were approved by the local ethical committee of the Technical University Munich (project number 1894/07) and by the Cambridgeshire Research Ethics Committee (07/MRE05/44).
Cell culture and treatments
CLL cells were cultured in RPMI 1640 media (Gibco, Life Technologies) completed with 10% heat-inactivated fetal calf serum (Biochrom GmbH, Berlin, Germany). To investigate responses to TLR9 ligation, cells were treated with 1 µM of phosphorothioated unmethylated CpG-ODN DSP30 (5′-TCGTCGCTGTCTCCGCTTCTTCTTGCC) (TIB MOLBIOL, Berlin, Germany). Wherever indicated, cells were cotreated with P505-15 or ibrutinib (Selleckchem, Houston, TX), respectively.
Secretome analysis
CLL cells were treated with or without CpG in the presence of 200 nM tetraacetyl-N-azidoacetyl-mannosamine. After 72 hours of culture, conditioned media were collected by centrifugation, and secreted proteins were assessed by protein enrichment with click sugars (SPECS) using mass spectrometric analysis as described previously.15
Additional methods are presented in the supplemental Data (available on the Blood Web site).
Results
ZAP-70 predicts response of CLL cells to CpG
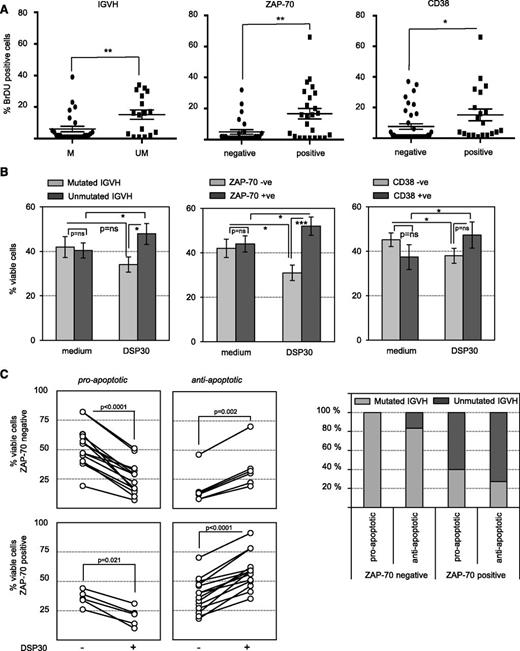
Dramatically divergent responses of CLL B cells to immunostimulatory CpG have been recognized for several years.11,12,14 In order to understand why the net outcome of CpG stimulation is proliferation in approximately half the cases, whereas induction of apoptosis is observed in the other patients, we analyzed the responses to CpG oligonucleotides (DSP30) in a cohort of 57 untreated CLL patients. In accordance with published data,12,13 we observed that induction of proliferation by CpG was strongly associated with CLL cells expressing unmutated IGVH genes. Because the majority of these CLL cells aberrantly express ZAP-70, its expression was also a strong predictive factor for CpG-induced proliferation. Expression of CD38 was also associated with 5-bromo-2′-deoxyuridine incorporation, albeit to a lesser extent (Figure 1A). In line with the increase in cell proliferation, CpG had antiapoptotic effects on IGVH-unmutated, Zap-70–positive and CD38-expressing CLL cells, whereas diametrical effects were observed in mutated cells lacking ZAP-70 and CD38 expression (Figure 1B). Notably, ZAP-70 expression discriminated most accurately the opposing viability response of CLL cells to CpG (P value for Zap-70 positive vs negative = .0003; P = .02 for mutated vs unmutated IGVH; and P = .1472 for CD38+ve vs CD38−ve). Focusing on CLL samples displaying a decrease or increase of at least 10% in cell viability upon TLR9 activation, we identified a small subgroup of ZAP-70–negative CLLs with an antiapoptotic response to CpG. Conversely, TLR9 induced apoptosis in a smaller subset of Zap-70–positive samples (Figure 1C, left panels). This different behavior was not because of differences in IGVH mutation status between the subgroups (Figure 1C, right panel).
ZAP-70 discriminates responses of CLL cells to TLR9 stimulation. (A) S-phase passage of TLR9-stimulated CLL cells assessed after 10 days of DSP30 treatment. Percentages of cells positive for 5-bromo-2′-deoxyuridine (BrdU) were analyzed in relation to IGVH mutational state (n = 48), ZAP-70 subsets (n = 56), or CD38 subsets (n = 56). Each dot represents an individual patient, and the horizontal bar represents the median ± standard deviation (SD). (B) Survival of DSP30-stimulated CLL cells assessed by determining the percentages of annexin-V/propidium iodide–negative cells in n = 57 purified patient samples treated with or without DSP30 for 72 hours. Results were analyzed in relation to IGVH mutational state (n = 48), ZAP-70 subsets (n = 57), or CD38 subsets (n = 56). Data are presented in a bar diagram as the mean ± standard error of the mean (SEM). (C) Patient samples were considered “responders” if difference in viability compared with untreated control was ≥10%. Responders (70% of all samples tested) were analyzed in relation to ZAP-70 subsets and discriminated between proapoptotic (n = 19) and antiapoptotic (n = 20) response to TLR9 stimulation. Each line shows an individual patient. Accordingly, the distribution of IGVH mutation status among these 4 subsets is depicted on the right panel. (A-C) Statistical analyses of the results were performed by 1-way analysis of variance followed by Student t test. P values are indicated by asterisks: ****P < .0001; ***P < .001; **P < .01; *P < .05.
ZAP-70 discriminates responses of CLL cells to TLR9 stimulation. (A) S-phase passage of TLR9-stimulated CLL cells assessed after 10 days of DSP30 treatment. Percentages of cells positive for 5-bromo-2′-deoxyuridine (BrdU) were analyzed in relation to IGVH mutational state (n = 48), ZAP-70 subsets (n = 56), or CD38 subsets (n = 56). Each dot represents an individual patient, and the horizontal bar represents the median ± standard deviation (SD). (B) Survival of DSP30-stimulated CLL cells assessed by determining the percentages of annexin-V/propidium iodide–negative cells in n = 57 purified patient samples treated with or without DSP30 for 72 hours. Results were analyzed in relation to IGVH mutational state (n = 48), ZAP-70 subsets (n = 57), or CD38 subsets (n = 56). Data are presented in a bar diagram as the mean ± standard error of the mean (SEM). (C) Patient samples were considered “responders” if difference in viability compared with untreated control was ≥10%. Responders (70% of all samples tested) were analyzed in relation to ZAP-70 subsets and discriminated between proapoptotic (n = 19) and antiapoptotic (n = 20) response to TLR9 stimulation. Each line shows an individual patient. Accordingly, the distribution of IGVH mutation status among these 4 subsets is depicted on the right panel. (A-C) Statistical analyses of the results were performed by 1-way analysis of variance followed by Student t test. P values are indicated by asterisks: ****P < .0001; ***P < .001; **P < .01; *P < .05.
TLR9-induced survival is associated with the degradation of Bim, but independent of Mcl-1
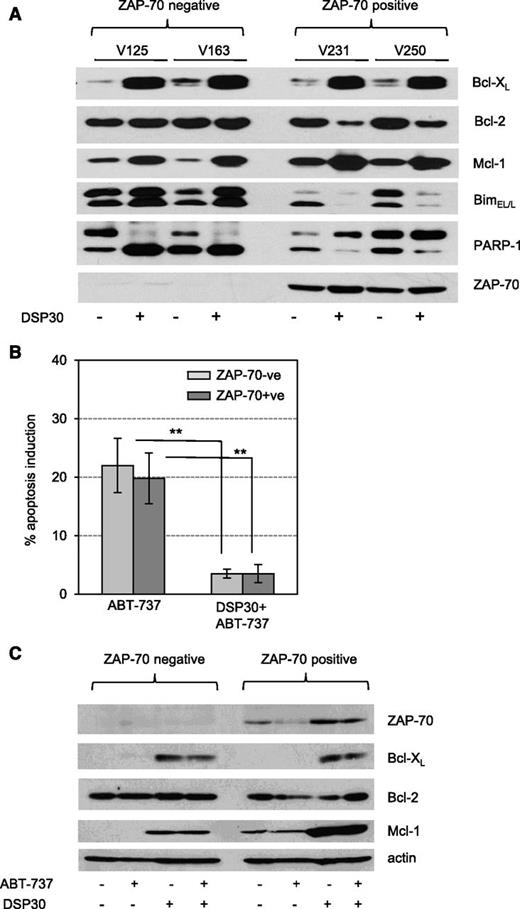
The diametrically opposing effects of CpG on Zap-70–positive and Zap-70–negative CLL survival prompted us to analyze the expression of apoptosis-regulating proteins in response to CpG. Surprisingly, we observed that Bcl-XL and Mcl-1 were induced by CpG in both subgroups (Figure 2A), although ZAP-70–negative cells died upon CpG treatment (Figure 1B). In addition, in Zap-70–positive cells, TLR9 stimulation decreased the expression of Bcl-2 in spite of an increased survival of CLL cells. However, this somehow unexpected regulation of antiapoptotic proteins was accompanied by a strong, inversely correlated regulation of BimEL/L (Figure 2A). Importantly, this increased expression of Mcl-1 is still likely to be functionally relevant because TLR9-activated cells were protected from the cytotoxic effects of ABT-737 (Figure 2B). Maintained high levels of Mcl-1 in CpG/ABT-737 treated cells antagonize the proapoptotic effect of ABT-737, in keeping with the known mechanism of resistance to ABT (Figure 2C).
Bim downmodulation is associated with prosurvival responses to TLR9 stimulation. (A) Expression profile of BH3 proteins in CLL cells treated with or without DSP30 for 72 hours. Data show western blot analysis of n = 4 representative patients samples out of n = 10 tested. Samples were encrypted and anonymized by V numbers. (B) Percentage of ABT-737–induced apoptosis was calculated in relation to the respective untreated controls. CLL cells were precultured with or without DSP30 for 24 hours and subsequently treated with 5 nM ABT-737 for 24 hours. Viability was assessed by determining the percentages of annexin-V/PI–negative cells. Data are presented as the mean ± SD of n = 18 patient samples in a bar diagram. P values are indicated by asterisks: **P < .01. (C) Expression profile of antiapoptotic proteins in DSP30-treated CLL cells with or without ABT-737 treatment (5 nM) for 24 hours. Data show representative western blot analysis of n = 4 tested patients samples.
Bim downmodulation is associated with prosurvival responses to TLR9 stimulation. (A) Expression profile of BH3 proteins in CLL cells treated with or without DSP30 for 72 hours. Data show western blot analysis of n = 4 representative patients samples out of n = 10 tested. Samples were encrypted and anonymized by V numbers. (B) Percentage of ABT-737–induced apoptosis was calculated in relation to the respective untreated controls. CLL cells were precultured with or without DSP30 for 24 hours and subsequently treated with 5 nM ABT-737 for 24 hours. Viability was assessed by determining the percentages of annexin-V/PI–negative cells. Data are presented as the mean ± SD of n = 18 patient samples in a bar diagram. P values are indicated by asterisks: **P < .01. (C) Expression profile of antiapoptotic proteins in DSP30-treated CLL cells with or without ABT-737 treatment (5 nM) for 24 hours. Data show representative western blot analysis of n = 4 tested patients samples.
Together, these data indicate that the opposite effects of CpG on cell survival are primarily reflected by the levels of proapoptotic Bim and not by Mcl-1 regulation.
TLR9 activation supports CLL survival via soluble factors
Because CLL cells can secrete numerous cytokines and chemokines,16 we assessed the secretion of soluble factors by CpG-stimulated CLL cells using a multiplex cytokine array. Notably, Zap-70–positive CLL produced significantly more factors than ZAP-70–negative cells (supplemental Figure 1A). Therefore, we hypothesized that stimulation of Zap-70–positive cells with CpG could promote survival by inducing the secretion of an antiapoptotic factor, whose secretion was restricted to this subgroup of CLL. In order to address this question, we harvested conditioned media from Zap-70–positive cells, either cultured in medium only or in the presence of CpG for 72 hours. As shown before, CpG protected Zap-70–positive cells from apoptosis (Figure 3A, left panel). Subsequently, ZAP-70–negative leukemic cells were cultured in these media, which were all supplemented with CpG. Analyses of cell viability indicated that TLR9 stimulation of ZAP-70–negative CLL induced apoptosis as expected. This proapoptotic effect was not affected by conditioned media from Zap-70–positive cells cultured without CpG (Figure 3A, star). In contrast, conditioned media from CpG-stimulated Zap-70–positive CLL cells (Figure 3A, diamond) completely antagonized the proapoptotic effect of CpG on ZAP-70–negative cells (Figure 3A, right panel). These data confirm that CpG induces an antiapoptotic response in Zap-70–positive CLL via secretion of soluble factors.
Autocrine secretion of soluble factors protects CLL cells from TLR9-mediated apoptosis. (A) Viability of Zap-70–positive CLL cells cultured with or without DSP30 for 72 hours. Line represents the mean ± SD of n = 3 prosurvival responder patient samples (left). Viability of ZAP-70–negative CLL cells cultured with or without DSP30 for 72 hours in fresh medium or in the respective conditioned media (CM) derived from Zap-70–positive samples (n = 3 for CM control and CM DSP30). Data are presented as the mean ± SD of n = 4 patient samples in a bar diagram (right panel). (B) Cytokines secreted by purified CLL cells, cultured with or without DSP30 for 72 hours, quantified by enzyme-linked immunosorbent assay (ELISA). Data are presented in a bar diagram as the mean ± SEM of n = 20 (interleukin [IL] 2), n = 12 (IL-15), and n = 28 patient samples for IL-21, BAFF, and IL-10. (C) DSP30-mediated changes in the expression of glycoproteins secreted by purified Zap-70–positive CLL assessed after 72 hours of culture. Results of mass spectrometric analysis are presented as the mean of n = 3 patients samples in a volcano plot. P values were calculated with Student t test for all proteins based on the log2 intensity ratios. Proteins with P ≤ .05 were considered as hits. (D) sIgM produced by CLL cells cultured with or without DSP30 for 48 hours, quantified by ELISA. Data are presented in a bar diagram as the mean ± SEM of n = 24 patient samples. P values for the entire figure are indicated by asterisks: ****P < .0001; ***P < .001; **P < .01; *P < .05.
Autocrine secretion of soluble factors protects CLL cells from TLR9-mediated apoptosis. (A) Viability of Zap-70–positive CLL cells cultured with or without DSP30 for 72 hours. Line represents the mean ± SD of n = 3 prosurvival responder patient samples (left). Viability of ZAP-70–negative CLL cells cultured with or without DSP30 for 72 hours in fresh medium or in the respective conditioned media (CM) derived from Zap-70–positive samples (n = 3 for CM control and CM DSP30). Data are presented as the mean ± SD of n = 4 patient samples in a bar diagram (right panel). (B) Cytokines secreted by purified CLL cells, cultured with or without DSP30 for 72 hours, quantified by enzyme-linked immunosorbent assay (ELISA). Data are presented in a bar diagram as the mean ± SEM of n = 20 (interleukin [IL] 2), n = 12 (IL-15), and n = 28 patient samples for IL-21, BAFF, and IL-10. (C) DSP30-mediated changes in the expression of glycoproteins secreted by purified Zap-70–positive CLL assessed after 72 hours of culture. Results of mass spectrometric analysis are presented as the mean of n = 3 patients samples in a volcano plot. P values were calculated with Student t test for all proteins based on the log2 intensity ratios. Proteins with P ≤ .05 were considered as hits. (D) sIgM produced by CLL cells cultured with or without DSP30 for 48 hours, quantified by ELISA. Data are presented in a bar diagram as the mean ± SEM of n = 24 patient samples. P values for the entire figure are indicated by asterisks: ****P < .0001; ***P < .001; **P < .01; *P < .05.
As a control, we repeated this experiment with supernatants from an exceptional Zap-70–positive patient, in which we did not observe an antiapoptotic response after CpG stimulation (supplemental Figure 1B, left panel). Supernatants from this patient were unable to rescue ZAP-70–negative patients from the proapoptotic effect of TLR9 stimulation (supplemental Figure 1B, right panel).
Because our data indicate that CpG mediates cell survival via the secretion of soluble factors, we analyzed the amount of antiapoptotic cytokines in the supernatants from ZAP-70–negative and ZAP-70–positive CLL. We used high-sensitivity ELISAs for this experiment and did not detect significant levels of IL-2, IL-15, IL-21, or BAFF in the supernatants of Zap-70–positive and ZAP-70–negative samples. In contrast, IL-10 secretion was induced by TLR9 stimulation, without significant differences between Zap-70–positive and ZAP-70–negative patients (Figure 3B).
To identify in an unbiased approach secreted, prosurvival factors, we analyzed the secretome of CLL cells by SPECS followed by mass spectrometric analysis.15 Supernatants were harvested from Zap-70–positive cell cultures after 72 hours of stimulation with CpG and compared with conditioned media from unstimulated Zap-70–positive cell cultures. We found 33 proteins significantly induced and secreted by TLR9 stimulation, whereas secretion of 5 proteins was reduced (Figure 3C). CpG strongly induced the secretion of metabolic enzymes, adhesion molecules, cytokines, and surface proteins in Zap-70–positive CLL (Table 1). Importantly, IgM was also induced 24-fold in Zap-70–positive TLR9-activated cells. By deducting proteins that were similarly regulated in a ZAP-70–negative sample, the number of significantly (greater than twofold) induced proteins further declined to 4 (Table 2). Of these, only CD44 and IgM are known to contribute functionally to the pathogenesis of CLL. In light of the significance of BCR signaling in CLL, we focused our attention on IgM. To exclude the possibility that CpG induced the secretion of unglycosylated IgM, which was not detected by SPECS, an ELISA from CpG-treated CLL cells was performed. Results from these experiments confirmed that TLR9 stimulation only induced the secretion of IgM in Zap-70–positive CLL, but not in ZAP-70–negative cells (Figure 3D).
Identified glycoproteins which were significantly increased according to their relative increase between TLR9 agonist–treated and –nontreated (Con) ZAP-70–positive CLL cells of 3 different patients
| Protein name . | Gene name . | Unique peptides . | Mean DSP30/Con . | P . |
|---|---|---|---|---|
| α-1,6-mannosylglycoprotein 6-β-N-acetylglucosaminyltransferase A | MGAT5 | 14 | 36.12 | 2.45E-02 |
| V-type proton ATPase subunit S1 | ATP6AP1 | 4 | 29.92 | 2.25E-02 |
| Protein CutA | CUTA | 3 | 29.15 | 2.06E-02 |
| Ig μ chain C region | IGHM | 10 | 24.22 | 9.63E-03 |
| Serglycin | SRGN | 6 | 12.98 | 6.83E-05 |
| 4F2 cell-surface antigen heavy chain | SLC3A2 | 10 | 12.38 | 5.67E-04 |
| Hypoxia upregulated protein 1 | HYOU1 | 5 | 12.06 | 4.74E-02 |
| Platelet-derived growth factor D | PDGFD | 5 | 9.59 | 3.23E-02 |
| l-selectin | SELL | 4 | 9.27 | 1.00E-02 |
| α-mannosidase 2 | MAN2A1 | 16 | 9.25 | 5.96E-04 |
| Fibromodulin | FMOD | 6 | 8.82 | 5.15E-03 |
| Attractin | ATRN | 8 | 8.69 | 7.30E-05 |
| HLA class I histocompatibility antigen, α chain E | HLA-E | 2 | 7.93 | 4.54E-02 |
| Transforming growth factor β receptor type 3 | TGFBR3 | 3 | 7.45 | 2.05E-04 |
| Receptor-type tyrosine-protein phosphatase η | PTPRJ | 7 | 7.36 | 3.59E-03 |
| CD44 antigen | CD44 | 10 | 6.78 | 4.87E-03 |
| Sulfhydryl oxidase 1 | QSOX1 | 16 | 6.72 | 4.28E-02 |
| Granulins | GRN | 16 | 6.67 | 4.04E-04 |
| Ig λ-2 chain C regions | IGLC2 | 3 | 6.21 | 5.29E-04 |
| Adipocyte enhancer-binding protein 1 | AEBP1 | 2 | 5.95 | 1.63E-02 |
| Calsyntenin-1 | CLSTN1 | 20 | 5.47 | 2.23E-03 |
| Semaphorin-7A | SEMA7A | 7 | 5.39 | 2.03E-03 |
| Dystroglycan | DAG1 | 4 | 5.17 | 6.43E-04 |
| Fc receptor-like protein 5 | FCRL5 | 7 | 4.93 | 1.14E-03 |
| Golgi apparatus protein 1 | GLG1 | 16 | 4.80 | 4.42E-02 |
| β-galactoside α-2,6-sialyltransferase 1 | ST6GAL1 | 7 | 4.54 | 4.07E-03 |
| Glucosidase 2 subunit β | PRKCSH | 6 | 4.19 | 1.39E-02 |
| CD48 antigen | CD48 | 4 | 3.94 | 1.81E-02 |
| Calsyntenin-3 | CLSTN3 | 11 | 3.72 | 1.99E-02 |
| Amyloid β A4 protein | APP | 12 | 3.40 | 4.80E-03 |
| Leucyl-cystinyl aminopeptidase, pregnancy serum form | LNPEP | 7 | 3.25 | 2.20E-03 |
| Endoplasmic reticulum aminopeptidase 1 | ERAP1 | 8 | 2.95 | 3.85E-02 |
| Neuroserpin | SERPINI1 | 3 | 2.51 | 3.14E-02 |
| CD82 antigen | CD82 | 3 | 0.42 | 3.60E-03 |
| SPARC | SPARC | 2 | 0.37 | 3.81E-03 |
| Xaa-Pro dipeptidase | PEPD | 6 | 0.29 | 9.21E-04 |
| C-X-C chemokine receptor type 4 | CXCR4 | 3 | 0.28 | 4.91E-02 |
| Glycogenin-1 | GYG1 | 3 | 0.10 | 2.32E-02 |
| Protein name . | Gene name . | Unique peptides . | Mean DSP30/Con . | P . |
|---|---|---|---|---|
| α-1,6-mannosylglycoprotein 6-β-N-acetylglucosaminyltransferase A | MGAT5 | 14 | 36.12 | 2.45E-02 |
| V-type proton ATPase subunit S1 | ATP6AP1 | 4 | 29.92 | 2.25E-02 |
| Protein CutA | CUTA | 3 | 29.15 | 2.06E-02 |
| Ig μ chain C region | IGHM | 10 | 24.22 | 9.63E-03 |
| Serglycin | SRGN | 6 | 12.98 | 6.83E-05 |
| 4F2 cell-surface antigen heavy chain | SLC3A2 | 10 | 12.38 | 5.67E-04 |
| Hypoxia upregulated protein 1 | HYOU1 | 5 | 12.06 | 4.74E-02 |
| Platelet-derived growth factor D | PDGFD | 5 | 9.59 | 3.23E-02 |
| l-selectin | SELL | 4 | 9.27 | 1.00E-02 |
| α-mannosidase 2 | MAN2A1 | 16 | 9.25 | 5.96E-04 |
| Fibromodulin | FMOD | 6 | 8.82 | 5.15E-03 |
| Attractin | ATRN | 8 | 8.69 | 7.30E-05 |
| HLA class I histocompatibility antigen, α chain E | HLA-E | 2 | 7.93 | 4.54E-02 |
| Transforming growth factor β receptor type 3 | TGFBR3 | 3 | 7.45 | 2.05E-04 |
| Receptor-type tyrosine-protein phosphatase η | PTPRJ | 7 | 7.36 | 3.59E-03 |
| CD44 antigen | CD44 | 10 | 6.78 | 4.87E-03 |
| Sulfhydryl oxidase 1 | QSOX1 | 16 | 6.72 | 4.28E-02 |
| Granulins | GRN | 16 | 6.67 | 4.04E-04 |
| Ig λ-2 chain C regions | IGLC2 | 3 | 6.21 | 5.29E-04 |
| Adipocyte enhancer-binding protein 1 | AEBP1 | 2 | 5.95 | 1.63E-02 |
| Calsyntenin-1 | CLSTN1 | 20 | 5.47 | 2.23E-03 |
| Semaphorin-7A | SEMA7A | 7 | 5.39 | 2.03E-03 |
| Dystroglycan | DAG1 | 4 | 5.17 | 6.43E-04 |
| Fc receptor-like protein 5 | FCRL5 | 7 | 4.93 | 1.14E-03 |
| Golgi apparatus protein 1 | GLG1 | 16 | 4.80 | 4.42E-02 |
| β-galactoside α-2,6-sialyltransferase 1 | ST6GAL1 | 7 | 4.54 | 4.07E-03 |
| Glucosidase 2 subunit β | PRKCSH | 6 | 4.19 | 1.39E-02 |
| CD48 antigen | CD48 | 4 | 3.94 | 1.81E-02 |
| Calsyntenin-3 | CLSTN3 | 11 | 3.72 | 1.99E-02 |
| Amyloid β A4 protein | APP | 12 | 3.40 | 4.80E-03 |
| Leucyl-cystinyl aminopeptidase, pregnancy serum form | LNPEP | 7 | 3.25 | 2.20E-03 |
| Endoplasmic reticulum aminopeptidase 1 | ERAP1 | 8 | 2.95 | 3.85E-02 |
| Neuroserpin | SERPINI1 | 3 | 2.51 | 3.14E-02 |
| CD82 antigen | CD82 | 3 | 0.42 | 3.60E-03 |
| SPARC | SPARC | 2 | 0.37 | 3.81E-03 |
| Xaa-Pro dipeptidase | PEPD | 6 | 0.29 | 9.21E-04 |
| C-X-C chemokine receptor type 4 | CXCR4 | 3 | 0.28 | 4.91E-02 |
| Glycogenin-1 | GYG1 | 3 | 0.10 | 2.32E-02 |
The Student t test value was calculated from the log2-transformed fold changes between control and DSP30 with a heteroscedastic Student t test.
Comparison of identified glycoproteins between patients V125 (ZAP-70 negative) and V250 (ZAP-70 positive) which were significantly increased by TLR9 stimulation relative to nontreated (Con) CLL cells
| Protein name . | Gene name . | Unique peptides . | LFQ ratio of V125 +DSP30 vs V125 control . | LFQ ratio of V250 control vs V125 control . | LFQ ratio of V250 +DSP30 vs V125 +DSP30 . |
|---|---|---|---|---|---|
| Ig μ chain C region;Ig μ heavy chain disease protein | IGHM | 10 | 0.00 | 4.10 | 30.20 |
| CD44 antigen | CD44 | 4 | 0.67 | 0.68 | 11.41 |
| Testican-2 | SPOCK2 | 4 | 0.00 | 63.17 | 2.55 |
| Dermcidin | DCD | 2 | 2.00 | 1.39 | 2.41 |
| Coagulation factor X | F10 | 2 | 0.72 | 1.88 | 1.98 |
| Endoplasmin | HSP90B1 | 5 | 1.17 | 1.17 | 1.74 |
| Protocadherin γ-C3 | PCDHGC3 | 3 | 0.66 | 1.06 | 1.65 |
| Semaphorin-4B | SEMA4B | 3 | 0.69 | 1.81 | 1.52 |
| Plexin domain-containing protein 2 | PLXDC2 | 3 | 1.69 | 1.20 | 1.45 |
| Fibronectin | FN1 | 12 | 1.57 | 1.72 | 1.37 |
| HLA class I histocompatibility antigen | HLA-A | 1 | 0.95 | 0.88 | 1.37 |
| Collagen α-1(VI) chain | COL6A1 | 3 | 0.66 | 0.82 | 1.36 |
| Coagulation factor XIII A chain | F13A1 | 2 | 0.00 | 0.89 | 1.33 |
| Apolipoprotein B-100;apolipoprotein B-48 | APOB | 6 | 1.01 | 0.68 | 1.30 |
| CD109 antigen | CD109 | 2 | 1.05 | 1.30 | 1.17 |
| Regucalcin | RGN | 2 | 1.29 | 1.02 | 1.14 |
| Contactin-1 | CNTN1 | 2 | 0.74 | 1.01 | 1.12 |
| Chloride intracellular channel protein 1 | CLIC1 | 9 | 0.99 | 0.86 | 1.07 |
| Insulin-like growth factor-binding protein 2 | IGFBP2 | 5 | 2.25 | 0.72 | 1.04 |
| Tyrosine-protein kinase receptor Tie-1 | TIE1 | 3 | 0.77 | 1.03 | 1.03 |
| Protein name . | Gene name . | Unique peptides . | LFQ ratio of V125 +DSP30 vs V125 control . | LFQ ratio of V250 control vs V125 control . | LFQ ratio of V250 +DSP30 vs V125 +DSP30 . |
|---|---|---|---|---|---|
| Ig μ chain C region;Ig μ heavy chain disease protein | IGHM | 10 | 0.00 | 4.10 | 30.20 |
| CD44 antigen | CD44 | 4 | 0.67 | 0.68 | 11.41 |
| Testican-2 | SPOCK2 | 4 | 0.00 | 63.17 | 2.55 |
| Dermcidin | DCD | 2 | 2.00 | 1.39 | 2.41 |
| Coagulation factor X | F10 | 2 | 0.72 | 1.88 | 1.98 |
| Endoplasmin | HSP90B1 | 5 | 1.17 | 1.17 | 1.74 |
| Protocadherin γ-C3 | PCDHGC3 | 3 | 0.66 | 1.06 | 1.65 |
| Semaphorin-4B | SEMA4B | 3 | 0.69 | 1.81 | 1.52 |
| Plexin domain-containing protein 2 | PLXDC2 | 3 | 1.69 | 1.20 | 1.45 |
| Fibronectin | FN1 | 12 | 1.57 | 1.72 | 1.37 |
| HLA class I histocompatibility antigen | HLA-A | 1 | 0.95 | 0.88 | 1.37 |
| Collagen α-1(VI) chain | COL6A1 | 3 | 0.66 | 0.82 | 1.36 |
| Coagulation factor XIII A chain | F13A1 | 2 | 0.00 | 0.89 | 1.33 |
| Apolipoprotein B-100;apolipoprotein B-48 | APOB | 6 | 1.01 | 0.68 | 1.30 |
| CD109 antigen | CD109 | 2 | 1.05 | 1.30 | 1.17 |
| Regucalcin | RGN | 2 | 1.29 | 1.02 | 1.14 |
| Contactin-1 | CNTN1 | 2 | 0.74 | 1.01 | 1.12 |
| Chloride intracellular channel protein 1 | CLIC1 | 9 | 0.99 | 0.86 | 1.07 |
| Insulin-like growth factor-binding protein 2 | IGFBP2 | 5 | 2.25 | 0.72 | 1.04 |
| Tyrosine-protein kinase receptor Tie-1 | TIE1 | 3 | 0.77 | 1.03 | 1.03 |
TLR9 agonists autonomously induce BCR signaling in ZAP-70–positive CLL cells
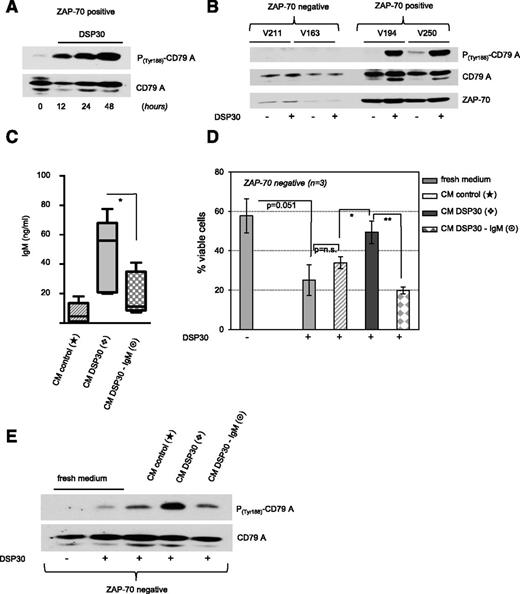
Previously reported data indicate that CLL-derived BCRs can recognize an internal epitope of their own BCR.17 Therefore, we hypothesized that the TLR9-dependent secretion of IgM in Zap-70–positive CLL could engage a BCR signal in the absence of any other exogenous stimulus, thereby acting as an autoantigen. To address this question, we analyzed the phosphorylation of CD79A on tyrosine 188 in Zap-70–positive cells in response to CpG stimulation. CD79A immunoreceptor tyrosine-based activation motif phosphorylation is an early event following engagement of the BCR and a prerequisite for the recruitment and activation of Src family kinases.18 TLR9-activated Zap-70–positive CLL cells showed a marked phosphorylation on tyrosine 188 of CD79A; importantly, this phosphorylation was evident after 12 hours and further increased with time, suggesting that signal transduction was regulated through a positive feedback loop (Figure 4A). Notably, TLR9 stimulation did not induce phosphorylation of CD79A immunoreceptor tyrosine-based activation motif in ZAP-70–negative CLL (Figure 4B).
Secreted IgM activates BCR signaling in TLR9-stimulated CLL. (A-B) Tyrosine-based activation of CD79A in DSP30-stimulated CLL cells cultured for the time as indicated. Comparison of ZAP-70–negative and ZAP-70–positive patient samples (encrypted and anonymized by V numbers) after 48 hours is depicted in panel B. (C) Concentrations of sIgM in CM derived from untreated (CM control) and DSP30-stimulated Zap-70–positive CLL cells before (CM DSP30) and after (CM DSP30-IgM) IgM depletion was performed. Data are presented in a bar diagram as the mean ± SEM of n = 4 patient samples. P values are indicated by asterisks: *P < .05. (D) Viability of DSP30-stimulated ZAP-70–negative B-CLL cells cultured in fresh media or in the respective CM derived from Zap-70–positive patient samples as shown in panel C. Data are presented as the mean ± SD of n = 3 patient samples in a bar diagram. P values are indicated by asterisks: **P < .01; *P < .05. (E) Activation of CD79A in DSP30-stimulated ZAP-70–negative B-CLL cells cultured in fresh media or in the respective CM derived from Zap-70–positive cells as shown in panel C. Data show western blot analysis of 1 representative patient sample out of n = 2 tested.
Secreted IgM activates BCR signaling in TLR9-stimulated CLL. (A-B) Tyrosine-based activation of CD79A in DSP30-stimulated CLL cells cultured for the time as indicated. Comparison of ZAP-70–negative and ZAP-70–positive patient samples (encrypted and anonymized by V numbers) after 48 hours is depicted in panel B. (C) Concentrations of sIgM in CM derived from untreated (CM control) and DSP30-stimulated Zap-70–positive CLL cells before (CM DSP30) and after (CM DSP30-IgM) IgM depletion was performed. Data are presented in a bar diagram as the mean ± SEM of n = 4 patient samples. P values are indicated by asterisks: *P < .05. (D) Viability of DSP30-stimulated ZAP-70–negative B-CLL cells cultured in fresh media or in the respective CM derived from Zap-70–positive patient samples as shown in panel C. Data are presented as the mean ± SD of n = 3 patient samples in a bar diagram. P values are indicated by asterisks: **P < .01; *P < .05. (E) Activation of CD79A in DSP30-stimulated ZAP-70–negative B-CLL cells cultured in fresh media or in the respective CM derived from Zap-70–positive cells as shown in panel C. Data show western blot analysis of 1 representative patient sample out of n = 2 tested.
To address whether the secreted IgM was the critical factor for TLR9-mediated survival of CLL cells and for CD79A phosphorylation, we generated conditioned media from CpG stimulated and unstimulated Zap-70–positive cells. Subsequently, IgM was depleted from the conditioned media by using an anti-IgM antibody and sepharose beads (Figure 4C). Similarly, as shown before (Figure 3A), conditioned media from CpG-stimulated Zap-70–positive cells antagonized the proapoptotic effect of TLR9 agonists on ZAP-70–negative cells. This effect was entirely abrogated by removal of sIgM from the conditioned media (Figure 4D). In accordance with this observation, the presence of IgM was responsible for the induction of CD79A phosphorylation in ZAP-70–negative CLL cells by conditioned media from CpG-treated Zap-70–positive cells (Figure 4E).
In summary, our data provide evidence that TLR9 agonists induce a prosurvival signal in Zap-70–positive CLL cells based on the secretion of IgM, which can autonomously induce a BCR signal. In support of this notion, we observed that TLR9 agonists phenocopied the previously reported anti-IgM–mediated downregulation of CXCR419 on the surface of CLL cells. This was evident in Zap-70–positive, but not Zap-70–negative, cells (supplemental Figure 1C).
Proximal activation of the BCR signalosome is mediated by TLR9
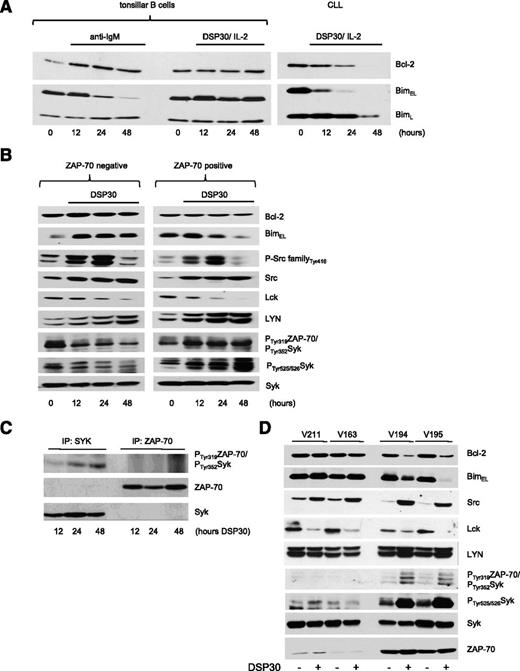
Our data demonstrate that the opposing effects of TLR9 stimulation, cell death or survival, in CLL are related to the expression of Bim (Figure 2A). Notably, in normal B cells, degradation of BimEL is a feature of BCR signaling, but not of TLR9 signaling (Figure 5A). In contrast, TLR9 activation induced degradation of both Bim isoforms in CLL cells (Figure 2) with BimL level generally declining at later time points than BimEL (Figure 5A). To assess whether TLR9 activation is able to fully engage a BCR signal in Zap-70–positive cells, which could ultimately lead to the degradation of BimEL, we analyzed the activation of receptor proximal kinases recruited to the BCR complex. Following CpG stimulation of CLL cells, we identified activation of Src family proteins indicated by an increased phosphorylation on tyrosine 41620 over 24 hours in both Zap-70–positive and Zap-70–negative cells (Figure 5B). Subsequently, Src family proteins were inactivated after 48 hours of TLR9 stimulation. Notably, total levels of Src also increased. Because these antibodies may cross-react with other Src family members, we separately assessed the expression of Lck and Lyn. TLR9 activation rapidly decreased Lck expression but moderately increased the expression of Lyn. The subsequent activation of Syk is a multistep process and involves the phosphorylation of tyrosine 352. In line with a previous report,21 an antibody recognizing both phosphorylation of Syk on tyrosine 352 and of ZAP-70 on tyrosine 31922 detected constitutive activation of kinase(s) in Zap-70–positive and Zap-70–negative samples. TLR9 activation increased and decreased this signal in Zap-70–positive and Zap-70–negative cells, respectively (Figure 5B). In order to discriminate whether ZAP-70 or Syk was phosphorylated in Zap-70–positive cells, ZAP-70 and Syk were immunoprecipitated, and immunoprecipitates were probed with this phospho-specific antibody. TLR9 activation of Zap-70–positive cells induced a slight phosphorylation of Syk on tyrosine 352, but not of ZAP-70 (Figure 5C). In addition to the phosphorylation of tyrosine 352, phosphorylation of tyrosines 525/526 in the kinase domain of Syk is required for its full activity.23 TLR9 activation induced a marked phosphorylation of these tyrosine residues only in Zap-70–positive CLL, but not in negative samples (Figure 5B). In summary, comparison of these signal events between Zap-70–positive and Zap-70–negative CLL cells showed that the induced, full activation of Syk was restricted to Zap-70–positive cells, whereas the induction and phosphorylation of Src was observed in all samples, indicating that Src phosphorylation is not sufficient to transmit a BCR signal in response to TLR9 stimuli (Figure 5B,D). Because the only difference we observed between pro- and antiapoptotic responses to TLR9 activation was the expression of ZAP-70, our data strongly suggest that ZAP-70 is functionally required for the full activation of Syk by TLR9 signals.
Syk, but not Src, activation by TLR9 signals is restricted to ZAP-70–positive cells. (A) BH3 proteins expressed in purified B cells from 1 tonsil or from CLL patients stimulated with anti-IgM or costimulated with DSP30/IL-2 for the time points as indicated. (B) Expression profile of BCR-related signaling molecules in ZAP-70–negative and ZAP-70–positive CLL cells stimulated with DSP30 over a time course of 48 hours. One out of 3 independent experiments are shown. (C) Syk or ZAP-70 was immunoprecipitated from DSP30-stimulated Zap-70–positive cells at the time points as indicated. Blots were probed with P-Tyr319/T352-ZAP-70/Syk antibody or with ZAP-70/Syk antibodies, respectively. (D) Expression profile of BCR-related signaling molecules in CLL cells treated with or without DSP30 for 48 hours. Data show western blot analysis of n = 4 representative patient samples out of n = 6 tested. Samples were encrypted and anonymized by V numbers.
Syk, but not Src, activation by TLR9 signals is restricted to ZAP-70–positive cells. (A) BH3 proteins expressed in purified B cells from 1 tonsil or from CLL patients stimulated with anti-IgM or costimulated with DSP30/IL-2 for the time points as indicated. (B) Expression profile of BCR-related signaling molecules in ZAP-70–negative and ZAP-70–positive CLL cells stimulated with DSP30 over a time course of 48 hours. One out of 3 independent experiments are shown. (C) Syk or ZAP-70 was immunoprecipitated from DSP30-stimulated Zap-70–positive cells at the time points as indicated. Blots were probed with P-Tyr319/T352-ZAP-70/Syk antibody or with ZAP-70/Syk antibodies, respectively. (D) Expression profile of BCR-related signaling molecules in CLL cells treated with or without DSP30 for 48 hours. Data show western blot analysis of n = 4 representative patient samples out of n = 6 tested. Samples were encrypted and anonymized by V numbers.
TLR9 activation engages an autonomous BCR signal in ZAP-70–positive cells, degrading Bim
To assess whether this activation of Syk by TLR9 signals in Zap-70–positive CLL cells is functionally required to autonomously drive a BCR signal, we investigated the effects of Syk inhibition on TLR9-mediated IgM secretion and cell survival. P505-15 is a small-molecule inhibitor with a very low nanomolar potency to inhibit Syk (50% inhibition concentration [IC50] for purified Syk = 6 nM).24 Analysis of secreted IgM indicated that inhibition of Syk fully abrogated TLR9-mediated secretion of IgM in Zap-70–positive CLL in a dose-dependent manner (IC50 = 17.7 nM). Similarly, inhibition of Btk by ibrutinib, which blocks Syk signaling further downstream, abrogated IgM production and secretion (IC50 = 5 nM; Figure 6A-B). In accordance with the antiapoptotic effects of secreted IgM on CLL cells (Figure 4), inhibition of Syk or Btk antagonized the antiapoptotic effect of TLR9 stimulation on Zap-70–positive CLL cells (the inhibitor dose to reach viability of unstimulated cells was 15.2 nM for P505-15 and 0.7 nM for ibrutinib; Figure 6C-D). Notably, Syk inhibition had only a minor effect on the viability of ZAP-70–negative cells (Figure 6E) or unstimulated Zap-70–positive cells (supplemental Figure 1D), indicating that Syk-mediated downstream signaling is constitutively almost negligible in ZAP-70–negative cells and minimal in Zap-70–positive cells. Lastly, Syk inhibition only abrogated TLR9-mediated degradation of BimEL in Zap-70–positive cells without affecting the expression of antiapoptotic proteins (Figure 6F).
BCR inhibitors completely abrogate TLR9 responses in ZAP-70–positive CLL. (A) IgM secretion by Zap-70–positive CLL cells stimulated with DSP30 for 48 hours in the presence of increasing doses P505-15 or ibrutinib, respectively. Curves represent the mean ± SEM of n = 3 patient samples relative to IgM levels in the absence of inhibitors. (B) sIgM production of Zap-70–positive CLL cells stimulated with DSP30 for 48 hours with or without cotreatment of 30 nM P505-15 or ibrutinib, respectively. Data are presented as the mean ± SEM of n = 10 patient samples in a bar diagram. Asterisks indicate P values: **P < .01. (C) Viability of Zap-70–positive CLL cells stimulated with DSP30 for 48 hours in the presence of increasing doses P505-15 or ibrutinib, respectively. Curves represent the mean ± SEM from n = 3 (DSP30 stimulated) patient samples relative to cells cultured without inhibitor. The dotted line indicates the dose of inhibitor required to reach viability of unstimulated cells. (D) Viability of Zap-70–positive CLL cells stimulated with DSP30 for 48 hours in the absence or presence of 30 nM P505-15 or ibrutinib, respectively. Data are presented as the mean ± SEM of n = 10 patient samples in a bar diagram. P values are indicated by asterisks: **P < .01; ***P < .001. (E) Viability of DSP30-stimulated CLL cells cultured with or without 30 nM P505-15 in ZAP-70 subgroups. Data are presented in a bar diagram as the mean ± SEM of n = 24 patient samples. P values are indicated by asterisks: ****P < .0001; ***P < .001; **P < .01; *P < .05. (F) Expression profile of BH3 proteins in CLL cells stimulated with DSP30 for 48 hours in the presence or absence of 30 nM P505-15. Data show western blot analyses of 2 representative patient samples out of n = 5 tested.
BCR inhibitors completely abrogate TLR9 responses in ZAP-70–positive CLL. (A) IgM secretion by Zap-70–positive CLL cells stimulated with DSP30 for 48 hours in the presence of increasing doses P505-15 or ibrutinib, respectively. Curves represent the mean ± SEM of n = 3 patient samples relative to IgM levels in the absence of inhibitors. (B) sIgM production of Zap-70–positive CLL cells stimulated with DSP30 for 48 hours with or without cotreatment of 30 nM P505-15 or ibrutinib, respectively. Data are presented as the mean ± SEM of n = 10 patient samples in a bar diagram. Asterisks indicate P values: **P < .01. (C) Viability of Zap-70–positive CLL cells stimulated with DSP30 for 48 hours in the presence of increasing doses P505-15 or ibrutinib, respectively. Curves represent the mean ± SEM from n = 3 (DSP30 stimulated) patient samples relative to cells cultured without inhibitor. The dotted line indicates the dose of inhibitor required to reach viability of unstimulated cells. (D) Viability of Zap-70–positive CLL cells stimulated with DSP30 for 48 hours in the absence or presence of 30 nM P505-15 or ibrutinib, respectively. Data are presented as the mean ± SEM of n = 10 patient samples in a bar diagram. P values are indicated by asterisks: **P < .01; ***P < .001. (E) Viability of DSP30-stimulated CLL cells cultured with or without 30 nM P505-15 in ZAP-70 subgroups. Data are presented in a bar diagram as the mean ± SEM of n = 24 patient samples. P values are indicated by asterisks: ****P < .0001; ***P < .001; **P < .01; *P < .05. (F) Expression profile of BH3 proteins in CLL cells stimulated with DSP30 for 48 hours in the presence or absence of 30 nM P505-15. Data show western blot analyses of 2 representative patient samples out of n = 5 tested.
Discussion
Steadily growing evidence over the past decade indicates that BCR signals play a key role in the development and progression of CLL, initially based on the observation of a restricted repertoire of BCRs in mutated and unmutated CLL, termed “stereotyped BCRs.”25-27 In addition to antigen-specific BCR signaling, a cell-autonomous, antigen-independent activation of the BCR has been identified in CLL, based on the recognition of an internal epitope of the heavy-chain CDR3 region by the BCR, which may facilitate intra- or intermolecular interactions between BCRs.17 Recently, it was elegantly demonstrated in the Eμ-TCL1 model that both antigen-dependent and antigen-independent BCR-signals can contribute to disease progression in vivo.28
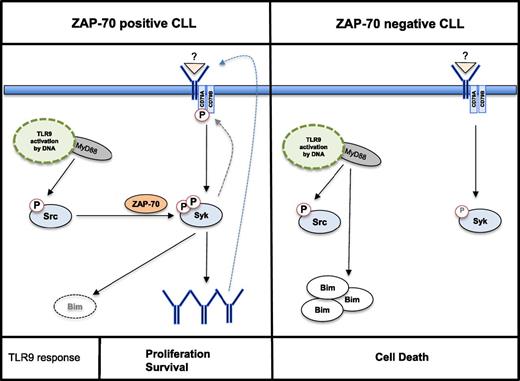
Our data now provide evidence that this survival and proliferation signal can autonomously be activated by an innate immune signal. Engagement of TLR9 is able to mount an auto-/paracrine feedback loop based on the activation of the BCR and the subsequent production, secretion, and autorecognition of IgM (Figure 7). By depleting IgM from TLR9 conditioned media of Zap-70–positive CLLs, we demonstrate that this IgM is proficient to also engage a BCR signal in ZAP-70–negative cells. Notably, exogenously supplied IgM, purified by affinity chromatography from Zap-70–positive, CpG-conditioned media failed to rescue ZAP-70–negative cells from TLR9-mediated apoptosis (data not shown). This finding suggests that additional factors may act in an autocrine, collaborative manner with secreted IgM, because Zap-70–positive CLLs produce significantly more cytokines and chemokines in response to TLR9 stimulation than negative cells (supplemental Figure 1A). Nevertheless, our data suggest that self-recognition of sIgM is the driving underlying mechanism, although we cannot fully exclude the possibility that binding to classical autoantigen(s), derived from CLL cells and captured in IgM:antigen complexes, contributes to TLR9-mediated survival of CLL cells. However, because IgM derived from Zap-70–positive, mostly unmutated, CLLs rescued ZAP-70–negative, mutated CLLs from apoptosis, recognition of another common antigen is highly unlikely. Also, binding of IgM:antigen complexes to Fcμ receptors appears to not play a significant role in TLR9-induced survival because the receptor is significantly downregulated in both ZAP-70 subgroups following stimulation with CpG (own unpublished data and reference29 ). Studies in nonmalignant B cells provide evidence of innate signaling pathways integrating into the BCR-signaling cascade to overcome anergy in the absence of specific T-cell help.30 However, the underlying mechanisms are still not fully understood. In nonmalignant B cells, activation of TLR9 stabilizes a preexisting trimeric complex of MyD88, Pyk2, and DOCK8, thereby creating binding sites for the activation of Src, which results in a subsequent activation of Syk kinase.31 For that reason, it was an unexpected finding that TLR9 stimulation mediated the activation of Src in all samples investigated, but Syk activation in its kinase domain was restricted to the subset of Zap-70–positive CLL. This finding indicates the involvement of a yet unknown mechanism leading to the full activation of Syk. We hypothesize that the TLR9-mediated activation of Syk is sufficient to push CLL cells over the threshold to produce and secrete IgM, which subsequently establishes and maintains a feedback loop by engaging the BCR.
Proposed model of TLR9 signaling in CLL cells. Activation of TLR9 signaling through CpG ODN mediates phosphorylation of Src in both Zap-70–positive and Zap-70–negative subsets of CLL. In Zap-70–positive cells, phosphorylation of Syk in the SH2 domain (tyrosine 352) and in the kinase domain (tyrosine 525/526) fully activates Syk, which mediates the production and secretion of autoreactive IgM. Subsequently, in an autocrine feedback loop, BCR signaling further engages Syk, and the proapoptotic Bim is degraded (left). In ZAP-70–negative CLL, TLR9 signaling mediates the activation of Src and accumulation of Bim. However, the signal is not transduced to Syk (right).
Proposed model of TLR9 signaling in CLL cells. Activation of TLR9 signaling through CpG ODN mediates phosphorylation of Src in both Zap-70–positive and Zap-70–negative subsets of CLL. In Zap-70–positive cells, phosphorylation of Syk in the SH2 domain (tyrosine 352) and in the kinase domain (tyrosine 525/526) fully activates Syk, which mediates the production and secretion of autoreactive IgM. Subsequently, in an autocrine feedback loop, BCR signaling further engages Syk, and the proapoptotic Bim is degraded (left). In ZAP-70–negative CLL, TLR9 signaling mediates the activation of Src and accumulation of Bim. However, the signal is not transduced to Syk (right).
Notably, our data suggest that the expression of ZAP-70 is of crucial importance for this activation of Syk because its expression most accurately predicted response to TLR9 stimuli. Following anti-IgM treatment, differences in BCR signal capacity have also been observed by analyzing Ca2+ flux. These studies have described a stronger association with the IGVH mutational status than with ZAP-70,32 suggesting that qualitative differences exist between anti-IgM treatment and BCR engagement following TLR9 activation. In support of our hypothesis, we observed that ZAP-70 was induced in constitutive ZAP-70–negative cells in which TLR9 activation paradoxically induced survival, but not cell death, accompanied by IgM secretion and Bim degradation (supplemental Figure 2).
Notably, Syk, but not ZAP-70, became phosphorylated on tyrosine 352 upon TLR9 stimulation, in agreement with previously published data.5 Because ZAP-70–negative CLLs are not signaling incompetent per se, activation of Syk can be achieved in the absence of ZAP-70 if the signal is derived directly from the BCR.33 However, it appears that TLR9 signaling to Syk relies on the expression of ZAP-70. It is therefore reasonable to speculate that the main role here of ZAP-70 is to assemble a signaling complex via its adaptor function of the interdomain B. To functionally demonstrate that ZAP-70 is the key molecule for the integration of innate into adaptive immune responses in CLL, we would need to directly interfere with its expression in primary CLL cells. Although we managed to sufficiently knock down ZAP-70 using RNA interference, the TLR9-mediated induction of ZAP-70 (Figures 2, 5, and 6) overcame the knock-down effects and prevented us from successfully performing such an experiment.
Results from our work underscore that TLR9 signaling may actively and independently from other costimulatory factors contribute to disease progression in Zap-70–positive CLL. It is generally assumed that anergic B cells are prone to apoptosis in order to prevent expansion of self-reactive B cells. In CLL, essentially all cells express high levels of Bcl-2, which may protect them from apoptosis in a constitutive (anergic) state. However, our data strongly question the causality of Bcl-2 and Mcl-1 in preventing cell death in TLR9-activated CLL because Bcl-2 levels significantly decreased in CpG-treated Zap-70–positive cells and Mcl-1 was induced in all clones tested. In contrast to Mcl-1 or BCL-XL, expression of Bim inversely correlated with cell viability in response to TLR9 stimuli. In light of our data demonstrating that TLR9 signals autonomously induce a BCR signal, this finding is also in line with previously published data indicating that Bim is regulated by BCR signals in CLL.34 In contrast to our data, Paterson et al did not find a significant decrease of Bim levels upon BCR ligation within 8 hours. This discrepancy, however, is likely explained by the kinetics of TLR9-mediated activation of the BCR.
Finally, it remains to be discussed how relevant these signaling alterations in Zap-70–positive CLL are from a clinical perspective. For several reasons, we believe that the integration of TLR9 signals into BCR signals may actively promote disease progression in vivo: (1) TLR9 signals can drive proliferation of CLL cells, a prerequisite for clonal evolution. (2) The absence of TIR8, which dampens TLR responses, was shown to accelerate disease progression in TCL1-transgenic mice.35 (3) In a previously reported phase 2 trial, CLL patients were treated with anti–Bcl-2 oligonucleotide, and an increase in serum IgM was observed in approximately half the patients. The authors did not find significant on-target effects of the drug, and a clinical response was only observed in a few patients.36 Because gene expression analyses from oligonucleotide-treated patients indicated an activation of TLR in CLL cells, these data suggest that TLR signals may also engage a BCR response in vivo. (4) Although not systematically studied, published data suggest that bacterial infections can promote disease onset and progression,37,38 which may be related to TLR9 activation by bacterial DNA.
Our data indicate that the integration of innate into adaptive immune responses may significantly contribute to disease progression in Zap-70–positive CLL. They further highlight the significance of the BCR for the pathogenesis of CLL and suggest that inhibitors of this pathway, such as Bruton agammaglobulinemia tyrosine kinase inhibitors, may in particular be beneficial to improve the prognosis of Zap-70–positive CLL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the Cambridge Blood and Stem Cell Biobank, which is supported by the Cambridge National Institute for Health Research Biomedical Research Centre, Wellcome Trust–Medical Research Council Stem Cell Institute, and the Cambridge Experimental Cancer Medicine Centre, United Kingdom, for kindly providing the CLL samples. The authors also thank Dr Marc Schmidt-Supprian, Technical University Munich, for constructive discussions and providing critical comments on the manuscript.
This work was supported by a grant from the Deutsche Krebshilfe (111253) (I.R.) and by Cancer Research UK (CRUK) (award C49940). I.R. is a CRUK senior clinical fellow.
Authorship
Contribution: M.W., M.O., A.M., and F.G. planned, performed, and analyzed the experiments; P.-H.K. performed and analyzed the secretome from supernatants; T.H. provided a molecular/genetic analysis of the samples; M.F., C.B., C.P., E.J.B., G.A.F., and I.R. provided conceptual ideas to the work; I.R. planned and analyzed experiments; and M.W., A.M., and I.R. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ingo Ringshausen, Department of Haematology, University of Cambridge, Cambridge Biomedical Campus, Hills Road, The Clifford Allbutt Building, Room 505, Cambridge CB2 0AH, United Kingdom; e-mail: ir279@cam.ac.uk.

![Figure 3. Autocrine secretion of soluble factors protects CLL cells from TLR9-mediated apoptosis. (A) Viability of Zap-70–positive CLL cells cultured with or without DSP30 for 72 hours. Line represents the mean ± SD of n = 3 prosurvival responder patient samples (left). Viability of ZAP-70–negative CLL cells cultured with or without DSP30 for 72 hours in fresh medium or in the respective conditioned media (CM) derived from Zap-70–positive samples (n = 3 for CM control and CM DSP30). Data are presented as the mean ± SD of n = 4 patient samples in a bar diagram (right panel). (B) Cytokines secreted by purified CLL cells, cultured with or without DSP30 for 72 hours, quantified by enzyme-linked immunosorbent assay (ELISA). Data are presented in a bar diagram as the mean ± SEM of n = 20 (interleukin [IL] 2), n = 12 (IL-15), and n = 28 patient samples for IL-21, BAFF, and IL-10. (C) DSP30-mediated changes in the expression of glycoproteins secreted by purified Zap-70–positive CLL assessed after 72 hours of culture. Results of mass spectrometric analysis are presented as the mean of n = 3 patients samples in a volcano plot. P values were calculated with Student t test for all proteins based on the log2 intensity ratios. Proteins with P ≤ .05 were considered as hits. (D) sIgM produced by CLL cells cultured with or without DSP30 for 48 hours, quantified by ELISA. Data are presented in a bar diagram as the mean ± SEM of n = 24 patient samples. P values for the entire figure are indicated by asterisks: ****P < .0001; ***P < .001; **P < .01; *P < .05.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/127/4/10.1182_blood-2015-05-646935/4/m_436f3.jpeg?Expires=1767719047&Signature=TVn66eu9M4vZrPrJxikHRT72qZJJberI3yzZ8fwtHdnd58~F16FfUDnOolQqH1GtLCDAa0h~-mABmOm8RZI4vIs7m2gidTiPEITI~FwZXaC3ne75jn8bMJgCxCv9akIaBLbeU9qvFUuQWPwK77YTPBjEJcZCjkFwxsnV~gp5qTB2KaQYvyCjt57Nink5U-~LlSPj-5vZsVi1-eWizdtNZlX58jiJh5gczv~nWNwesWh2-sz7TSQp9H6HLxwATL1c1b2F958WqARghcdFoJE7bbqgl~keHOx8rbXUSt5HfZZNJk6dtubFc2UqIrIyf5kiqDjwwDJYUPaWkZEmgw21sg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal