Key Points
Aberrations in genome maintenance and DNA repair genes including POT1 occur at a high frequency in Sézary syndrome.
Candidate driver genes and affected pathways in Sézary syndrome show extensive heterogeneity but overlap with other mature T-cell lymphomas.
Abstract
Sézary syndrome (SS) is a leukemic variant of cutaneous T-cell lymphoma (CTCL) and represents an ideal model for study of T-cell transformation. We describe whole-exome and single-nucleotide polymorphism array–based copy number analyses of CD4+ tumor cells from untreated patients at diagnosis and targeted resequencing of 101 SS cases. A total of 824 somatic nonsynonymous gene variants were identified including indels, stop–gain/loss, splice variants, and recurrent gene variants indicative of considerable molecular heterogeneity. Driver genes identified using MutSigCV include POT1, which has not been previously reported in CTCL; and TP53 and DNMT3A, which were also identified consistent with previous reports. Mutations in PLCG1 were detected in 11% of tumors including novel variants not previously described in SS. This study is also the first to show BRCA2 defects in a significant proportion (14%) of SS tumors. Aberrations in PRKCQ were found to occur in 20% of tumors highlighting selection for activation of T-cell receptor/NF-κB signaling. A complex but consistent pattern of copy number variants (CNVs) was detected and many CNVs involved genes identified as putative drivers. Frequent defects involving the POT1 and ATM genes responsible for telomere maintenance were detected and may contribute to genomic instability in SS. Genomic aberrations identified were enriched for genes implicated in cell survival and fate, specifically PDGFR, ERK, JAK STAT, MAPK, and TCR/NF-κB signaling; epigenetic regulation (DNMT3A, ASLX3, TET1-3); and homologous recombination (RAD51C, BRCA2, POLD1). This study now provides the basis for a detailed functional analysis of malignant transformation of mature T cells and improved patient stratification and treatment.
Introduction
Primary cutaneous T-cell lymphomas (CTCL) represent a heterogeneous group of mature T-cell lymphomas. Targeted treatment options for advanced stages of CTCL are limited and associated with modest and short-lived responses.1,2 Sézary syndrome (SS) is a leukemic variant of CTCL and represents an ideal model for defining the molecular pathways involved in the malignant transformation of mature T cells.
Recent studies3-9 have revealed marked genomic heterogeneity in SS illustrated by extensive copy number variants (CNVs) and single-nucleotide variants (SNVs) affecting many genes, including known cancer genes, and selection for genes involved in T-cell receptor (TCR), JAK-STAT, and NF-κB signaling3,5,6,9 and epigenetic regulation.3,7,8
Furthermore, nodal T-cell lymphomas (TCL) show considerable genomic overlap with CTCL.10-13 Although adult T-cell leukemia lymphoma (ATLL) is associated with HTLV-1 transformation, both ATLL and CTCL are mature T-cell lymphomas of skin-homing memory CD4+ T cells with marked clinical and phenotypic overlap. A recent comprehensive genomic study of ATLL has also shown striking similarities at the genomic level with high rates of CNV.13 However the underlying basis for genomic instability, reflected in the high prevalence of CNVs detected in mature T-cell lymphomas, including CTCL, has yet to be clarified.
We have performed a discovery screen using next-generation sequencing (NGS) to analyze enriched tumor cell populations and matched normal DNA from samples obtained at diagnosis from untreated patients using whole-exome sequencing and SNP arrays. This was followed with a prevalence screen in a large cohort of SS samples using targeted resequencing.
Patients, materials, and methods
Samples
All patients fulfilled the WHO-EORTC diagnostic criteria for SS.2 Patient samples were obtained from the nationally approved CTCL research tissue bank (National Research Ethics Committee: 07/H10712/111+5); healthy control samples were obtained with the approval of the Guy’s and St Thomas’ Hospital Research Ethics Committee (EC01/301). Written and informed consent were obtained from all patients and volunteers. Discovery samples: DNA was extracted from CD4+-enriched peripheral blood mononuclear cells (PBMCs) using RosetteSep (Stemcell Technologies, Cambridge, UK) and matched primary fibroblasts from skin explants obtained from 10 untreated patients with SS at diagnosis. Targeted capture samples: DNA was extracted from PBMCs of 101 SS and 32 healthy control samples (supplemental Table 1, available on the Blood Web site).
Whole-exome sequencing (WES) and targeted capture
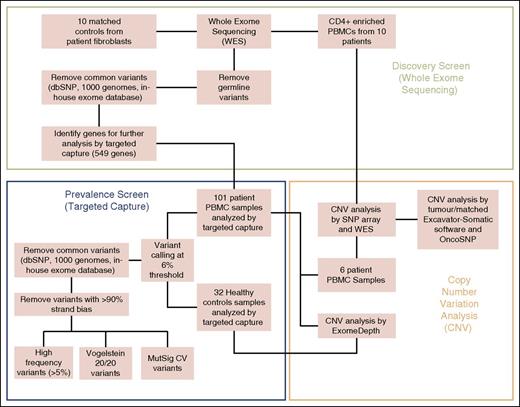
The workflow overview is summarized in Figure 1. Paired-end sequencing library preparation was performed according to manufacturer’s instructions and sequenced on an Illumina Hi-Seq2000 with reads aligned to Hg19 using Novoalign v2.07.11 and postalignment processing performed by picard tools.
Workflow overview of experimental methods. Discovery screen (whole-exome sequencing), prevalence screen (targeted capture), and CNV analysis.
Workflow overview of experimental methods. Discovery screen (whole-exome sequencing), prevalence screen (targeted capture), and CNV analysis.
For WES, Varscan2 Somatic was used to separate tumor variants from patient-matched fibroblasts. ANNOVAR was used for variant annotation.14 Somatic and nonsynonymous variants were selected based on exclusion of variants in dbSNP, the 1000-genomes project, exome variant server, in-house exome database, and genes reported to be error prone in NGS analysis because of sequence repeats and high GC content.15
For targeted capture, Varscan2 and ANNOVAR were also used but the threshold on the minimum allele frequency for calling tumor variants was calibrated to account for the heterogeneity of tumor samples derived from PBMCs (supplemental Methods). Mpileup2cns was used for SNV and INDEL identification with ≥20× depth, ≥15 phred score, ≥6% minimum variant frequency, and read frequency ≤90% in either direction. Variants from 32 nonmatched healthy controls were used to identify tumor-specific variants and exclude sequencing artifacts. Variants selected from WES and targeted capture data were validated by Sanger sequencing on original tumor and additional skin, lymph node, and tumor-derived cDNA samples from the same patients.
Mutational pattern analysis
Several types of mutational pattern analysis were conducted using custom in-house Perl scripts. These included proportions of different types of variant (synonymous, nonsynonymous), SNV base change patterns, and mutation context (motif) analysis upstream and downstream at 3 bp.
Identification of SNV drivers
Several parallel criteria were used to identify genes affected by SNVs. These included MutSigCV,16 the 20/20 rule17 (see supplemental Methods for details), and simple frequency filtering of >5% after removing genes previously identified as problematic.15 We also compared the list of candidate driver genes to those present in the network of cancer genes.18
Gene copy number analysis
Tumor-specific CNVs were identified through integrative analysis of discovery and targeted capture data generated using exome/targeted sequencing and SNP array technologies. Data from WES were analyzed by Excavator v2.2 in matched pairs. HumanOmni5Exome arrays were analyzed using OncoSNP, v1.4. Raw data (BAF and LRR) required for OncoSNP was extracted using Illumina Genome Studio software. Data from WES and SNP array were combined for final analysis (n = 16). Remaining prevalence samples (n = 91) were analyzed with ExomeDepth software19 using the targeted capture data and 32 healthy controls (Figure 1). This analysis was restricted to targeted capture genes (n = 549) but allowed deeper resolution. The genotype array data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE80650 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc= GSE80650).
Pathway analysis
To investigate for significant perturbations at the pathway level, we performed a gene set enrichment analysis on WES and TC SNVs (supplemental Methods) using the MSigDB repositories. Pathway-level perturbations were quantified using 2 inter-related metrics. One metric, “fraction of pathway genes mutated,” captures the proportion of pathway genes involved in nonsynonymous SNVs or indels across all patients. The second metric, “pathway perturbation frequency score,” captures how often each pathway is perturbed as a proportion of all samples (both uncorrected and corrected for pathway size), assuming perturbation occurs if at least one of the pathway’s genes is mutated.
Results
Whole-exome sequencing of CD4+-enriched cells and matched controls (discovery screen)
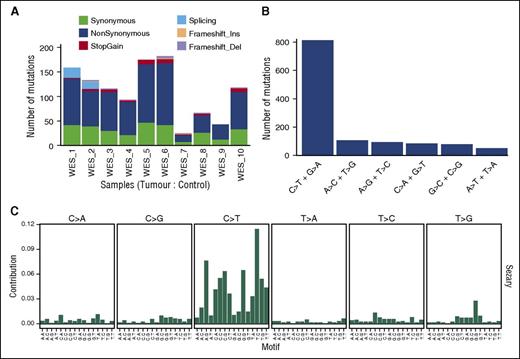
For 10 CD4+-enriched/control-matched DNA samples, we obtained depth >20 reads covering 82% to 95% of the target region across all samples, with a median of 91.12%. The most frequent type of variant effect (Figure 2A) was nonsynonymous (63%) followed by synonymous (26%) and stop-gain (3.5%). After filtering, we identified 824 somatic, nonsynonymous variants (750 genes; supplemental Table 2) from which we selected 549 genes for targeted capture analysis. Overall mutation rates for filtered somatic tumor variants were between 0.54 and 4.2 mutations per megabase with total nonsynonymous variants per tumor from 23 to 182, with a median of 98 comparable with other NGS studies of CTCL.3,4,7 Furthermore this rate is similar to rates reported for other non-Hodgkin lymphomas13 and distinct from tumors with a higher median (130-160) associated with specific carcinogens such as lung cancer and melanoma.16 Two samples showed low levels of nonsynonymous variants (23 and 41 SNVs) with the youngest patient having the lowest number (supplemental Table 2). The most common type of nucleotide change (Figure 2B) was C>T and G>A (61%), in keeping with what has been observed already in many types of cancers.20 Specifically, 42% of the C>T variants occurred at NpCpG sites, reflecting age-related spontaneous deamination at methylated CpG sites, and 27.5% occurred at NpCpC sites, but there were <1% CC>TT mutations (14 of 1520 SNVs affecting 6% of samples). Although interpretation is limited by our sample size, mutation context analysis shows consistency with several trinucleotide signatures (Figure 2C) identified in a recent study.20
Summary of somatic tumor mutations in 10 whole-exome sequences. (A) Types and ratios of all somatic mutations detected in the discovery panel. (B) Distribution of all SNV base changes from the panel. (C) Trinucleotide analysis showing frequency of bases immediately 5′ and 3′ of the mutated bases.
Summary of somatic tumor mutations in 10 whole-exome sequences. (A) Types and ratios of all somatic mutations detected in the discovery panel. (B) Distribution of all SNV base changes from the panel. (C) Trinucleotide analysis showing frequency of bases immediately 5′ and 3′ of the mutated bases.
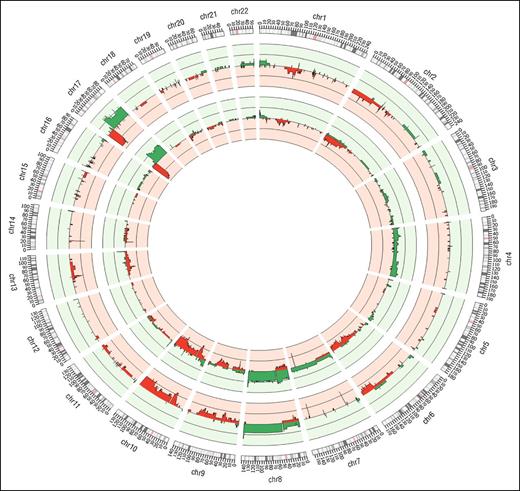
Copy number analysis of WES sequence data from the discovery panel (Figure 3, outer track) revealed recurrent (>1) tumor-specific CNVs consisting of large (>1 MB) and focal (<1 MB) regions of amplification and deletion and confirmed using SNP array (Figure 3, inner track). Noticeably, 2 deleted regions on chromosomes 7 and 14 containing the γ and α TCR genes occurred in all samples, reflecting clonal TCR gene rearrangements.
Summary of CNVs identified genome-wide for 10 WES samples. The outermost track represents results from WES samples anaylzed using Excavator; the middle track represents 16 SNP array samples analyzed with OncoSNP. In all the tracks, red denotes losses and green denotes gains.
Summary of CNVs identified genome-wide for 10 WES samples. The outermost track represents results from WES samples anaylzed using Excavator; the middle track represents 16 SNP array samples analyzed with OncoSNP. In all the tracks, red denotes losses and green denotes gains.
Analysis of SNP array and WES CNV data confirms and extends previous findings in SS using array CGH and cytogenetic techniques,21-25 including large complex chromosomal abnormalities such as isochromosome 17q (loss of 17p >70% and gain of 17q >50% of tumor samples),26-28 and recurrent focal CNVs often affecting individual genes already subject to SNVs. Other frequently observed large CNVs were losses on 1p, 2p, 13p, and 10q, and gains on 8, consistent with previous reports.3,21-25 However, array data showed gains on chromosome 4 that were not observed in the WES data, although these have been previously reported in SS.21
Targeted capture sequencing of 101 SS samples from patients (prevalence screen)
In the targeted custom capture of 549 genes—depth was between 149 and 848 reads starting from the list of all variants in tumors—we filtered out those also present in the healthy control panel. To enrich for somatic variants, we further filtered out variants present in dbSNP, the 1000 Genomes Project, Exome Variant Server (National Heart, Lung, and Blood Institute Exome Sequencing Project), and our in-house exome database. We also filtered out variants in genes reported to be error-prone in NGS analysis because of sequence repeats and high GC content.15 From this list we focused on the final subgroup of nonsynonymous variants including Indels, stop gain/loss, splice variants, and indels, for a total of 1520 variants. There were between 2 and 93 variants per tumor, with a median of 13 variants per tumor (supplemental Table 3).
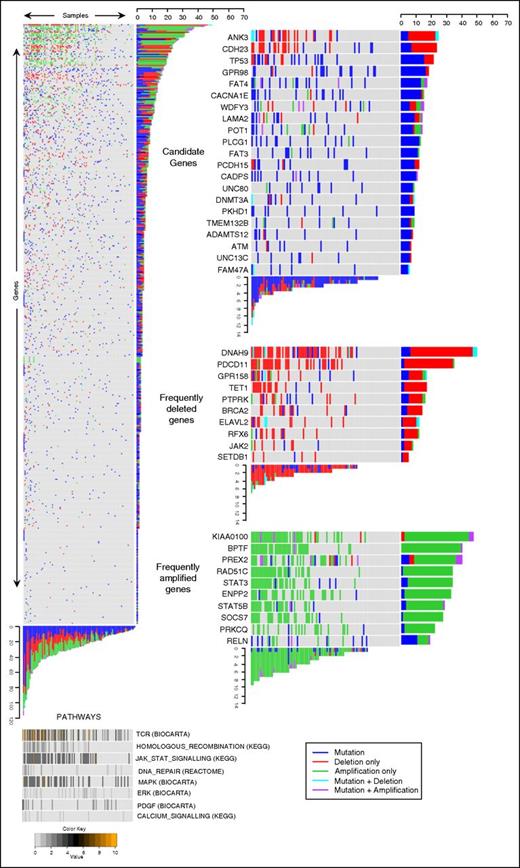
Analysis of CNVs in the targeted capture samples revealed a similar distribution of CNVs to the discovery WES samples. Overall, 453 of 549 genes in the targeted capture were affected by CNVs in at least 1 tumor (supplemental Table 4). Illustration of CNVs and mutations occurring in each gene are reported in Figure 4 and supplemental Figure 2.
Genomic data of 549 genes from 101 Sézary tumors identifies candidate driver genes. Heat map showing all genes (y-axis) and all tumors (x-axis) (left); pathways identified as frequently perturbed are aligned below the main panel (bottom). The color code represents the percentage of mutationally perturbed genes in each tumor sample for each pathway. Candidate driver genes showing high frequencies of SNVs (top right), frequently deleted genes (middle right), and frequently amplified genes (bottom right) are subsetted from the main panel.
Genomic data of 549 genes from 101 Sézary tumors identifies candidate driver genes. Heat map showing all genes (y-axis) and all tumors (x-axis) (left); pathways identified as frequently perturbed are aligned below the main panel (bottom). The color code represents the percentage of mutationally perturbed genes in each tumor sample for each pathway. Candidate driver genes showing high frequencies of SNVs (top right), frequently deleted genes (middle right), and frequently amplified genes (bottom right) are subsetted from the main panel.
Identification of potential driver genes
In view of the marked genomic heterogeneity, we applied different parallel criteria (supplemental Methods) to identify 21 potential driver genes based on SNVs and 42 genes based on high rates of CNVs (Figure 4; supplemental Tables 4 and 5).
We identified 51 genes with SNVs occurring in >5% of tumors. Analysis of all SNVs using MutSigCV16 confirmed that 5 of these 51 genes (DNMT3A, FAM47A, POT1, CADPS, TP53) were mutated more often than expected by chance (q < 0.1) and all except FAM47A have been implicated as driver gene mutations in other cancer types. Two genes (PREX2 and PCLO) had MutSigCV values close to significance (q = 0.11). In this data set, ATM and both TP53 and DNMT3A identified with MutSigCV, have been previously defined as driver genes based on specific criteria applied to COSMIC and the Cancer Genome Atlas and functional validation.17,29 In contrast CSMD1, CSMD3, PCLO, and CNTNAP516 have been identified as likely false-positive cancer genes, although overexpression of CSMD3 has been associated with growth advantage in epithelial cells.30 Finally, correlation of these putative driver genes with data sets annotating candidate driver genes18 identified 21 genes as potential or established driver genes. Analysis of SNVs using the “20/20” rule17 identified 16 of these 21 genes (supplemental Table 5) as either potential oncogenes (5 genes including FAM47A, PLCG1, and GPR158) or tumor-suppressor genes (11 genes including POT1, ANK3, UNC13C, ATM, DNMT3A, and TP53).
The overall frequency of SNVs affecting these 21 potential driver genes in our prevalence data set ranged from 5.5% to 19% (6-21), with 7 mutated genes affecting >10% of tumors. These consisted of known tumor suppressor genes TP53 and FAT3/FAT4 (upstream regulators of the Hpo pathway31 ) and putative oncogenes such as PLCG1,6,32 as well as GPR98, CADPS, and CACNAIE,33 whose potential functional role in cancer has yet to be established. Of these potential driver genes, 15 had identical recurrent variants, with 2 genes having more than one recurrent variant, namely GPR98 (2) and PLCG1 (5).
Two tumor samples were notable for having relatively few SNVs detected in the whole-exome study (supplemental Table 2). Both tumors had aberrations affecting only a few of our potential driver genes including identical variants reported in COSMICv71 namely POT1R117C, JAK3A573V, and PREX2 in one tumor and PREX2, DNMT3A, STAT5BN642H, FAT4R3615W, and GPCR158R757C in the other tumor, suggesting that these gene mutations could be sufficient for tumor development.
We identified 42 genes with at least 1 SNV and CNVs affecting >10% of tumors as additional potential driver genes. CNVs affected all 21 potential driver genes identified based on analysis of SNVs. We also identified a higher prevalence of aberrations (14%-48% of tumor samples) for other putative driver genes such as DNAH9, ENPP2, ELAVL2, RFX6, PDCD11, GPR158, PTPRK, PRKCQ, BRAC2, TET1, RAD51C, and PREX2. Notably few tumor samples had SNVs affecting individual JAK and STAT genes, but overall 55% of tumors had combined SNVs and CNVs affecting these genes including regulators of STAT3 such as SOCS7.
Overall, 510 of 549 genes in our targeted capture had SNVs and/or CNVs reported in recent studies of SS and mycosis funcgoides (MFs) (supplemental Table 6). Specifically, SNVs affecting 15 of our 21 potential driver genes have been detected in recent studies of SS and/or MF.3-9 Identical gene variants have also been functionally validated in these and other studies, namely TP53,34 , POT1,35 PLCG1,3,6,36-38 ATM,39 JAK3,40,41 STAT3,42 and STAT5B43-46 (Table 1). In addition, identical variants without functional validation have been reported in COSMICv71 for 4 other genes from our 21 potential drivers, namely FAT3R4213C, FAT4R3615W, GPR158R757C, and UNC13CR2037H/G2150R.17
Identical gene variants reported in other studies
| Gene . | Variant . | Functional validation . | References . |
|---|---|---|---|
| JAK3 | A573V | Yes | 40, 41 |
| STAT3 | Y640F | Yes | 42 |
| STAT5B | Y665F | Yes | 43, 44 |
| STAT5B | N642H | Yes | 43, 45, 46 |
| STAT5B | E150Q | Yes | 46 |
| PLCG1 | S345F | Yes | 3-6, 8, 9, 13, 36, 37 |
| PLCG1 | S520F | Yes | 4, 6, 8 |
| PLCG1 | D342N | Yes | 3, 37, 38 |
| PLCG1 | R48W | Not done | 3, 4, 7, 13 |
| PLCG1 | E1163K | Not done | 3, 4, 7, 8, 13 |
| PLCG1 | D1165H | Not done | 4, 13 |
| POT1 | R117C | Yes | 35 |
| TP53 | S127F | Yes | >10 papers |
| TP53 | H20R | Not done | 5 |
| TP53 | R37X | Not done | 5 |
| ATM | E2423K | Yes | 39 |
| CSMD1 | A408V | Not done | 4 |
| ENPEP | V97L | Not done | 7 |
| LRP1B | R790Q | Not done | 4 |
| Gene . | Variant . | Functional validation . | References . |
|---|---|---|---|
| JAK3 | A573V | Yes | 40, 41 |
| STAT3 | Y640F | Yes | 42 |
| STAT5B | Y665F | Yes | 43, 44 |
| STAT5B | N642H | Yes | 43, 45, 46 |
| STAT5B | E150Q | Yes | 46 |
| PLCG1 | S345F | Yes | 3-6, 8, 9, 13, 36, 37 |
| PLCG1 | S520F | Yes | 4, 6, 8 |
| PLCG1 | D342N | Yes | 3, 37, 38 |
| PLCG1 | R48W | Not done | 3, 4, 7, 13 |
| PLCG1 | E1163K | Not done | 3, 4, 7, 8, 13 |
| PLCG1 | D1165H | Not done | 4, 13 |
| POT1 | R117C | Yes | 35 |
| TP53 | S127F | Yes | >10 papers |
| TP53 | H20R | Not done | 5 |
| TP53 | R37X | Not done | 5 |
| ATM | E2423K | Yes | 39 |
| CSMD1 | A408V | Not done | 4 |
| ENPEP | V97L | Not done | 7 |
| LRP1B | R790Q | Not done | 4 |
Several specific variants have been reported previously in CTCL and other malignancies. Functionally validated specific variants are indicated.
Analysis of signaling pathways affected by SNVs
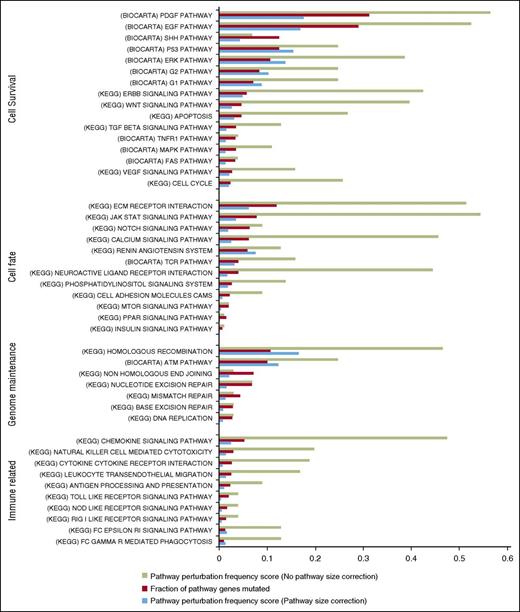
Our gene set enrichment analysis highlighted aberrations affecting numerous pathways involved in cell fate, cell survival, genome maintenance, and immune-related functions in both the TC and WES data sets (Figure 5; supplemental Figure 3). Notably, several pathways have SNVs affecting the same gene(s), specifically JAKs, STATs, PLCG1, and TP53. This analysis showed enrichment for genetic aberrations involving many of the putative driver genes affecting pathways including homologous recombination (RAD51C, BRAC2, POLD1: 45%) and DNA repair (ATM, TP53, BRAC2: 32%).
Gene-set enrichment analysis identifies key pathways. Highly perturbed pathways from the KEGG and BIOCARTA repositories were grouped according to related functions. The proportion of tumors showing perturbations in a given pathway is shown in green. Proportion of genes in each pathway with a perturbation across all affected tumors is shown in red bar. Pathway perturbation frequency score (blue bar) shows how often a pathway is perturbed but taken in consideration of the pathway size.
Gene-set enrichment analysis identifies key pathways. Highly perturbed pathways from the KEGG and BIOCARTA repositories were grouped according to related functions. The proportion of tumors showing perturbations in a given pathway is shown in green. Proportion of genes in each pathway with a perturbation across all affected tumors is shown in red bar. Pathway perturbation frequency score (blue bar) shows how often a pathway is perturbed but taken in consideration of the pathway size.
Gene perturbations (SNVs and CNVs; supplemental Figure 4) were also grouped into families with related functions including DNA repair (at least 1 perturbation 64%; >1 perturbation 36%), global epigenetic regulation (at least 1 perturbation 42%; >1 perturbation 14%), and programmed cell death (at least 1 perturbation 64%; >1 perturbation 37%) in line with well-known hallmarks of cancer.47
Lack of correlation with clinical outcome
A pairwise analysis of gene mutations using Bonferroni adjustment failed to identify any SNVs and CNVs, which either occurred together or were mutually exclusive. In addition, we did not detect any correlation between mutational load (including total SNVs and CNVs) and overall survival. Analysis of individual genes in the targeted capture data set identified 10 genes affected by SNV, CNV, or a combination of both, which were associated with a worse overall survival (supplemental Table 7). Only one of these 10 genes (RELN) was identified as a potential driver gene. However in view of lack of power, these results should be interpreted cautiously because the likelihood of chance occurrence is 1 in 5.
Validation of potential driver gene mutations
Sanger resequencing was performed on variants from 55 genes from the Discovery Panel (supplemental Table 8). A total of 97 of 101 variants were validated, consistent with other NGS cancer studies.48-52 Similar proportions of variants were successfully validated on the prevalence screen data (134/139). Highly recurrent gene mutations in the targeted capture analysis were also validated in multiple and different tissue samples (blood, lesional skin, and lymph node) from the same patients at diagnosis and at disease progression, further supporting their role as candidate drivers (supplemental Table 8). In contrast for those patients who achieved a complete clinical remission after reduced intensity allogeneic transplantation, we could not detect specific gene variants, identified in the diagnostic samples, in the post-transplant tissue samples consistent with the absence of the original T-cell clone and a complete molecular remission.
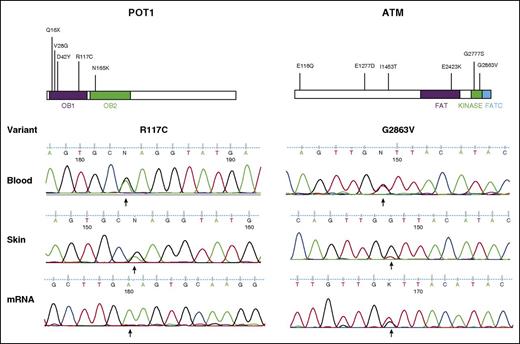
The presence of POT1R117C and ATMG2863V variants were confirmed in additional blood and skin samples and at the transcriptional level in mRNA from enriched CD4+ tumor cells (Figure 6 and supplemental Table 8). Interestingly, for POT1R117C, predominant expression of the mutant was detected over the wild-type allele. This is likely attributable to LOH affecting the wild-type allele. This was confirmed by sequencing genomic DNA from the same CD4+-enriched tumor cells, in which POTR117C was also predominantly detected (data not shown).
Validation of candidate driver genes POT1 and ATM. Schematics of each protein are shown with the locations of mutations identified (top). Sanger validation of SNVs in lesional skin, diagnostic blood samples, and mRNA isolated from CD4+-enriched tumor cells (bottom).
Validation of candidate driver genes POT1 and ATM. Schematics of each protein are shown with the locations of mutations identified (top). Sanger validation of SNVs in lesional skin, diagnostic blood samples, and mRNA isolated from CD4+-enriched tumor cells (bottom).
Finally, we sought to identify variants from our study that had been detected and/or functionally validated in previous studies. Several such variants (Table 1) were identified including PLCG1(S345F, S520F, D342N), 3,6,36-38JAK3A573V,40,41STAT3Y640F,42STAT5B(Y665F N642H),43-45ATME2423K,39 and POT1R117C,35. Other candidate driver genes from our study including PTPRK and FAT3 (as opposed to proven drivers such as TP53) have also been functionally validated based on analysis of different variants.18
Discussion
Our analysis of a large series of SS patient samples has identified novel variants and CNVs predicted to be potential drivers. Specifically, we found a high frequency of perturbations in POT1, ATM, and BRCA2, which are involved in genome maintenance. Dysregulation of genome maintenance processes may contribute to the high prevalence of structural variation observed in CTCL. We also detected mutations of JAK-STAT, DNMT3A, TP53, and PLCG1, genes previously reported as likely drivers in other lymphomas17,53 and CTCL.3-5,7-9 These putative driver gene mutations were present in diagnostic blood, skin, and node samples and samples collected at disease progression time points, but that were absent in samples from those patients who achieved a complete remission after stem cell transplant.
Genomic instability is a feature of SS with complex copy number variation reported using different techniques.3,21,22,24 More than 50% of tumors had SNVs and/or CNVs affecting genes involved in DNA repair and telomere maintenance. Notably, a significant number of tumors (23%) had mutations and/or loss of genes involved in telomere maintenance such as POT1 and ATM. POT1 is part of the multiprotein Shelterin complex responsible for telomere length and loss of POT1 function increases chromosomal instability.54 All the POT1 variants occurred in the oligonucleotide/oligosaccharide binding (OB) domains and loss of OB function has been shown to cause extensive telomere elongation55 and frequent telomere fusions.56 Recurrent POT1 mutations have also been detected in a subset of patients with CLL (5%)57 and ATLL.13 In addition, the POT1R117C variant has recently been identified as the cause of an inherited cancer syndrome in which loss of function causes an age-related increase in telomere length and genomic instability, contributing to the development of malignancies including lymphomas.35 ATM is a PI3 kinase involved in the recognition of DNA double-strand breaks and recruitment of telomerase, and copy number losses have recently been reported in SS.3,58 The ATME2423K variant is associated with loss of function in non–small-cell lung cancer.39 POT1 also represses the ATM damage response checkpoint.56 Other genes involved in telomere maintenance include ATRX and TEP1, both with somatic mutations. We also detected frequent SNVs/CNVs affecting genes involved in homologous recombination such as RAD51C, BRCA2, and POLD1, a component of the DNA polymerase δ complex,59 and loss of function TP53 mutations, which are described in CTCL.60,61 Previous mouse models showed that combined defects of telomerase and cell-cycle genes are associated with the development of mature T-cell lymphomas.62 Loss of cell-cycle control (TP53), telomere maintenance (POT1/ATM), and DNA repair initiation (BRCA2) could contribute to the genomic instability, which is a consistent feature of SS.
Overall, 40% of tumors had somatic mutations affecting genes involved in TCR/NF-κB signaling. We detected recurrent PLCG1 gene variants in 11 patients including several variants reported previously3-9,37 in MF and SS, as well as PTCL, AITL, and ATLL.13,36 The PLCG1S345F variant has been shown to induce expression of both NFAT via IP3 activation and NF-κB via DAG activation of PKC signaling. This mutation is predicted to impair the auto-inhibitory function of PLCG1, which limits TCR signaling downstream of receptor ligation.6,63 In addition, a further recurrent variant (PLCG1D342N) has been shown to increase inositol phosphate production in COS-7 cells.38 It is not yet clear whether the other recurrent PLCG1 variants identified affect this same catalytic function and whether these variants are sufficient alone to enable constitutive TCR signaling without costimulatory signals, but recent studies in SS and ATLL have detected activating CD28 mutations and CTLA4-CD28 and ICOS-CD28 gene fusions.3,13 Although we did not detect abnormalities of CD28, key findings in our study included mutations of other TCR/ NF-κB signaling genes, notably PRKCQ (20% of cases) as well as NFATC2, NFκB1, and PAK7. PRKCQ belongs to the PKC family of serine/threonine kinases, is highly expressed in T cells, and has a pivotal role downstream of PLCG1 in transducing TCR and costimulatory CD28 signals.13 In all but 2 cases, PRKCQ aberrations were independent of PLCG1 mutations. In ATLL, studies have identified gain-of-function PRKCB mutations and associated downstream activating mutations of CARD11, leading to enhanced NF-κB activation.13 CARD113-5,7,8 activating mutations and PRKCQ3,4 SNVs and CNVs have also been detected recently in SS.
Constitutive activation of NF-κB is described in CTCL,64,65 and recurrent gain-of-function mutations affecting the TNFRSF1B gene in MF/SS have been shown to enhance noncanonical NF-κB signaling.5 In PTCL, the t(5;9)(q33;22) results in an ITK-SYK fusion kinase, which induces constitutive TCR activation,66 and LCK mutations have been documented in lymphoma.67 These findings now provide compelling support for the hypothesis that the survival of malignant T cells in SS and other mature T-cell lymphomas is at least partly dependent on TCR and NF-κB signaling: In mature T-cell lymphomas such as SS, selection for activating mutations of PLCG1, PRKCQ, PREX2, and CARD11 is likely to enhance cell survival if accompanied by appropriate costimulatory signals and resistance to TNFRSF-mediated apoptosis.68-72
Although we did not detect a high frequency of SNVs affecting individual JAK STAT genes, the presence of activating JAK1/3, STAT3, and STAT5A/B mutations and copy number gains of 17q including STAT3 and STAT5 could explain constitutive STAT3 activation in some cases of SS.63,65,73-75 STAT5B mutations have recently been described in γδ T-cell lymphomas10 and JAK/STAT mutations have now been documented in other extranodal and nodal T-cell lymphomas including SS.8,42,44,76 Recent studies have also shown that constitutive STAT3 expression in ALCL can be the result of gene fusions.77 Previously we reported constitutive STAT3 protein expression in 10 patients with SS, all of whom were included in this study.76 Overall, 6 of these cases showed copy number gains but no SNVs of JAKs, STAT3, or SOCS7, suggesting that other upstream events can lead to aberrant STAT3 signaling in SS. There is growing interest in the role of GPCRs in malignancy, and one, SIPR1, is involved in noncanonical activation of JAK STAT signaling in lymphoid cells via PI3K signaling.78 Whether other GPCRs are implicated in JAK STAT signaling is unclear, but SNVs affecting 2 GPCRs (98, 158) were detected in 30% of tumors. One variant (GPR158R757C) has been reported in COSMIC, and recent studies in ATLL also detected a high prevalence of GPCR aberrations including SNVs affecting GPR183.13
Overall, 40% of tumors had either SNVs and/or CNVs affecting genes involved in epigenetic regulation including ASLX3, TET1, TET2, and DNMT3A, which have been described in myeloid malignancies and lymphomas.79,80 These include inactivating mutations of TET1/2, ASLX3, and DNMT3A, which have a role in DNA methylation, and IDH2 mutations affecting histone methylation in AITL.81 Both histone acetylation and methylation are known to be critical for T-cell differentiation and memory. Loss of epigenetic regulation in SS is reflected by promoter hypermethylation of multiple genes82 and clinical responses to HDAC inhibitors such as Romidepsin.83 In addition, recent studies in both CTCL and PTCL have shown mutations of ARID1A/B involved in chromatin remodeling.3,84 These findings suggest that chromatin modification plays a key role in malignant transformation of mature T cells as recently described for B-cell non-Hodgkin lymphoma.85
Analysis of our data sets revealed that 42% of the C>T variants occurred at NpCpG sites, which could be consistent with at least 5 of 21 recently described signatures including age-related deamination of methylated cytosines.21,86,87 Although UV-specific TP53 mutations (CC>TT transversions at pyrimidine sites) have been described previously in MF,61 we only detected very rare CC-TT transversions in SS. MF is considered to be derived from skin resident memory T cells,88 which may be exposed to environmental UV, and MF patients are often treated with phototherapy. In contrast, SS is thought to derive from central memory T cells. Further studies of larger data sets are required to define the mutational signatures associated with SS and other CTCL variants including MF.
In conclusion, our findings illustrate that the genomic landscape of SS is markedly heterogeneous. We suggest that the high prevalence of perturbations in genes maintaining genome integrity is a likely cause of the loss of genome stability in SS. Furthermore, there is selection for gene mutations/structural variation contributing to deregulation of key pathways regulating T-cell homeostasis, cell survival, and global epigenetic processes. These findings provide the basis for detailed functional analyses to define novel therapeutic targets for CTCL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This research was supported by grants from The British Skin Foundation, Skin Tumour Unit Fund 461, Guy’s and St Thomas’ Charity, and the National Institute for Health Research (NIHR) Biomedical Research Centre based at Guy's and St Thomas' NHS Foundation Trust and King's College London.
The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health. C.F., R.M.B., and R.T. were supported by the King’s Bioscience Institute and the Guy’s and St Thomas' Charity Prize Programme in Biomedical and Translational Science.
Authorship
Contribution: W.J.W., V.M.P., R.M.B., A.B., N.B., F.B., K.D., J.F., S.A.D., C.C.C., A.A.G., R.M.H., N.M., E.A.N., A.P., C.A.R., E.G.S., R.T., A.Y., C.Z.B., S.F., and I.T. performed experiments and data analysis; W.J.W., V.P., A.L., M.A.S., and E.d.R. performed bioinformatic analysis; W.J.W., V.P., A.L., F.B., and E.d.R. produced figures and tables; W.J.W., V.P., A.L., E.d.R., T.J.M., and S.J.W. wrote the manuscript; and T.J.M. and S.J.W. led the project.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tracey J. Mitchell, St John’s Institute of Dermatology, Division of Genetics and Molecular Medicine, Tower Wing, King’s College London, London SE1 9RT, United Kingdom; e-mail: tracey.mitchell@kcl.ac.uk.
References
Author notes
T.J.M. and S.J.W. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal