Key Points
Epigenetic silencing of miR-124a leads to constitutive STAT3 and activation of downstream Pim1.
Pim1 kinase signaling is constitutively activated in ATL cells and represents a novel therapeutic target.
Abstract
Human T-cell leukemia virus type 1 (HTLV-1)–associated adult T-cell leukemia and T-cell lymphoma (ATL) are aggressive diseases with poor prognoses, limited therapeutic options, and no curative treatment. In this study, we used a mouse model of ATL and restored expression of the microRNA, miR-124a, to identify in vivo downstream effectors responsible for its tumor-suppressive functions in ATL cells. Our results revealed that STAT3, a direct target of miR-124a, is constitutively activated in HTLV-I–transformed cells and ATL cells, and activating STAT3 mutations were detected in 25.5% of primary ATL patients. Interestingly, we found that the STAT3 downstream kinase effector, Pim1, is constitutively activated in ATL cells. The dependence of ATL cells to Pim1 activity was demonstrated using 2 Pim1 small inhibitors, SMI-4a and AZD1208. These studies indicated that HTLV-I–transformed and ATL cells, but not normal peripheral blood mononuclear cells, are highly sensitive to AZD1208, and the inhibition of Pim1 signaling triggers an apoptotic signal in leukemic cells. Finally, preclinical testing of AZD1208 in a mouse model of ATL resulted in significant prevention of tumor growth in vivo. In conclusion, our studies suggest that constitutive activation of the STAT3-Pim1 pathway represents a novel therapeutic target for the treatment of ATL.
Introduction
Human T-cell leukemia virus type 1 (HTLV-I) is the etiologic agent of adult T-cell leukemia/lymphoma (ATL),1,2 a disease classified into 4 subtypes referred to as smoldering, chronic, acute, or lymphoma type.3 Although our understanding of HTLV-I molecular pathogenesis has made significant progress, this has failed to translate into effective therapeutic options4 and the 4-year disease survival rates for acute and lymphoma ATL are only 11% and 16%, respectively.5 A recent report demonstrates that the survival times for smoldering ATL have actually worsened over time,6 stressing the importance of novel approaches to treating ATL.
Spontaneous proliferation of ATL cells from chronic or smoldering patients in vitro is dependent upon cytokines autocrine/paracrine loops.7,8 Progression to the acute type is associated with ligand-independent growth and the constitutive activation of the Janus-activated kinases/signal transducer and activator of transcription (JAK/STAT) pathway.9 The importance of JAK/STAT signaling in leukemia has been documented10 and the importance of the IL-2R common γ-chain as potential therapeutic approach for ATL has been reported.11 Although constitutive JAK3 activation is required for the proliferation and survival of ATL cells,12 current JAK3 inhibitors have serious overimmunosuppression side effects and, although they block STAT5 activation in most T-cell subpopulations, JAK3 inhibitors are less effective in T-regulator cells (Tregs), a major target and reservoir for HTLV-I in vivo.13
In this study, we show that miR-124a–mediated loss of STAT3 significantly reduced ATL tumor cell proliferation in vivo. The use of S3I-201, a specific STAT3 inhibitor,14 demonstrated antiproliferative and apoptotic effects in ATL cells. Moreover, we found activating mutations in 25.5% of ATL patients. STAT3 expression strongly correlated with Pim1 expression in primary ATL patients, suggesting that Pim1 plays an important role in ATL pathogenesis. Consistent with this notion, constitutive activation of Pim1 and its downstream targets were detected in ATL cells, which were strictly dependent upon Pim1 signaling as treatment with the Pim1 inhibitors SMI-4a or AZD1208, and potently inhibited growth and induced apoptosis. Finally, AZD1208 consistently prevented tumor growth in a mouse model of 2ATL, suggesting that Pim1 activation represents a novel attractive therapeutic target for the treatment of ATL.
Materials and methods
miR-124a stable cell line production
The pre–miR-124a was inserted in place of the TurboRFP marker in the pTRIPZ inducible lentiviral vector (Thermo Scientific). Stable lines were made with virus using the vesicular stomatitis virus-glycoprotein and pDLN6 packaging system after puromycin selection.
Mutagenesis and luciferase assays
293T were transfected using Polyfect (QIAGEN). The STAT3 3′UTR was cloned into a modified pGL3-Promoter luciferase vector (Promega). pre–miR-124a was cloned into the pCDNA3.1 vector and mutated using Site-Directed Mutagenesis Kit (Stratagene).
Cell lines and treatments
The HTLV-I–transformed cell lines (MT4, C8166, HUT102, and MT2), ATL-like cell lines (ED-40515(–), Tl-Om1, MT1, ATL-T, and ATL25), and ALL cell lines (Nalm-20, Nalm-6, Tom-1, Tanoue, Molt4, PEER, RCH-ACV, and KE-37) were grown in RPMI 1640 with 10% fetal bovine serum. The HTLV-I–immortalized cell lines (LAF and 1185), and the ATL-like cell lines ATL43T, ATL55T, KOB, KK1, and LM-Y1 were grown in media with 20% serum and 50 U/mL IL-2. 293T (ATCC) were grown in Dulbecco’s modified Eagle medium with 10% fetal bovine serum. Cell lines were treated with 5′-Azacytidine (Sigma-Aldrich), VI-S3I-201 (Calbiochem), and AZD1208 and SMI-4a (Selleckchem).
Patient samples
ATL patients have been used in previous studies, and all samples were obtained after informed consent, and in agreement with the regulations for the protection of human subjects and after internal institutional review board approval.15,16 Patients’ characteristics are provided in supplemental Tables 2 and 3 (available on the Blood Web site). Control samples consisted of peripheral blood mononuclear cells (PBMCs) from healthy, noninfected (HTLV-I–negative) individuals.
RNA expression
RNA was extracted with TRIzol (Invitrogen), DNase I–treated, reverse-transcribed with RNA-to-cDNA synthesis kit (Applied Biosystems), and used in assays with a StepOnePlus Real-time PCR System (Applied Biosystems), Primers are provided in supplemental Table 1. Mature miRNA expression was detected using the miScript PCR system (QIAGEN). RT2 Profiler PCR array-Human Cancer Pathway Finder was used for gene expression (SABiosciences).
Protein expression
Cell lysates were probed with the following antibodies: STAT3(sc-482), STAT5(sc-835), Actin(sc-1615), Cyclin A(sc-751), Cyclin B1(sc-752), Cyclin E(sc-198), Cyclin D1(sc-718), Mcl-1(sc-819), Bcl-xL(sc-7195), Survivin(sc-8807), p21(sc-397), SOCS3(sc-9023), Caspase3(sc-7148) (Santa Cruz), Pim1(ab66767), α-Tubulin(ab7291) (Abcam), p-p27-Thr157(AF1555) (R&D Systems), p-STAT3-Tyr705(9138), p-Bcl2-Ser70(BL134), p-4E-BP1-Thr37/46(2855), p-70S6 Kinase-Thr389(9234) (Cell Signaling), c-myc (11667149001) (Roche), p-STAT5a/b-Tyr694/Tyr699(05-495) (Upstate), and pBad-Ser112(34022) (Owl Laboratories).
Animal studies
All animal studies were performed by Advance Bioscience Laboratories Inc. (ABL) (Rockville, MD). Protocols were reviewed and approved by ABL institutional review board ACUC#328. To study the role of miR-124 in ATL tumor growth, 6-to 8-week-old, female NOG (NOD/Shi-scid/IL-2Rγnull) mice were used. Before injection of ATL cells, mice were given doxycycline 2 mg/mL (Vibramycin Monohydrate) in their drinking water with 2% sucrose for 7 days. The water was thereafter changed every day. Each animal received 2.5 × 107 ATL cells control (ED-40515(–)-TripZ) and miR124 modified (ED-40515(–)-miR-124) in the left and the right flanks, respectively. For the Pim kinase inhibitor AZD1208 study, mice were injected with ED-40515(–) cells and given vehicle control or AZD1208 (30 mg/kg) by oral gavage once daily.
Bisulfite treatment and methylation analysis
Genomic DNA was treated with bisulfite using the MethylCode Bisulfite Conversion Kit (Invitrogen) and used for methylation-specific polymerase chain reaction (MSPCR) and bisulfite genomic sequencing (BGS) reactions. For BGS, amplified products were cloned into the pGEM-T Easy Kit (Promega), where a least 5 colonies were sequenced,
Immunohistochemistry
Mouse specimens were cut at 5 microns and fixed with formalin, and immunohistochemistry was performed using an EXPOSE Rabbit-specific HRP/DAB Detection IHC Kit (Abcam), with STAT3 (Spring Bioscience) and Pim1 (Abcam) antibodies, counterstaining with Mayer’s hematoxylin (Lillie’s modification) and Bluing reagent (ScyTek).
Cell proliferation and flow cytometry
Cell viability and proliferation were measured by cell counts, Cell Proliferation Kit II (XTT) (Roche), BrdU Cell Proliferation Assay (Millipore), propidium iodine staining, CFDA SE Cell Tracer kit (Invitrogen), or Vybrant Apoptosis Assay Kit #2 (Invitrogen), according to manufacturer’s instructions, and analyzed on a LSR II flow cytometer.
Results
MicroRNA miR-124a inhibits cellular proliferation in ATL cells in vitro and tumor growth in a mouse model of adult T-cell leukemia
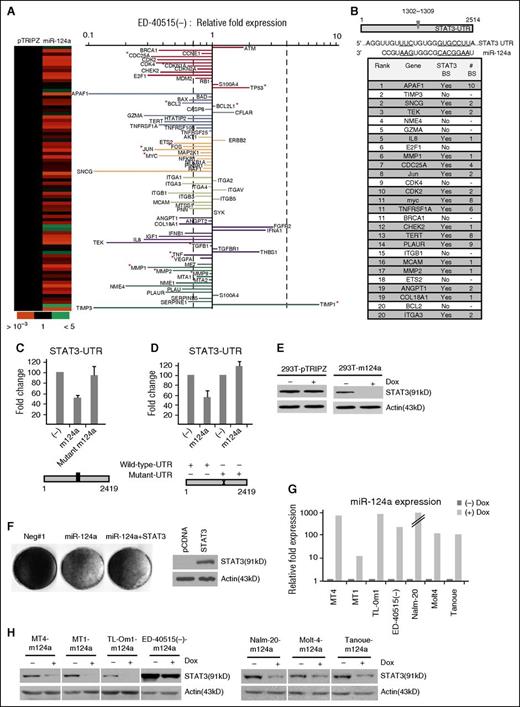
Profiling of miRNA expression revealed several miRNAs deregulated between primary ATL and PBMCs.17 Among the top downregulated miRNAs, we found low levels of mature miR-124a in uncultured ATL patient samples (Figure 1A). Substantial decreases in pre–miR-124a expression were observed in in vitro ATL- and ALL-derived cells (supplemental Figure 1A-B). To investigate the role of miR-124a, we generated miR-124a Tet-On inducible stable cell lines in ATL cell, ED-40515(–) and Tl-Om1, and in the ALL-derived cell, Nalm-20. Adding doxycycline restored miR-124a expression (Figure 1B and supplemental Figure 1C) and was associated with reductions in cellular proliferation (Figure 1C and supplemental Figure 1D-G) and reduced expression of cyclin A, B1, and E (Figure 1D-E and supplemental Figure 1E-F). We also observed a decreased expression of survivin, Bcl-2, and BclxL, and an increase in Bax expression (Figure 1D-F). We next sought to determine the source of miR-124a downregulation in ATL cells. Because amplification of the genomic DNA encompassing the miR-124a locus found no alteration (supplemental Figure 2A), we performed MSPCR and BGS of miR-124a CpG islands. These results indicated that the miR-124a locus is highly methylated in ATL cells (supplemental Figure 2B-E). Consistent with these data, treatment with the demethylation agent 5-azacytidine restored expression of pre–miR-124a (supplemental Figure 2F). To determine the physiologic relevance of miR-124a, we investigated whether expression of miR-124a plays a role in ATL tumor cell growth or survival in a mouse model of ATL. ED-40515(–) miR-124a or pTRIPZ Tet-On cells were injected in NOG mice. miR-124a was induced for 3 weeks and animals were sacrificed for tumor growth examination (Figure 1G). miR-124a expression was confirmed by reverse-transcription PCR from excised tumors (Figure 1H). These studies showed that in ATL cells, expression of miR-124a was associated with a significant reduction in tumor formation and growth in vivo (Figure 1I-J) and represent a potent tumor suppressor gene in ATL cells.
Overexpression of miR-124a in ATL cells inhibits tumor growth in vivo. (A) Mature miRNA detection of miR-124a in ATL patient samples (n = 17) vs healthy noninfected donor. miR-24 served as an internal control. (B-C) Induction of miR-124a in ED-40515(–) cells inhibits cell proliferation. Pre–miR-124a expression was detected by real-time PCR (B) after doxycycline (Dox) addition. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as an internal control. Cells were counted every 48 hours after receiving 2 μg/mL Dox (every 48 h) (C). Results represent at least 2 independent experiments. (D) Loss of proliferation/cell cycle markers (cyclins A, B1, and E, and survivin) in miR-124a–expressing ATL cells, 6 days after receiving 2 μg/mL Dox every other day. Tubulin was used as a loading control. (E) ED-40515(–) cells expressing miR-124a enter cell cycle arrest after induction with 2 μg/mL Dox every day for 5 days. (F) Cell survival markers (Bax, Bcl-xl, and Bcl-2) are altered in ED-40515(–)–miR-124a expressing cells 5 days after receiving 2 μg/mL Dox every day. Tubulin served as a loading control. (G) ED-40515(–)–pTRIPZ (n = 12) or miR-124a (n = 12) TET-On cells were injected into the right or left flank of NOG mice. Pictures are representative of 2 mice simultaneously receiving pTRIPZ (left) or miR-124a (right) TET-On ED-40515(–) cells. (H) Real-time PCR detection of pre–miR-124a in a tumor model of ATL. GAPDH served as a control. (I) In vivo growth of NOG mice engrafted with ED-40515(–) pTRIPZ (left) or miR-124a (right) cells (plotted as the average tumor volume (mm3) (calculated as the [width2 × length]/2) per every 7 days). (J) The percentage of tumors with volumes of 200 m3 or larger was plotted against the days after implantation into mice. Volumes were calculated every 7 days, up to 21 days. Statistics are calculated using the N-1 2-proportion test, with the number of tumors in each group (n = 12) 200 m3 or larger, considered positive.
Overexpression of miR-124a in ATL cells inhibits tumor growth in vivo. (A) Mature miRNA detection of miR-124a in ATL patient samples (n = 17) vs healthy noninfected donor. miR-24 served as an internal control. (B-C) Induction of miR-124a in ED-40515(–) cells inhibits cell proliferation. Pre–miR-124a expression was detected by real-time PCR (B) after doxycycline (Dox) addition. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as an internal control. Cells were counted every 48 hours after receiving 2 μg/mL Dox (every 48 h) (C). Results represent at least 2 independent experiments. (D) Loss of proliferation/cell cycle markers (cyclins A, B1, and E, and survivin) in miR-124a–expressing ATL cells, 6 days after receiving 2 μg/mL Dox every other day. Tubulin was used as a loading control. (E) ED-40515(–) cells expressing miR-124a enter cell cycle arrest after induction with 2 μg/mL Dox every day for 5 days. (F) Cell survival markers (Bax, Bcl-xl, and Bcl-2) are altered in ED-40515(–)–miR-124a expressing cells 5 days after receiving 2 μg/mL Dox every day. Tubulin served as a loading control. (G) ED-40515(–)–pTRIPZ (n = 12) or miR-124a (n = 12) TET-On cells were injected into the right or left flank of NOG mice. Pictures are representative of 2 mice simultaneously receiving pTRIPZ (left) or miR-124a (right) TET-On ED-40515(–) cells. (H) Real-time PCR detection of pre–miR-124a in a tumor model of ATL. GAPDH served as a control. (I) In vivo growth of NOG mice engrafted with ED-40515(–) pTRIPZ (left) or miR-124a (right) cells (plotted as the average tumor volume (mm3) (calculated as the [width2 × length]/2) per every 7 days). (J) The percentage of tumors with volumes of 200 m3 or larger was plotted against the days after implantation into mice. Volumes were calculated every 7 days, up to 21 days. Statistics are calculated using the N-1 2-proportion test, with the number of tumors in each group (n = 12) 200 m3 or larger, considered positive.
Gene expression array reveals miR-124a as a master regulator of STAT3-activated proliferation, survival, and metastatic genes in ATL cells
We next sought to identify target genes of miR-124a. ED-40515(–) miR-124a Tet-On and TripZ control cells were screened using the RT2 profiler PCR array Human cancer pathway finder (Figure 2A). Our results suggested a majority of genes were suppressed by miR-124a expression, including cell cycle regulators CDC25A, CDKN1A, CDK2, CDK4, and E2F1; apoptosis APAF1 and Bcl2; transcription factors Ets2, Fos, Jun, and Myc; signaling molecules IL-8, tumor necrosis factor, and TEK; and cell migration factors such as MMP1, MMP2, and TIMP3 (Figure 2A). In contrast, a few genes were upregulated by miR-124a and included ATM, p53, TIMP1 and IFNA1 (Figure 2A). In silico promoter analyses revealed STAT3 as a common regulator of the most highly downregulated genes (Figure 2B), suggesting that STAT3 may be a key mediator of miR-124a’s effect in ATL cells. Consistent with a previous study,18 we found that miR-124a directly targets STAT3 in luciferase assays using the full-length 3′-UTR of STAT3 (Figure 2C). In contrast, a mutated miR-124a or STAT3-UTR abolished the repressive effect (Figure 2C-D). We then generated a miR-124a Tet-inducible 293T line, which caused loss of endogenous STAT3 protein (Figure 2E). Similar to ATL cells, induction of miR-124a caused a severe reduction in 293T cell proliferation (data not shown), which was partially restored by overexpressing a STAT3 cDNA expression vector before miR-124a induction (Figure 2F). This suggests that some of the antiproliferative effects of miR-124a are related to inhibition of STAT3, and that additional miR-124a targets may have an effect that reduces proliferation in cancer cells. Because miRNA functions are cell line– and context-dependent, we made several TET-miR-124a–inducible HTLV-I and ALL cell lines (Figure 2G). All TET-miR-124a leukemic cell lines demonstrated a significant reduction of endogenous STAT3 protein levels upon induction of miR-124a (Figure 2I). This is important, because a role for miR-124a-STAT3 has never been investigated in HTLV-I–transformed cells.
miR-124a alters cancer-related genes while specifically targeting STAT3. (A) Heat map (left) and fold-expression (right) of genes altered by miR-124a–induced 72 hours. Fold change is calculated compared with pTRIPZ–induced ED-40515(–) cells. Bold, dotted lines mark a onefold loss or gain in fold expression. Genes with “*” are known to be altered by STAT3. (B) Illustration of the miR-124a–binding sites in the 3′-UTR of STAT3 (top). In silico analysis of the top 20 altered genes in ED-40515(–) TET-On miR-124a cells (shaded areas are possible STAT3 binding sites). (C-D). pCDNA, miR-124a/pCDNA, or mutant miR-124a/pCDNA (C-mutated miR-124a sequence) were transfected into 293T cells along with wild-type or mutant (D) STAT3-UTR-pGL3 and the RL-TK plasmid. Forty-eight hours after transfection, cell lysates were measured for firefly (STAT3 3′UTR) and renilla (RL-TK, internal control) activity. All luciferase assays was performed at least twice. (E) Detection of STAT3 in stable 293T-pTRIPZ or –miR-124a cells induced 72 hours with 2 μg/mL Dox. (F) 293T-pCDNA and –wild-type-STAT3 cells were established under puromycin selection. Cells were then transfected with either 50 nM miR-124a oligo or a control oligo (Negt#1). One week after transfection, cells were stained for cell growth (left). Results represent 1 of 2 experiments performed. The overexpression of STAT3 was confirmed (right). (G-H) Stable miR-124a lines were established in MT4, MT1, Tl-Om1, ED-40515(–), and Nalm-20, Molt4, Tanoue cell lines. miR-124a expression was confirmed after 72 hours’ induction with 2 μg/mL Dox (G), along with loss of STAT3 (H).
miR-124a alters cancer-related genes while specifically targeting STAT3. (A) Heat map (left) and fold-expression (right) of genes altered by miR-124a–induced 72 hours. Fold change is calculated compared with pTRIPZ–induced ED-40515(–) cells. Bold, dotted lines mark a onefold loss or gain in fold expression. Genes with “*” are known to be altered by STAT3. (B) Illustration of the miR-124a–binding sites in the 3′-UTR of STAT3 (top). In silico analysis of the top 20 altered genes in ED-40515(–) TET-On miR-124a cells (shaded areas are possible STAT3 binding sites). (C-D). pCDNA, miR-124a/pCDNA, or mutant miR-124a/pCDNA (C-mutated miR-124a sequence) were transfected into 293T cells along with wild-type or mutant (D) STAT3-UTR-pGL3 and the RL-TK plasmid. Forty-eight hours after transfection, cell lysates were measured for firefly (STAT3 3′UTR) and renilla (RL-TK, internal control) activity. All luciferase assays was performed at least twice. (E) Detection of STAT3 in stable 293T-pTRIPZ or –miR-124a cells induced 72 hours with 2 μg/mL Dox. (F) 293T-pCDNA and –wild-type-STAT3 cells were established under puromycin selection. Cells were then transfected with either 50 nM miR-124a oligo or a control oligo (Negt#1). One week after transfection, cells were stained for cell growth (left). Results represent 1 of 2 experiments performed. The overexpression of STAT3 was confirmed (right). (G-H) Stable miR-124a lines were established in MT4, MT1, Tl-Om1, ED-40515(–), and Nalm-20, Molt4, Tanoue cell lines. miR-124a expression was confirmed after 72 hours’ induction with 2 μg/mL Dox (G), along with loss of STAT3 (H).
Constitutive activation of STAT3 is required for the proliferation and survival of human leukemic cells
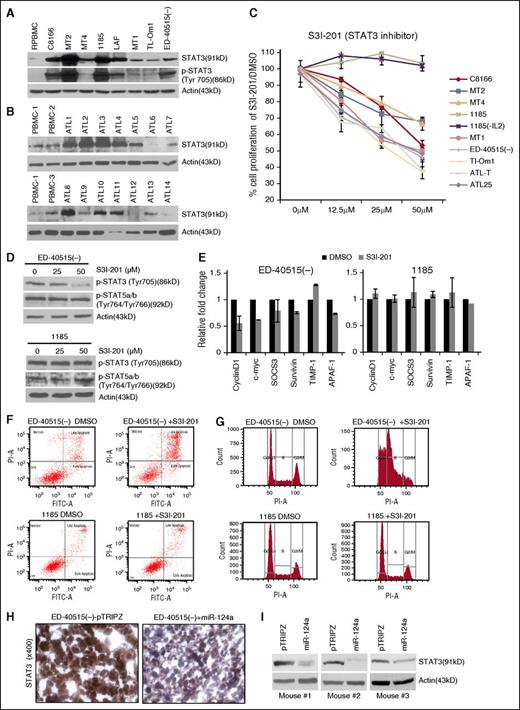
Constitutive activation of the JAK/STAT pathway is a hallmark of HTLV-I–transformed and ATL cells,9 but, to date, most studies have focused on inhibition of JAKs and STAT5 signaling. We found increased expression of STAT3 and pTyr705 STAT3 in HTLV-I–transformed cells (Figure 3A) and upregulated expression of STAT3 in 8 of 14 freshly isolated uncultured ATL samples, which was consistent with previous reports19 (Figure 3B). To determine the biological significance of STAT3, we used the STAT3 small inhibitor S31-201 (NSC74859).14 Incubation with S31-201 caused a significant reduction in proliferation of all HTLV-I and ATL cells (Figure 3C). For reasons unknown at this time, 1185 cells were resistant (Figure 3C). Resistance was independent from IL-2 signaling because 1185 cells remained resistant to S31-201, even after IL-2 withdrawal from media (Figure 3C). Consistent with these results, treatment of ED-40515(–) but not 1185 cells with S31-201 was associated with decreased levels of pTyr703 STAT3 (Figure 3D). We then confirmed that in the presence of S31-201, phosphorylated STAT5 levels were unaffected in ATL cells (Figure 3D). Furthermore, STAT3-dependent target genes identified in the miR-124a-cancer array, were transcriptionally deregulated in ED-40515(–) cells, which underwent cell cycle arrest and apoptosis after treatment with S31-201, whereas these effects were not observed in 1185 cells (Figure 3E-G). Overall, these results suggest that constitutive STAT3 signaling is required for ATL cell growth and survival, and although STAT3 may represent a therapeutic target for ATL, STAT3 inhibitors had significant toxicity toward normal PBMCs (not shown). These observations prompted us to seek a downstream target of STAT3 to achieve better specificity toward cancer cells.
HTLV-I/ATL cells over-express STAT3, which is required for cell survival. (A-B) Increased expression of STAT3 and p-STAT3 (Tyr705) in HTLV-I and ATL cell lines and increased STAT3 expression in primary ATL patient samples (n = 14). Resting noninfected PBMCs (R.PBMCs, n = 3) served as controls. Actin served as a loading control. (C) The STAT3 inhibitor (S3I-201) decreases cellular proliferation in HTLV-I/ATL-lines. Cells were treated with 0, 12.5, 25, or 50 μM S3I-201 or dimethyl sulfoxide (DMSO) for 72 hours. The average growth curve is representative of the percentage of proliferation between S3I-201 and DMSO-treated cells. Each cell line was treated at least twice for standard deviation. 1185 (-IL2) cells were washed in phosphate-buffered saline and resuspended in media without IL2, followed by treatment with S3I-201 for 72 hours. (D) p-STAT3 (Tyr705) or p-STAT5a/b (Tyr764/Tyr766) in ED-40515(–) or 1185 cells treated with 0, 25, or 50 μM S3I-201 for 24 hours. (E) ED-40515(–) or 1185 cells were treated with 50 μM S3I-201 or DMSO for 24 hours. Expression of CyclinD1, c-myc, SOCS3, survivin, TIMP-1, and APAF-1 were analyzed by real-time quantitative PCR. Standard deviation was calculated from at least 2 independent experiments, with GAPDH as a control. (F-G) ED-40515(–) or 1185 cells were treated with 50 μM S3I-201 for 72 hours. Annexin V/PI (F) or PI staining for cell cycle (G) was analyzed. (H-I). Mouse tumor tissue was immunohistochemistry-stained (H) or in vivo lysates used for STAT3 protein expression (I) from 3 ED-40515(–) TET-On tumors. Images were taken at room temperature on a Nikon Eclipse 80i microscope (Nikon Instruments, Inc., Melville, NY) and a Nikon DSFI1 camera, with a 40× objective lens.
HTLV-I/ATL cells over-express STAT3, which is required for cell survival. (A-B) Increased expression of STAT3 and p-STAT3 (Tyr705) in HTLV-I and ATL cell lines and increased STAT3 expression in primary ATL patient samples (n = 14). Resting noninfected PBMCs (R.PBMCs, n = 3) served as controls. Actin served as a loading control. (C) The STAT3 inhibitor (S3I-201) decreases cellular proliferation in HTLV-I/ATL-lines. Cells were treated with 0, 12.5, 25, or 50 μM S3I-201 or dimethyl sulfoxide (DMSO) for 72 hours. The average growth curve is representative of the percentage of proliferation between S3I-201 and DMSO-treated cells. Each cell line was treated at least twice for standard deviation. 1185 (-IL2) cells were washed in phosphate-buffered saline and resuspended in media without IL2, followed by treatment with S3I-201 for 72 hours. (D) p-STAT3 (Tyr705) or p-STAT5a/b (Tyr764/Tyr766) in ED-40515(–) or 1185 cells treated with 0, 25, or 50 μM S3I-201 for 24 hours. (E) ED-40515(–) or 1185 cells were treated with 50 μM S3I-201 or DMSO for 24 hours. Expression of CyclinD1, c-myc, SOCS3, survivin, TIMP-1, and APAF-1 were analyzed by real-time quantitative PCR. Standard deviation was calculated from at least 2 independent experiments, with GAPDH as a control. (F-G) ED-40515(–) or 1185 cells were treated with 50 μM S3I-201 for 72 hours. Annexin V/PI (F) or PI staining for cell cycle (G) was analyzed. (H-I). Mouse tumor tissue was immunohistochemistry-stained (H) or in vivo lysates used for STAT3 protein expression (I) from 3 ED-40515(–) TET-On tumors. Images were taken at room temperature on a Nikon Eclipse 80i microscope (Nikon Instruments, Inc., Melville, NY) and a Nikon DSFI1 camera, with a 40× objective lens.
Pim1 kinase is a central mediator of STAT3 signaling in leukemic cells and is overexpressed in ATL cells in vivo
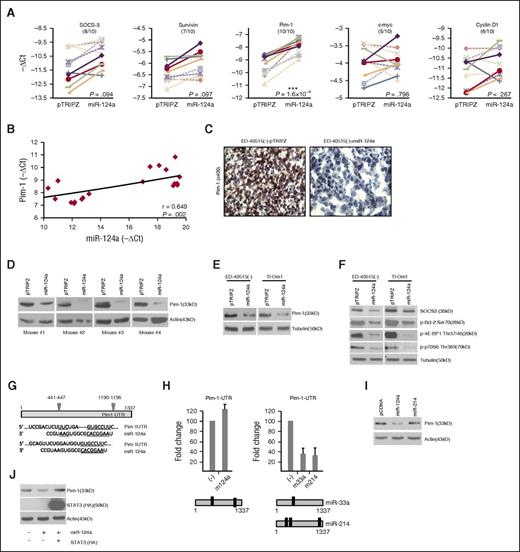
Analyses confirmed a significant loss of STAT3 protein expression in tumors excised from mice engrafted with ED-40515(–) Tet-miR-124a compared with pTRIPZ (Figure 3H-I). We then sought to identify downstream effectors of STAT3 responsible for reducing tumor growth. Expression of STAT3 target genes involved in proliferation and survival were analyzed in ED-40515(–) Tet-miR-124a and pTRIPZ tumor samples (Figure 4A and supplemental Figure 3A). A loss in the gene expression of SOCS-3 and survivin was found in 8 of 10 and 7 of 10 samples, respectively, with no significant decrease in cyclin D1 and c-myc (Figure 4A and supplemental Figure 3B). Surprisingly, we found that the gene encoding Pim1 had the greatest loss of expression in all miR-124a tumor samples (Figure 4A) and a slight correlation between Pim1 and miR-124a expression was observed (Figure 4B). This is interesting because a role for Pim1 kinase has not been previously linked to HTLV-I or ATL-transformed cells, although it has been implicated in other leukemias.20 We confirmed loss of Pim1 protein expression in tumors from ED-40515(–) TET-mIR-124a mice (Figure 4C-D) and the suppressive effects of miR-124a on Pim1 in vitro, TET-miR-124a ATL lines ED-40515(–), and Tl-Om1 (Figure 4E). Pim1 loss was accompanied by decreased phosphorylation of substrates, Bcl-2, 4E-BP1, and p70S6 Kinase, and decreased expression of SOCS3, a target of Pim1 regulation (Figure 4F). Importantly, Pim2 and Pim3 expression were not significantly affected in in vitro TET-miR-124a–expressing leukemic lines and ED-40515(–) TET-miR-124a tumor samples (supplemental Figure 3C-D). Together our results suggested that in ATL cells, Pim1 is a major target of the miR-124a/STAT3 signaling axis.
The STAT3 targeted gene, Pim1, is significantly lost in ATL tumors established from mice. (A) Real-time PCR analysis in ED-40515(–) pTRIPZ or miR-124a TET-On tumors. Each line represents the expression of the target gene (-ΔCt) between pTRIPZ and miR-124a in the corresponding mice. P values are calculated using a 2-tailed Student t test between the -ΔCt values for pTRIPZ vs miR-124a (n = 10). (B) Correlation graph plotting Pim1 expression (-ΔCt; y-axis) vs miR-124a expression (-ΔCt; x-axis) in ED-40515(–) tumors. The Spearman correlation coefficient is indicated along with corresponding P value. (C-D) Pim1 expression is decreased by immunohistochemistry staining (C) or western blot (D) in ED-40515(–) pTRIPZ or miR-124a TET-On tumors. Images were taken at room temperature on a Nikon Eclipse 80i microscope (Nikon Instruments, Inc.; Melville, NY) and a Nikon DSFI1 camera, with a 40× objective lens. Pim1 (E) and Pim1 target genes SOCS3, p-Bcl2 (Ser70), p-4EBP1 (Thr37/46), and p-p70S6K (Thr389) (F) were analyzed by western blot in miR-124a TET-On, ED-40515(–), and Tl-Om1 induced with 2 µg/mL Dox for 72 hours. (G) Illustration of the miR-124a binding sites in the 3′-UTR of Pim1. Pim1 UTR luciferase (H) or Pim1 western blot (I) was performed in 293T cells. pCDNA and miR-124a/pCDNA (left) or pCDNA and miR-33a/pCDNA or miR-214/pCDNA (right) were transfected into 293T cells, along with the RL-TK plasmid. Forty-eight hours after transfection, cell lysates were measured for firefly (Pim1 UTR) and renilla (RL-TK, internal control) activity. All luciferase was performed at least twice. (J) Exogenous STAT3 expression rescues miR-124a–mediated inhibition of Pim1. pCDNA (control) or STAT3-HA–tagged lines were established under puromycin selection in 293T cells, transefected with miR-124a or negative oligos, and analyzed several days later for Pim1 expression. Stable STAT3 expression was detected by HA antibodies.
The STAT3 targeted gene, Pim1, is significantly lost in ATL tumors established from mice. (A) Real-time PCR analysis in ED-40515(–) pTRIPZ or miR-124a TET-On tumors. Each line represents the expression of the target gene (-ΔCt) between pTRIPZ and miR-124a in the corresponding mice. P values are calculated using a 2-tailed Student t test between the -ΔCt values for pTRIPZ vs miR-124a (n = 10). (B) Correlation graph plotting Pim1 expression (-ΔCt; y-axis) vs miR-124a expression (-ΔCt; x-axis) in ED-40515(–) tumors. The Spearman correlation coefficient is indicated along with corresponding P value. (C-D) Pim1 expression is decreased by immunohistochemistry staining (C) or western blot (D) in ED-40515(–) pTRIPZ or miR-124a TET-On tumors. Images were taken at room temperature on a Nikon Eclipse 80i microscope (Nikon Instruments, Inc.; Melville, NY) and a Nikon DSFI1 camera, with a 40× objective lens. Pim1 (E) and Pim1 target genes SOCS3, p-Bcl2 (Ser70), p-4EBP1 (Thr37/46), and p-p70S6K (Thr389) (F) were analyzed by western blot in miR-124a TET-On, ED-40515(–), and Tl-Om1 induced with 2 µg/mL Dox for 72 hours. (G) Illustration of the miR-124a binding sites in the 3′-UTR of Pim1. Pim1 UTR luciferase (H) or Pim1 western blot (I) was performed in 293T cells. pCDNA and miR-124a/pCDNA (left) or pCDNA and miR-33a/pCDNA or miR-214/pCDNA (right) were transfected into 293T cells, along with the RL-TK plasmid. Forty-eight hours after transfection, cell lysates were measured for firefly (Pim1 UTR) and renilla (RL-TK, internal control) activity. All luciferase was performed at least twice. (J) Exogenous STAT3 expression rescues miR-124a–mediated inhibition of Pim1. pCDNA (control) or STAT3-HA–tagged lines were established under puromycin selection in 293T cells, transefected with miR-124a or negative oligos, and analyzed several days later for Pim1 expression. Stable STAT3 expression was detected by HA antibodies.
Because miR-124a has 2 putative binding sites in the 3′UTR of Pim1, we tested whether miR-124a could directly target Pim1 independently of STAT3 (Figure 4G). Luciferase assays with the Pim1 3′-UTR found no decrease by miR-124a (Figure 4H). microRNAs miR-33a (a known Pim1 target)21 and miR-214 (a putative high-seed match to the Pim1 3′UTR), both targeted Pim1 (Figure 4H). Although the effects of miR-33a were previously reported, this is the first report demonstrating that miR-214 can directly target the Pim1 3′UTR. However, miR-214 did not appreciably lower Pim1 expression to the levels demonstrated by miR-124a (Figure 4I). Therefore our results suggest that miR-124a is unable to target Pim1 directly and the effects reported in this study are indirect. Furthermore, transfection of cells with miR-124a in the absence or presence of STAT3 demonstrated that decreased Pim1 expression was no longer observed when STAT3 was provided in trans, suggesting that miR-124a’s effect on Pim1 is in fact mediated through STAT3 (Figure 4J).
Primary ATL patients are characterized by a high rate of STAT3-activating mutations
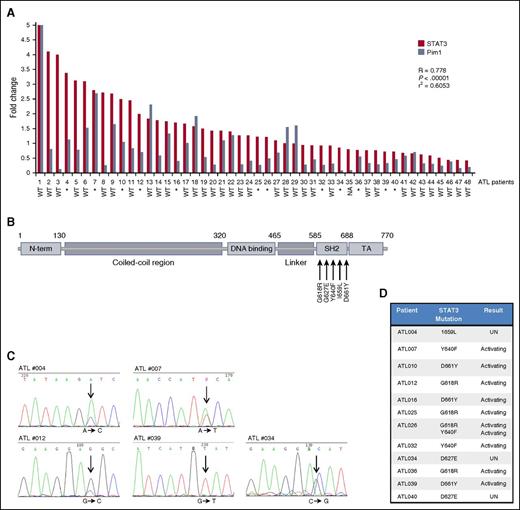
STAT3 and Pim1 expression were readily detected in primary ATL patients and healthy donors (supplemental Figure 4A). Interestingly, Pim1 significantly correlated with STAT3 expression, suggesting that in ATL patients STAT3 expression regulates the level of Pim1 expression (Figure 5A). The SH2 domain of STAT3 is a hotspot for activating mutations in cancer (Figure 5B). Given the large number of activating STAT3 mutations found in large granular lymphocytic (LGL) leukemia22 and the recent genetic profiling of ATL,23 we sequenced the hotspot region in ATL patients. We found that 25.5% of primary ATL patients had a mutation in the SH2 domain of STAT3 (Figure 5A-D and supplemental Figure 4B). ATL patients harbored several well-characterized, activating mutations—Y640F, D661Y, and G618R—known to correlate with increased STAT3 transcriptional activity.22 An I659L mutation that had been previously found only in LGL leukemia was also observed in 1 ATL patient, and our analyses also uncovered 2 patients with D627E mutations.
Primary ATL patient samples are characterized by a high rate of STAT3 mutations and a strong correlation between STAT3 and Pim1. (A). Correlation of STAT3 and Pim1 in ATL patients. Real-time PCR analysis of STAT3 (red) and Pim1 (steel blue) expression in primary ATL patients (n = 48). Statistical analysis was carried out using Spearman’s correlation coefficient. Corresponding P values and linear regression values are indicated. GAPDH amplification served as an internal control. Mutational status of the STAT3 hotspot region is indicated below the corresponding patient. *, mutated; WT, wild-type; NA, not available. (B) Illustration of the STAT3 protein indicating the SH2 domain where frequent ATL STAT3 mutations were found. (C) A representational example of STAT3 mutations found in individual ATL patients. Arrows highlight the specific point mutation in the sequence, with the corresponding nucleotide change below. (D) Primary ATL patients have a 25.5% percentage of STAT3 mutations. Table demonstrating individual STAT3 mutations found in ATL patients. The consequence of each mutation, if known, is highlighted. UN, unknown.
Primary ATL patient samples are characterized by a high rate of STAT3 mutations and a strong correlation between STAT3 and Pim1. (A). Correlation of STAT3 and Pim1 in ATL patients. Real-time PCR analysis of STAT3 (red) and Pim1 (steel blue) expression in primary ATL patients (n = 48). Statistical analysis was carried out using Spearman’s correlation coefficient. Corresponding P values and linear regression values are indicated. GAPDH amplification served as an internal control. Mutational status of the STAT3 hotspot region is indicated below the corresponding patient. *, mutated; WT, wild-type; NA, not available. (B) Illustration of the STAT3 protein indicating the SH2 domain where frequent ATL STAT3 mutations were found. (C) A representational example of STAT3 mutations found in individual ATL patients. Arrows highlight the specific point mutation in the sequence, with the corresponding nucleotide change below. (D) Primary ATL patients have a 25.5% percentage of STAT3 mutations. Table demonstrating individual STAT3 mutations found in ATL patients. The consequence of each mutation, if known, is highlighted. UN, unknown.
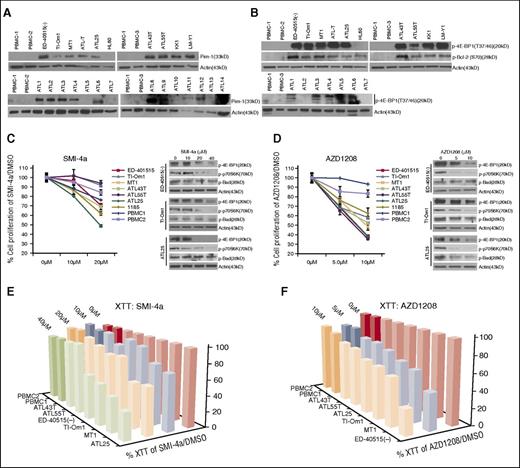
Pim1 is constitutively activated in ATL cells, and targeting Pim1 with small inhibitors SMI-4a and AZD-1208 induces apoptosis and reduces tumor growth in a xenograft mouse model of ATL
Constitutive activation of the Pim1 kinase signaling pathway in ATL cells was characterized by high levels of Pim1 expression and activation of its downstream targets p4E-BP1 (T37/46) and pBcl2 (S70) (Figure 6A-B). Importantly, increased protein expression of Pim1 was also detected in 10 of 14 (71%) freshly isolated uncultured ATL patient samples but not in PBMC from healthy donors (0/3) (Figure 6A). Furthermore, the Pim1 signaling pathway was constitutively activated in primary ATL patients, as shown by an increased phosphorylation of 4E-BP1 (T37/46) (Figure 6B). We also found that HTLV-I cell lines harbored high levels of Pim1, but exogenous expression of viral proteins, Tax and HBZ, did not appreciably alter Pim1 expression (supplemental Figure 4C). To confirm the biological relevance of Pim1, we investigated the effect of Pim1 inhibitors, SMI-4a24 and AZD1208,25 on ATL cells. Treatment with either SMI-4a or AZD1208 was associated with a significant reduction in cell proliferation (Figure 6C-D), independently confirmed by XTT assays (Figure 6E-F and supplemental Figure 4D-F). Inhibition of Pim1 activity by SMI-4a and AZD1208 in ATL cells was verified by reduced levels of p4E-BP1, p-p70S6K, and p-Bad (Figure 6C-D). Our studies suggest that SMI-4a exhibits high nonspecific toxicity toward PBMCs. In contrast, AZD1208 was more specific toward ATL cells. Of note, levels of p-Bad were less affected by treatment with SMI-4a, when compared with AZD1208, suggesting that AZD1208 may be a strong candidate for treatment of ATL disease.
Pharmacological Pim kinase inhibitors inhibit ATL cell proliferation. (A) Pim1 is overexpressed in ATL-derived cell lines and primary ATL patient samples. Normal PBMCs (n = 3) and HL60, an AML line that does not express Pim1, were used as controls. Primary acute/lymphoma ATL patient samples (n = 14) were used. (B) Overexpression of Pim1 target genes, p-4EBP1 (Thr37/46), and p-Bcl2 (Ser70) in ATL-derived cell lines and ATL patient samples (n = 7). (C-D) Loss of proliferation in ATL-derived cell lines by the Pim-kinase inhibitors, Smi-4a (C), and AZD1208 (D). Cell counts were repeated at least twice. Results represent the percentage of cells alive after 5 days of Pim inhibitor treatment, compared with 5 days treated with DMSO. For Smi-4a treatment, cells were treated with 0, 10, or 20 μM Smi-4a, and for AZD1208, cells were treated with 0, 5, or 10 μM AZD1208. Normal PBMCs (n = 2) were used as a control. Western blots indicate loss of Pim1 targets, p-4EBP1 (Thr37/46), p-p70S6K (Thr389), and loss of pBad (Ser20) (negligible for Smi-4a) after 24 hours with 0, 10, 20, or 40 μM Smi-4a; 0, 5, or 10 μM AZD1208; or DMSO control. (E-F). Loss of cell viability in ATL-derived cell lines treated with the Pim-kinase inhibitors, Smi-4a (E) and AZD1208 (F). XTT cellular metabolic assays were performed on ATL lines treated with Pim inhibitors. Results represent the percentage of XTT-positive cells after 5 days of Pim inhibitor treatment, compared with 5 days treated with DMSO. Bars represent the average of at least 2 independent experiments.
Pharmacological Pim kinase inhibitors inhibit ATL cell proliferation. (A) Pim1 is overexpressed in ATL-derived cell lines and primary ATL patient samples. Normal PBMCs (n = 3) and HL60, an AML line that does not express Pim1, were used as controls. Primary acute/lymphoma ATL patient samples (n = 14) were used. (B) Overexpression of Pim1 target genes, p-4EBP1 (Thr37/46), and p-Bcl2 (Ser70) in ATL-derived cell lines and ATL patient samples (n = 7). (C-D) Loss of proliferation in ATL-derived cell lines by the Pim-kinase inhibitors, Smi-4a (C), and AZD1208 (D). Cell counts were repeated at least twice. Results represent the percentage of cells alive after 5 days of Pim inhibitor treatment, compared with 5 days treated with DMSO. For Smi-4a treatment, cells were treated with 0, 10, or 20 μM Smi-4a, and for AZD1208, cells were treated with 0, 5, or 10 μM AZD1208. Normal PBMCs (n = 2) were used as a control. Western blots indicate loss of Pim1 targets, p-4EBP1 (Thr37/46), p-p70S6K (Thr389), and loss of pBad (Ser20) (negligible for Smi-4a) after 24 hours with 0, 10, 20, or 40 μM Smi-4a; 0, 5, or 10 μM AZD1208; or DMSO control. (E-F). Loss of cell viability in ATL-derived cell lines treated with the Pim-kinase inhibitors, Smi-4a (E) and AZD1208 (F). XTT cellular metabolic assays were performed on ATL lines treated with Pim inhibitors. Results represent the percentage of XTT-positive cells after 5 days of Pim inhibitor treatment, compared with 5 days treated with DMSO. Bars represent the average of at least 2 independent experiments.
A dose response of AZD1208 in ED-40515(–) cells showed a complete loss of cellular proliferation with increasing doses of AZD1208 (Figure 7A), which was confirmed by carboxyfluorescein diacetate succinimidyl ester measurements (Figure 7B). The inhibition of ATL proliferation was accompanied by a loss of antiapoptotic factors Bcl2 and surviving, and induction of caspase 3–dependent apoptosis (Figure 7C-D). These results supported the notion that Pim1 represents a biologically important target for ATL. The absence of a curative treatment of ATL prompted us to initiate in vivo studies using a clinically relevant dose of AZD1208, which is currently being studied in phase 1 clinical trials for acute myeloid leukemia and malignant lymphomas. NOG mice were injected with ATL patient–derived ED-40515(–) cells and received daily oral gavage of AZD1208 or vehicle for 14 days. Our results demonstrated that AZD1208 significantly inhibits ATL tumors, as shown by reduced tumor volume and tumor weight by almost threefold (Figure 7E-G).
AZD1208 prevents in vivo tumor growth in an ATL mouse model. (A) Dose response of ED-40515(–) with AZD1208, demonstrates complete loss of proliferation in treated cells. Cell counts (left) and XTT assays (right) were repeated at least twice. Results represent the percentage of cells alive after 5 days of AZD1208 (0, 1, 3, 5, 10, 20, or 50 μM) compared with 5 days treated with DMSO. (B) Carboxyfluorescein diacetate succinimidyl ester staining of ED-40515(–) cells treated with or without AZD1208. DMSO and AZD1208 cells were stained and analyzed with carboxyfluorescein diacetate succinimidyl ester on day 0, and then re-analyzed on Day 5. (C) AZD1208 induces apoptosis in ATL cells with minimal cell death in normal PBMCs. Annexin V/PI staining was performed 5 days after treatment (percentage of cells PI+/Annexin V+ is indicated). (D) Loss of Pim1 activity is associated with deregulation of prosurvival and apoptotic proteins (caspase3, Bcl2, survivin, and Mcl-1, along with Pim1-regulated proteins) in ED-40515(–) cells after 5 days of AZD1208 (10 μM) or DMSO treatment. (E-G) In vivo growth of NOG mice engrafted with ED-40515(–) cells treated with AZD1208. Mice were treated every day with either vehicle (n = 7) or AZD1208 (n = 7) at a dose of 30 mg/kg. (E) Representative images of tumors extracted from mice. (F) Growth curves of in vivo tumor growth, plotted as the average tumor volume (mm3) (calculated as the [width2 × length]/2) per every 2 days. P values are calculated using a 2-sided Student t test between the tumor volumes for vehicle vs AZD1208-treated. (G) Tumor weight (in grams) of ED-40515(–) tumors taken at the time of sacrifice. P values are calculated using a 2-sided Student t test between the tumor weight for vehicle vs AZD1208-treated. The mean tumor weight is indicated with a bar and green square, with the standard error of the mean indicated.
AZD1208 prevents in vivo tumor growth in an ATL mouse model. (A) Dose response of ED-40515(–) with AZD1208, demonstrates complete loss of proliferation in treated cells. Cell counts (left) and XTT assays (right) were repeated at least twice. Results represent the percentage of cells alive after 5 days of AZD1208 (0, 1, 3, 5, 10, 20, or 50 μM) compared with 5 days treated with DMSO. (B) Carboxyfluorescein diacetate succinimidyl ester staining of ED-40515(–) cells treated with or without AZD1208. DMSO and AZD1208 cells were stained and analyzed with carboxyfluorescein diacetate succinimidyl ester on day 0, and then re-analyzed on Day 5. (C) AZD1208 induces apoptosis in ATL cells with minimal cell death in normal PBMCs. Annexin V/PI staining was performed 5 days after treatment (percentage of cells PI+/Annexin V+ is indicated). (D) Loss of Pim1 activity is associated with deregulation of prosurvival and apoptotic proteins (caspase3, Bcl2, survivin, and Mcl-1, along with Pim1-regulated proteins) in ED-40515(–) cells after 5 days of AZD1208 (10 μM) or DMSO treatment. (E-G) In vivo growth of NOG mice engrafted with ED-40515(–) cells treated with AZD1208. Mice were treated every day with either vehicle (n = 7) or AZD1208 (n = 7) at a dose of 30 mg/kg. (E) Representative images of tumors extracted from mice. (F) Growth curves of in vivo tumor growth, plotted as the average tumor volume (mm3) (calculated as the [width2 × length]/2) per every 2 days. P values are calculated using a 2-sided Student t test between the tumor volumes for vehicle vs AZD1208-treated. (G) Tumor weight (in grams) of ED-40515(–) tumors taken at the time of sacrifice. P values are calculated using a 2-sided Student t test between the tumor weight for vehicle vs AZD1208-treated. The mean tumor weight is indicated with a bar and green square, with the standard error of the mean indicated.
Discussion
Although the growth and survival of HTLV-I–transformed cells rely upon the activation of signaling pathways such as JAK/STAT, NF-κB, NOTCH1, and transforming growth factor-β,9,15,26,27 an effective therapy for ATL has yet to be discovered. In this study, we revealed the molecular basis for miR-124a–associated tumor suppressor effects in HTLV-I–transformed cells. miRNAs are important regulators in HTLV-I. miR-223 and miR-150 have been shown to regulate STAT1 activity28 ; miR-31 mediates NF-κB–inducing kinase activity in HTLV-I cells,29 whereas miR-28-3p regulates HTLV-I viral replication.30 A tumor suppressor effect for miR-124a has been suggested for some cancers,31-33 but to date, miR-124a had not been studied in ATL. Our data demonstrate that miR-124a is a potent tumor suppressor in ATL by targeting the STAT3-Pim1 axis. We found that expression of miR-124a is downregulated in HTLV-I–transformed cells and freshly isolated uncultured ATL cells through promoter hypermethylation. We also found that restoring miR-124a expression in ATL cells significantly decreases in vitro cellular proliferation and causes a significant reduction in tumor formation in vivo. Although miR-124 inhibition of STAT3 has been shown,34 to our knowledge, this is the first study reporting miR-124a regulation of STAT3 in human leukemia/lymphomas. Using a cancer profiler array along with in silico promoter analyses of genes downregulated by miR-124a, we identified STAT3 as a common regulator. S3I-201 altered the expression of genes in a similar fashion to those found in the cancer array, strengthening the link between STAT3 and miR-124a.
We found several mutations in the STAT3 SH2 domain, a region known to cause constitutive activation of the STAT3 protein. A recent study reported STAT3 mutations in 21% of ATL patients.23 This was similar to the level found in our study (25.5%); however, we also found 2 novel STAT3 mutations, I659L and D627E, not previously reported in ATL patients. This may relate to the ethnic origin of our samples (Caribbean vs Japanese23 ). The majority of STAT3 mutations have been studied in LGL leukemia, but mutations have also been found in B-cell, natural killer–cell, and other T-cell lymphomas.35 Though STAT3 mutational status did not correlate with higher STAT3/Pim1 activation, we only focused our research on the hotspot region located in the SH2-domain of STAT3. Recent studies in LGL leukemia, demonstrate that there may be additional activating mutations in the coiled-coil and DNA-binding domains.36 The novel loss of miR-124a inhibition of STAT3, along with the large amount of activating STAT3 mutations in primary ATL, point to the relevance of the STAT3 pathway in ATL leukemogenesis. These results, accompanied by the severe loss of proliferation after STAT3 targeting, demonstrate that constitutive activation of STAT3 is required in ATL cells.
Because the moderate to high micromolar activities of S31-201 have hampered clinical development, we aimed at identifying STAT3 targets responsible for tumor suppressor effects in ATL cells. To this end, we analyzed STAT3 transcriptional target genes in miR-124a tumor samples and controls. Results from these experiments identified the Pim1 kinase as a consistently downregulated target in tumors with miR-124a–mediated reduced STAT3 expression. We also wanted to know if the miR-124a-STAT3-Pim1 axis was important in other leukemia, because it has been previously demonstrated that miR-124 could inhibit tumor growth of ALL cells.37 Establishment of Nalm-20 mir-124a-TET-On cells showed loss of proliferation and STAT3/Pim1 protein expression (supplemental Figures 1 and 5). Studies have shown constitutive activation of STAT1 and STAT5 in ALL patients, whereas AML patients harbored STAT1, STAT3, and STAT5 activation.38 Like ATL, the miR-124a-STAT3-Pim1 axis may be more important in AML patients, or other leukemias that harbor high STAT3 activity.39 Despite this, recent data have shown that SMI-4a was effective in killing T-cell lymphoblastic leukemia/lymphoma (pre–T-LBL/T-ALL) lines.24
In this study, we show for the first time a high level of Pim1 expression in all patient-derived ATL lines tested (100%) and in a high percentage of freshly isolated, uncultured ATL samples (71%). Notably, constitutive phosphorylation Pim1 targets 4E-BP1 (T37/46) and Bcl2 (S70) were also detected in all ATL cells, suggesting a global constitutive activation of the Pim1 signaling pathway in ATL cells. To demonstrate the biological significance of our findings and the role of activated Pim1 in HTLV-I–transformed cells, we treated ATL cells with the Pim1 inhibitors SMI-4a and AZD1208, the latter of which is currently in phase 1 clinical trials. In vitro experiments indicated that SMI-4a was effective but associated with significant nonspecific toxicity toward PBMCs of healthy donors. In contrast, the Pim1 inhibitor AZD1208 demonstrated lower toxicity against PBMCs and better specificity toward ATL cells. AZD1208 was associated with reduced p-p70S6K, p-4E-BP1, and pBad in ATL cells. This suggests that AZD1208 inhibits 2 important functions of Pim1, translational regulation and apoptosis, in ATL cells. Finally, we tested the efficacy of AZD1208 in a preclinical murine model of ATL and demonstrated a significant reduction of tumor weight and volume in all animals. AZD1208 has been shown to be effective in reducing tumor growth in mice harboring AML tumors.25 In summary, re-expression of miR-124a in a murine model of ATL revealed a critical function of the STAT3/Pim1 signaling axis in HTLV-I–transformed ATL cells. Constitutive activation of the Pim1 pathway is a promising novel therapeutic target for ATL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Felipe Prosper (Hematology and Cell Therapy, Clínica Universitaria, Spain) for providing ALL cell lines. ATL-like cell lines ATL-T, ATL-25, ATL43T, ATL55T, KOB, KK1, and LM-Y1 were provided by M. Matsouka, Kyoto University, Japan. ATL-like cell lines ED-40515(–), Tl-Om1, and MT1 were provided by M. Maeda, Kyoto University, Japan. They also thank Brandi Miller for editorial assistance.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This study was supported by the National Cancer Institute, National Institutes of Health (CA106258, CA141386) (C.N.); and a grant from the National Center for Research Resources (P20RR016475) (M.B.)
Authorship
Contribution: M.B. designed research studies, conducted experiments, acquired and analyzed data, and wrote the manuscript; L.L. provided reagents; and C.N. designed research studies, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christophe Nicot, Department of Pathology and Laboratory Medicine, MS 3045, University of Kansas Medical Center, 3901 Rainbow Blvd, Kansas City, KS 66160; e-mail: cnicot@kumc.edu.

![Figure 1. Overexpression of miR-124a in ATL cells inhibits tumor growth in vivo. (A) Mature miRNA detection of miR-124a in ATL patient samples (n = 17) vs healthy noninfected donor. miR-24 served as an internal control. (B-C) Induction of miR-124a in ED-40515(–) cells inhibits cell proliferation. Pre–miR-124a expression was detected by real-time PCR (B) after doxycycline (Dox) addition. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as an internal control. Cells were counted every 48 hours after receiving 2 μg/mL Dox (every 48 h) (C). Results represent at least 2 independent experiments. (D) Loss of proliferation/cell cycle markers (cyclins A, B1, and E, and survivin) in miR-124a–expressing ATL cells, 6 days after receiving 2 μg/mL Dox every other day. Tubulin was used as a loading control. (E) ED-40515(–) cells expressing miR-124a enter cell cycle arrest after induction with 2 μg/mL Dox every day for 5 days. (F) Cell survival markers (Bax, Bcl-xl, and Bcl-2) are altered in ED-40515(–)–miR-124a expressing cells 5 days after receiving 2 μg/mL Dox every day. Tubulin served as a loading control. (G) ED-40515(–)–pTRIPZ (n = 12) or miR-124a (n = 12) TET-On cells were injected into the right or left flank of NOG mice. Pictures are representative of 2 mice simultaneously receiving pTRIPZ (left) or miR-124a (right) TET-On ED-40515(–) cells. (H) Real-time PCR detection of pre–miR-124a in a tumor model of ATL. GAPDH served as a control. (I) In vivo growth of NOG mice engrafted with ED-40515(–) pTRIPZ (left) or miR-124a (right) cells (plotted as the average tumor volume (mm3) (calculated as the [width2 × length]/2) per every 7 days). (J) The percentage of tumors with volumes of 200 m3 or larger was plotted against the days after implantation into mice. Volumes were calculated every 7 days, up to 21 days. Statistics are calculated using the N-1 2-proportion test, with the number of tumors in each group (n = 12) 200 m3 or larger, considered positive.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/127/20/10.1182_blood-2015-11-685032/4/m_2439f1.jpeg?Expires=1769782181&Signature=fl6sDjvR557MSiae4oFNI9oEYCq5RYinoD3MVlY-y~ZjF-DXFN9xo4DA89kNTIggwURDXcbuQF6k7VYH42l-rvm9hXtf24f~SPdxc5myHmTiH2dZ8Oyz~aU9lL0BaOufsy2q6EoVVRwsA7KGrpl~gclRC1yvGgCSKtfmluRXDTp9HsInJg43pjr96jpiCnC7r9HpmWJJi-xqdtN59Ky9rr0cYkIDjwDjfjzoNE1i5fCZsFgFR~C61cr9I5Keq6OziERJX0FqJEQhRnEL8sRp5-Ki6qp4B3rwxdJAtfysghY10hkt2W5UmVqNZgEWwcP~Gu7ZjYxRH39Dz7pfCyd60A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 7. AZD1208 prevents in vivo tumor growth in an ATL mouse model. (A) Dose response of ED-40515(–) with AZD1208, demonstrates complete loss of proliferation in treated cells. Cell counts (left) and XTT assays (right) were repeated at least twice. Results represent the percentage of cells alive after 5 days of AZD1208 (0, 1, 3, 5, 10, 20, or 50 μM) compared with 5 days treated with DMSO. (B) Carboxyfluorescein diacetate succinimidyl ester staining of ED-40515(–) cells treated with or without AZD1208. DMSO and AZD1208 cells were stained and analyzed with carboxyfluorescein diacetate succinimidyl ester on day 0, and then re-analyzed on Day 5. (C) AZD1208 induces apoptosis in ATL cells with minimal cell death in normal PBMCs. Annexin V/PI staining was performed 5 days after treatment (percentage of cells PI+/Annexin V+ is indicated). (D) Loss of Pim1 activity is associated with deregulation of prosurvival and apoptotic proteins (caspase3, Bcl2, survivin, and Mcl-1, along with Pim1-regulated proteins) in ED-40515(–) cells after 5 days of AZD1208 (10 μM) or DMSO treatment. (E-G) In vivo growth of NOG mice engrafted with ED-40515(–) cells treated with AZD1208. Mice were treated every day with either vehicle (n = 7) or AZD1208 (n = 7) at a dose of 30 mg/kg. (E) Representative images of tumors extracted from mice. (F) Growth curves of in vivo tumor growth, plotted as the average tumor volume (mm3) (calculated as the [width2 × length]/2) per every 2 days. P values are calculated using a 2-sided Student t test between the tumor volumes for vehicle vs AZD1208-treated. (G) Tumor weight (in grams) of ED-40515(–) tumors taken at the time of sacrifice. P values are calculated using a 2-sided Student t test between the tumor weight for vehicle vs AZD1208-treated. The mean tumor weight is indicated with a bar and green square, with the standard error of the mean indicated.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/127/20/10.1182_blood-2015-11-685032/4/m_2439f7.jpeg?Expires=1769782181&Signature=RWBNoMlbE6CMvSRMuz69TTyADLdEDvtOBWDzIEhuAkOFxrIyO915HVp1HWphL2FsQQTy111LIQpeTY96iY88wlvG61wMMJvjXCqdO2PGsFD5QsOkPTiPXywvrtqlrVX11XM-qcGvaf69hbXDjZ-HUQbXo8fCEV684bgLiBpQ3JpMml60Vp4o6aRJOSm7Fw~GaNMrWJ~x8BHyD9LeXn4gayoUi3OdHf5nMGcJHoHWFl~T~6f7ZbgmImCkKWE6sfWB2hChmYa4QWeqbRQszGeZaprEnpavzVSupEMRp3rYZcpC7upRbBRN7xJEOWyrc6v3TajcORUJIyibcsuYQjE3hg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal