Key Points
CD19-targeted CAR-T-cell therapy of patients with MLL-rearranged B-ALL effectively induced marrow remission of B-ALL.
Patients with MLL-rearranged B-ALL who attain CR after CD19 CAR-T-cell therapy may be at risk for relapse with clonally related AML.
Abstract
Administration of lymphodepletion chemotherapy followed by CD19-specific chimeric antigen receptor (CAR)–modified T cells is a remarkably effective approach to treating patients with relapsed and refractory CD19+ B-cell malignancies. We treated 7 patients with B-cell acute lymphoblastic leukemia (B-ALL) harboring rearrangement of the mixed lineage leukemia (MLL) gene with CD19 CAR-T cells. All patients achieved complete remission (CR) in the bone marrow by flow cytometry after CD19 CAR-T-cell therapy; however, within 1 month of CAR-T-cell infusion, 2 of the patients developed acute myeloid leukemia (AML) that was clonally related to their B-ALL, a novel mechanism of CD19-negative immune escape. These reports have implications for the management of patients with relapsed and refractory MLL-B-ALL who receive CD19 CAR-T-cell therapy.
Introduction
Immunotherapy with T cells that are genetically modified to express a CD19-specific chimeric antigen receptor (CAR) represents a promising approach for patients with refractory B-cell acute lymphoblastic leukemia (B-ALL).1-3 Early-phase clinical trials have shown high rates of complete remission (CR) after CD19 CAR-T-cell therapy, even in patients who have failed multiple chemotherapy regimens and/or allogeneic hematopoietic stem cell transplantation (HCT). Durable remissions can be achieved when functional CAR-T cells persist sufficiently long to eradicate all malignant cells in the recipient; however, leukemic relapse may occur with premature loss of CD19 CAR-T cells or because of the emergence of leukemia that has lost CD19 expression despite persistence of functional CD19 CAR-T cells.4
Rearrangements of the mixed lineage leukemia (MLL) gene on chromosome 11q23 confer a poor prognosis on patients with B-ALL treated with chemotherapy.5-7 We found that CD19 CAR-T-cell therapy is highly effective for patients with MLL-B-ALL, with 7 of 7 patients achieving CR by flow cytometry of blood and bone marrow. However, we observed leukemia relapse in 2 patients associated with acquisition of a myeloid phenotype and loss of expression of B lymphoid lineage antigens, a novel mechanism of relapse that is distinct from the isolated loss of CD19 expression noted previously.4
Study design
Patients received lymphodepletion chemotherapy, followed by infusion of autologous CD19 CAR-T cells, according to 2 clinical trial protocols (NCT01865617, NCT02028455). T cells were modified using a CAR construct incorporating a truncated human epidermal growth factor receptor as a marker of transgene expression, which enabled enumeration of CAR-T cells.8 Evaluation of leukemic blasts by flow cytometry, fluorescence in situ hybridization (FISH), G-banded karyotyping, and chromosomal genomic array testing (CGAT) was performed in Clinical Laboratory Improvement Amendments–certified laboratories, and deep sequencing of the IGH gene was performed by Adaptive Biotechnologies (Seattle, WA). Quantitative polymerase chain reaction for vector sequence was used to enumerate integrated CAR transgene in CAR-T cells and myeloblasts. These studies were conducted with approval of the institutional review boards of Fred Hutchinson Cancer Research Center and Seattle Children’s Hospital and in accordance with the Declaration of Helsinki.
Results and discussion
After receiving CD19-targeted CAR-T cell therapy, all patients with MLL-B-ALL achieved CR in the marrow by flow cytometry; however, 2 of 7 patients relapsed, both with a myeloid phenotype leukemia approximately 1 month after CAR-T cell infusion. Pathology findings are summarized in Table 1 and Figure 1.
Pathology findings in 2 patients who received CD19 CAR-T-cell therapy for CD19+MLL-rearranged B-ALL
| Day in relation to CAR-T-cell infusion . | Assay . | Tissue . | Result . |
|---|---|---|---|
| Case 1 | |||
| Pre-CAR-T cells | Flow cytometry | BMMC | Abnormal lymphoblasts expressing CD45, CD19, CD22, CD38, HLA-DR, CD15, CD33, CD13 (dim), and TdT (dim) |
| Karyotype | BMMC | 46,XX,t(4;11)(q21;q23),add(9)(p12)[cp20] | |
| FISH | BMMC | MLL rearranged | |
| IGH gene sequencing | BMMC | Index clone identified | |
| Day 22 after CAR-T cells | Flow cytometry | BMMC | No abnormal lymphoblasts |
| Karyotype | BMMC | 46,XX,t(4;11)(q21;q23),+6,add(9)(p13),-10/46,sl,del(1)(p34.1;q36.3),-6,+8[cp9]/80,slx2,-X,-1,-2,-4,-6,-6,-6,-11,-14,-15,-17,-18 | |
| FISH | BMMC | MLL rearranged | |
| IGH gene sequencing | BMMC | 19,507 copies of the index clone/1 × 106 BMMC | |
| Relapse | Flow cytometry | PBMC | Abnormal monocytic population expressing CD13 (dim), CD64, HLA-DR (dim), CD15, CD33, CD71, and MPO. No abnormal B cells. |
| FISH | PBMC | MLL rearranged | |
| IGH gene sequencing | PBMC | 70,789 copies of the index clone/1 × 106 PBMC | |
| Case 2 | |||
| Pre-CAR-T cells | Flow cytometry | BMMC | Abnormal lymphoblasts expressing CD19, CD38, CD58, CD22 (dim), HLA-DR, CD34, and CD45 |
| Karyotype | BMMC | 46,XX,ins(11;10)(q23;p12p1?1.2) | |
| FISH | BMMC | MLL rearranged | |
| IGH gene sequencing | BMMC | Index clone identified | |
| Day 21 after CAR-T cells | Flow cytometry | BMMC | No abnormal lymphoblasts |
| IGH gene sequencing | BMMC | Index clone not identified | |
| Relapse | Flow cytometry | BMMC | Abnormal myeloblasts expressing CD4, CD56, CD64, CD13, CD33, CD38, HLA-DR, CD34, CD45 and CD71. No abnormal B cell population. |
| Karyotype | BMMC | 46,XX,ins(11;10)(q23;p12p1?1.2) | |
| FISH | BMMC | MLL rearranged | |
| IGH gene sequencing | BMMC | Index clone not identified |
| Day in relation to CAR-T-cell infusion . | Assay . | Tissue . | Result . |
|---|---|---|---|
| Case 1 | |||
| Pre-CAR-T cells | Flow cytometry | BMMC | Abnormal lymphoblasts expressing CD45, CD19, CD22, CD38, HLA-DR, CD15, CD33, CD13 (dim), and TdT (dim) |
| Karyotype | BMMC | 46,XX,t(4;11)(q21;q23),add(9)(p12)[cp20] | |
| FISH | BMMC | MLL rearranged | |
| IGH gene sequencing | BMMC | Index clone identified | |
| Day 22 after CAR-T cells | Flow cytometry | BMMC | No abnormal lymphoblasts |
| Karyotype | BMMC | 46,XX,t(4;11)(q21;q23),+6,add(9)(p13),-10/46,sl,del(1)(p34.1;q36.3),-6,+8[cp9]/80,slx2,-X,-1,-2,-4,-6,-6,-6,-11,-14,-15,-17,-18 | |
| FISH | BMMC | MLL rearranged | |
| IGH gene sequencing | BMMC | 19,507 copies of the index clone/1 × 106 BMMC | |
| Relapse | Flow cytometry | PBMC | Abnormal monocytic population expressing CD13 (dim), CD64, HLA-DR (dim), CD15, CD33, CD71, and MPO. No abnormal B cells. |
| FISH | PBMC | MLL rearranged | |
| IGH gene sequencing | PBMC | 70,789 copies of the index clone/1 × 106 PBMC | |
| Case 2 | |||
| Pre-CAR-T cells | Flow cytometry | BMMC | Abnormal lymphoblasts expressing CD19, CD38, CD58, CD22 (dim), HLA-DR, CD34, and CD45 |
| Karyotype | BMMC | 46,XX,ins(11;10)(q23;p12p1?1.2) | |
| FISH | BMMC | MLL rearranged | |
| IGH gene sequencing | BMMC | Index clone identified | |
| Day 21 after CAR-T cells | Flow cytometry | BMMC | No abnormal lymphoblasts |
| IGH gene sequencing | BMMC | Index clone not identified | |
| Relapse | Flow cytometry | BMMC | Abnormal myeloblasts expressing CD4, CD56, CD64, CD13, CD33, CD38, HLA-DR, CD34, CD45 and CD71. No abnormal B cell population. |
| Karyotype | BMMC | 46,XX,ins(11;10)(q23;p12p1?1.2) | |
| FISH | BMMC | MLL rearranged | |
| IGH gene sequencing | BMMC | Index clone not identified |
BMMC, bone marrow mononuclear cells; PBMC, peripheral blood mononuclear cells.
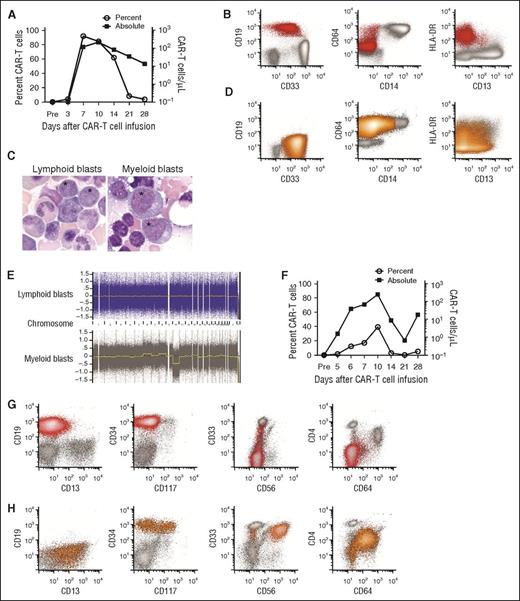
Emergence of CD19−myeloid phenotype blasts after effective CD19 CAR-T-cell therapy for CD19+ MLL-rearranged B-ALL. (A-E) Data from the patient in case 1. (F-H) Data from the patient in case 2. (A) The percentage of CD19-specific CAR-T cells in CD3+ T cells (open circles) and the absolute CAR-T-cell count (squares) in blood on the indicated days after CAR-T cell infusion are shown for patient 1. CAR-T cells were identified as viable CD45+/CD3+/epidermal growth factor receptor-positive events in a lymphoid forward scatter/side scatter gate by flow cytometry, and the absolute count was determined by multiplying the absolute lymphocyte count by the percentage of CAR-T cells in a lymphoid gate. (B) Flow cytometry of peripheral blood demonstrating CD19+ lymphoblasts (red) before CAR-T-cell therapy. The blasts expressed a low level of CD33 and were largely CD64−, CD14−, and CD4− (not shown). Flow plots are gated on mononuclear cells. (C) The abnormal blasts (*) before CAR-T-cell therapy were morphologically distinct from the abnormal blasts after CAR-T-cell therapy. (D) Flow cytometry of peripheral blood obtained on day 35 after CAR-T-cell infusion showing abnormal blasts (orange) without expression of CD19, CD20, CD22, CD24, or cytoplasmic CD79a (not shown). The abnormal blasts were CD33hi, CD64+, and CD4+ (not shown), and CD14− and CD34− (not shown), consistent with AML with monocytic differentiation. (E) CGAT identified multiple genomic aberrations in the monoblasts isolated after CAR-T-cell infusion, which were not present in the CD19+ lymphoblasts isolated before CAR-T-cell infusion. (F) The percentage of CD19-specific CAR-T cells in CD3+ T cells (open circles) and the absolute CAR-T-cell count (squares) in blood on the indicated days after CAR-T-cell infusion are shown for patient 2. (G) All plots show mononuclear cells. Flow cytometry of bone marrow before CAR-T-cell therapy demonstrating CD19+ abnormal lymphoblasts (red). At diagnosis, the patient’s abnormal blasts also expressed CD34, CD22 (not shown) without CD4, CD10 (not shown), CD13, significant CD33, CD56, CD64, or CD117. (H) Flow cytometry of bone marrow on day 30 after CAR-T-cell infusion, showing no abnormal CD19+ blasts or normal B-cell precursors, but abnormal myeloblasts (orange) that express CD34 with CD4, CD13, bright CD33, CD64 (intermediate), and CD117 (subset), with aberrant expression of CD56 on a major subset.
Emergence of CD19−myeloid phenotype blasts after effective CD19 CAR-T-cell therapy for CD19+ MLL-rearranged B-ALL. (A-E) Data from the patient in case 1. (F-H) Data from the patient in case 2. (A) The percentage of CD19-specific CAR-T cells in CD3+ T cells (open circles) and the absolute CAR-T-cell count (squares) in blood on the indicated days after CAR-T cell infusion are shown for patient 1. CAR-T cells were identified as viable CD45+/CD3+/epidermal growth factor receptor-positive events in a lymphoid forward scatter/side scatter gate by flow cytometry, and the absolute count was determined by multiplying the absolute lymphocyte count by the percentage of CAR-T cells in a lymphoid gate. (B) Flow cytometry of peripheral blood demonstrating CD19+ lymphoblasts (red) before CAR-T-cell therapy. The blasts expressed a low level of CD33 and were largely CD64−, CD14−, and CD4− (not shown). Flow plots are gated on mononuclear cells. (C) The abnormal blasts (*) before CAR-T-cell therapy were morphologically distinct from the abnormal blasts after CAR-T-cell therapy. (D) Flow cytometry of peripheral blood obtained on day 35 after CAR-T-cell infusion showing abnormal blasts (orange) without expression of CD19, CD20, CD22, CD24, or cytoplasmic CD79a (not shown). The abnormal blasts were CD33hi, CD64+, and CD4+ (not shown), and CD14− and CD34− (not shown), consistent with AML with monocytic differentiation. (E) CGAT identified multiple genomic aberrations in the monoblasts isolated after CAR-T-cell infusion, which were not present in the CD19+ lymphoblasts isolated before CAR-T-cell infusion. (F) The percentage of CD19-specific CAR-T cells in CD3+ T cells (open circles) and the absolute CAR-T-cell count (squares) in blood on the indicated days after CAR-T-cell infusion are shown for patient 2. (G) All plots show mononuclear cells. Flow cytometry of bone marrow before CAR-T-cell therapy demonstrating CD19+ abnormal lymphoblasts (red). At diagnosis, the patient’s abnormal blasts also expressed CD34, CD22 (not shown) without CD4, CD10 (not shown), CD13, significant CD33, CD56, CD64, or CD117. (H) Flow cytometry of bone marrow on day 30 after CAR-T-cell infusion, showing no abnormal CD19+ blasts or normal B-cell precursors, but abnormal myeloblasts (orange) that express CD34 with CD4, CD13, bright CD33, CD64 (intermediate), and CD117 (subset), with aberrant expression of CD56 on a major subset.
Case 1
A 52-year-old woman with relapsed B-ALL with t(4;11) was referred for CD19 CAR-T-cell therapy 5 months after initial diagnosis. Abnormal B lymphoid blasts represented 87.5% of the BMMCs. The karyotype was 46,XX,t(4;11)(q21;q23),add(9)(p12)[cp20], and FISH studies showed that 83.5% of BMMCs harbored MLL rearrangement. The patient received lymphodepletion chemotherapy with cyclophosphamide 60 mg/kg and fludarabine 25 mg/m2 × 3 and 2 × 106 CAR-T cells/kg. She developed severe cytokine release syndrome (CRS; peak IL-6 serum concentration 185.16 pg/mL), manifested by fever, hypotension, and a grand mal seizure on day 4 after CAR-T-cell infusion associated with CAR-T-cell expansion in blood (Figure 1A). On day 22, there was no detectable B-ALL in bone marrow by flow cytometry; however, karyotyping and FISH studies identified persistent MLL rearrangement. Deep sequencing of the IGH gene in BMMC revealed persistence of the malignant clone identified before CAR-T cell therapy. On day 35, circulating blasts, which now expressed myeloperoxidase, CD4, and CD64 without CD19 or other B-cell lineage antigens, were noted, consistent with acute myeloid leukemia (AML; Figure 1B-D). FISH for MLL rearrangement and IGH deep sequencing established that the B-ALL and AML were clonally related, and CGAT (Figure 1E) demonstrated clonal evolution in the myeloid blasts. After reinduction, the patient achieved CR and underwent allogeneic HCT. CD19 CAR-T cells were identified in the patient’s blood, and B-cell aplasia persisted until the time of HCT. She developed relapsed AML and died 147 days after CD19 CAR-T-cell infusion without evidence of B-ALL.
Case 2
An 18-month-old girl was referred for CD19 CAR-T-cell therapy 14 months after an initial diagnosis of MLL-rearranged infant B-ALL. B lymphoid blasts made up 20% of the marrow, and the karyotype was 46,XX,ins(11;10)(q23;p12p1?1.2)[10]/46,XX[10]. She received cyclophosphamide 1.5 g/m2, followed by 1 × 107 CD19 CAR-T cells/kg. On day 4, she developed severe CRS (peak IL-6 serum concentration, 110 pg/mL), with fever, hypotension, and encephalopathy associated with CAR-T cell expansion in blood (Figure 1F). On day 21, there was no evidence of B-ALL in the marrow by flow cytometry or deep sequencing of the IGH gene; however, on day 30, an abnormal myeloid population without B lineage antigens was identified, making up 5.5% of the marrow leukocytes (Figure 1G-H). The karyotype was 46,XX,ins(11;10)(q23;p12p1?1.2)[10]/46,XX[10], confirming persistent MLL rearrangement. CAR-T cells were detected in blood, and there was B-cell aplasia at the diagnosis of AML. On day 76, after failed reinduction chemotherapy, FISH studies confirmed MLL rearrangement in isolated myeloblasts, monocytes, and CD34+/CD38− stem cells. Deep sequencing of the IGH gene was negative for the rearrangement previously noted in the lymphoid blasts, suggesting myeloid relapse occurred from an immature stem cell clone.
Discussion
MLL rearrangement has been incorporated in the World Health Organization classification of hematopoietic neoplasms to describe unique entities of acute leukemia.9 Although MLL rearrangement may be associated with aberrant low-level “cross-lineage” myeloid antigen expression on lymphoid blasts in B-ALL or lymphoid antigen expression on myeloid blasts in AML, a change in phenotype to that of a distinct lineage is rare.10,11 Transition from a lymphoid to myeloid phenotype has been reported after intensive chemotherapy and HCT, but this study is the first to identify acquisition of a clonally related myeloid phenotype associated with CD19-negative escape after CD19 CAR-T-cell immunotherapy. Of the 7 patients who received CD19 CAR-T cell therapy for MLL-B-ALL, we identified 2 in whom the relapsed leukemia had lost expression of lymphoid B-lineage antigens, including CD19, and acquired expression of myeloid antigens. In each case, the initial diagnosis by morphology and immunophenotyping was consistent with B-ALL, and upon relapse met the World Health Organization definition of AML. This phenomenon was not observed in the non-MLL rearranged ALL cases (n = 62). In conjunction with recent in vivo studies of immune escape from CD19 CAR-T-cell therapy that identified dependence of lymphoid to myeloid lineage switching on the B-ALL genotype,12 the rare occurrence of lymphoid to myeloid phenotype in 2 patients with MLL-B-ALL suggests these patients may be susceptible to this novel mechanism of CD19-negative escape from CAR-T-cell therapy.
Acquisition of a myeloid phenotype distinguished these cases from a previous report of CD19-negative relapse of B-ALL occurring by selection of alternatively spliced CD19 isoforms.4 Secondary therapy-related AML was also excluded by the identification of cytogenetic abnormalities by conventional karyotyping, FISH, and CGAT that were shared between lymphoid blasts before and myeloid blasts after CAR-T cell therapy, and in 1 case, an identical rearranged IGH gene sequence that was identified in both lymphoid and myeloid blasts. Lentiviral integration was not detected in isolated myeloid blasts from either patient, excluding origin of the myeloid leukemia from oncogenic transformation associated with lentiviral integration.
Different mechanisms have been proposed to contribute to leukemic lineage switch, and the roles of bipotential progenitors, cell reprogramming or de-differentiation, and selection of rare phenotypically distinct clones are the subject of ongoing debate.10 The leukemic blasts in the cases we report changed from lymphoid to myeloid phenotype, apparently using 2 distinct mechanisms: retention of the IGH rearrangement in relapsed myeloid blasts (case 1) suggests a contribution from cell reprogramming or de-differentiation of previously committed B lymphoid blasts, whereas absence of the IGH rearrangement in myeloid blasts (case 2) is consistent with myeloid differentiation of a noncommitted precursor or selection of a preexisting myeloid clone after CD19 CAR-T-cell therapy. In both our cases, the absence of flow cytometry evidence of acute leukemia early after CAR-T-cell therapy, followed by explosive presentation of AML, suggests that CD19 CAR-T-cell therapy provided a selective advantage to a rare myeloid clone, regardless of whether the myeloid clone was present before CAR-T-cell therapy or had arisen by de-differentiation from a lymphoid blast after CAR-T-cell infusion. The importance of CD19 CAR-T-cell therapy in the acquisition of a myeloid phenotype is highlighted by the observation that multiple previous cycles of intensive chemotherapy did not induce AML in these patients. Although anti-CD19 immunologic pressure may have enabled outgrowth of a CD19-negative myeloid clone, it has also been proposed that a leukemic growth factor may contribute to lineage switch.10,13 After CAR-T-cell therapy, both patients who developed phenotypic switch had severe CRS with high serum concentrations of IL-6 and other cytokines, whereas those without a phenotypic switch did not experience severe CRS. In vitro IL-6 supplementation was found to be a key factor in driving myeloid differentiation of a t(4;11) MLL-B-ALL line,14 suggesting that high serum cytokine levels during CRS might contribute to myeloid differentiation of a lymphoid clone, outgrowth of a myeloid leukemic clone, or both.
These data suggest caution is warranted when treating patients with MLL-B-ALL, using CD19-targeted therapies. Although CD19 CAR-T-cell therapy resulted in CR in all patients, the rapid emergence of CD19-negative AML in 2 of 7 patients suggests it may not provide definitive therapy for MLL-B-ALL, and that judicious consolidation with early allogeneic HCT should be considered in this subset of patients.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
R.G. received a Conquer Cancer Foundation/ASCO Career Development Award. Additional support for NCT02028455 was provided by Alex’s Lemonade Stand and St. Baldrick’s/Stand Up 2 Cancer. S.R.R. and M.C.J. are recipients of National Institutes of Health, National Cancer Institute grant CA136551-07. C.J.T. is a Damon Runyon Clinical Investigator.
Authorship
Contribution: All authors contributed to experimental design and edited the manuscript. R.G. and C.J.T. wrote the manuscript.
Conflict-of-interest disclosure: This study was supported in part by research funding from Juno Therapeutics. S.R.R. and M.C.J. are cofounders of Juno Therapeutics. S.R.R., M.C.J., D.G.M., and C.J.T. receive research funding from Juno Therapeutics. D.G.M. and C.J.T. serve on advisory boards for Juno Therapeutics.
Correspondence: Cameron J. Turtle, Program in Immunology, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave N, Seattle, WA 98109; e-mail: cturtle@fredhutch.org.