Key Points
The NAE inhibitor pevonedistat induces Chk1/Wee1 activation and the intra-S checkpoint, limiting its anti-AML efficacy.
The HDAC inhibitor belinostat potentiates the in vitro and in vivo activity of pevonedistat in AML by disrupting the DDR.
Abstract
Two classes of novel agents, NEDD8-activating enzyme (NAE) and histone deacetylase (HDAC) inhibitors, have shown single-agent activity in acute myelogenous leukemia (AML)/myelodysplastic syndrome (MDS). Here we examined mechanisms underlying interactions between the NAE inhibitor pevonedistat (MLN4924) and the approved HDAC inhibitor belinostat in AML/MDS cells. MLN4924/belinostat coadministration synergistically induced AML cell apoptosis with or without p53 deficiency or FLT3–internal tandem duplication (ITD), whereas p53 short hairpin RNA (shRNA) knockdown or enforced FLT3-ITD expression significantly sensitized cells to the regimen. MLN4924 blocked belinostat-induced antiapoptotic gene expression through nuclear factor–κB inactivation. Each agent upregulated Bim, and Bim knockdown significantly attenuated apoptosis. Microarrays revealed distinct DNA damage response (DDR) genetic profiles between individual vs combined MLN4924/belinostat exposure. Whereas belinostat abrogated the MLN4924-activated intra-S checkpoint through Chk1 and Wee1 inhibition/downregulation, cotreatment downregulated multiple homologous recombination and nonhomologous end-joining repair proteins, triggering robust double-stranded breaks, chromatin pulverization, and apoptosis. Consistently, Chk1 or Wee1 shRNA knockdown significantly sensitized AML cells to MLN4924. MLN4924/belinostat displayed activity against primary AML or MDS cells, including those carrying next-generation sequencing–defined poor-prognostic cancer hotspot mutations, and CD34+/CD38−/CD123+ populations, but not normal CD34+ progenitors. Finally, combined treatment markedly reduced tumor burden and significantly prolonged animal survival (P < .0001) in AML xenograft models with negligible toxicity, accompanied by pharmacodynamic effects observed in vitro. Collectively, these findings argue that MLN4924 and belinostat interact synergistically by reciprocally disabling the DDR in AML/MDS cells. This strategy warrants further consideration in AML/MDS, particularly in disease with unfavorable genetic aberrations.
Introduction
Despite the recent introduction of agents targeting mutant oncoproteins implicated in acute myelogenous leukemia (AML), for example, FLT3 inhibitors,1 outcomes with relapsed/refractory disease or adverse prognostic factors remain grim.2 Consequently, new approaches are urgently needed. Histone deacetylase (HDAC) inhibitors (HDACIs) are epigenetic agents that modify chromatin structure and regulate expression of differentiation- and cell death–related genes.3 However, HDACIs also acetylate diverse nonhistone proteins.3 Recently, attention has focused on HDACI-mediated DNA damage response (DDR) disruption.4 For example, HDACIs downregulate genes involved in checkpoints5,6 and DNA repair7,8 including homologous recombination (HR) and nonhomologous end-joining (NHEJ) repair.9 Several HDACIs, including vorinostat, romidepsin, and belinostat, have been approved for cutaneous T-cell lymphoma or peripheral T-cell lymphoma,10 and pracinostat was granted orphan drug status in AML.11 Whether HDACIs can improve established antileukemic agent efficacy remains uncertain.12
Nuclear factor–κB (NF-κB) represents a family of transcription factors involved in diverse cellular processes including cell proliferation, survival, among others,13 and plays an important role in AML stem cell survival.14 We and others have shown that HDACIs activate NF-κB in leukemia cells15 through a DNA damage-induced ataxia telangiectasia mutated (ATM)–NF-κB essential modulator (NEMO)–dependent process.16 Notably, preventing NF-κB activation (eg, by IκB kinase [IKK] inhibitors17 or proteasome inhibitors,18 which block degradation of the NF-κB–inhibitory protein IκBα)19 dramatically potentiates HDACI lethality. Although IKK inhibitors (eg, LC1)20 are at early stages of development, these findings have prompted trials combining HDACIs with proteasome inhibitors (eg, bortezomib) in AML.21 However, minimal proteasome inhibitor activity in AML22 may limit their use in this disease. Alternatively, the first-in-class NEDD8-activating enzyme (NAE) inhibitor MLN4924 has recently been shown to inhibit NF-κB in AML23 and diffuse large B-cell lymphoma (DLBCL) cells24 by blocking IκBα degradation.
The ubiquitin-proteasome system (UPS) represents 1 of the major degradative pathways that rid cells of unwanted or misfolded proteins. Protein ubiquitination is mediated by cullin-ring E3 ligases (CRLs), which require activation by neddylation to disrupt inhibitory associations with cullin-associated and neddylation-dissociated 1 (CAND1).25 Neddylation involves conjugation of the ubiquitin-like protein NEDD8 to target proteins, an event catalyzed by NAEs. Neddylation inhibition perturbs multiple proteins associated with both NF-κB and DDR pathways,25 prompting the development of NAE inhibitors such as MLN4924, currently in multiple trials. MLN4924 induces AML23 and DLBCL24 cell death in association with NF-κB inactivation, reactive oxygen species induction, DNA reduplication, and DNA damage.26,27 MLN4924 also potentiates the activity of chemotherapeutic agents in solid tumors,28,29 bortezomib in multiple myeloma,30 and ara-C in leukemia.31 Notably, MLN4924, unlike bortezomib,22 has single-agent activity in AML/myelodysplastic syndrome (MDS), with overall response rates of 17%.32 Collectively, these findings provide a theoretical rationale for combining MLN4924 and HDACIs in AML.
Currently, information concerning whether and by what mechanisms MLN4924 might interact with HDACIs is lacking. Here we report that MLN4924 and the HDACI belinostat interact synergistically in diverse AML cell types, including those harboring adverse prognostic genetic mutations and primitive leukemic progenitors, in association with reciprocal effects on NF-κB activation, the intra-S checkpoint, and DNA repair (eg, HR and NHEJ). These findings support further pursuit of an HDAC/NAE coinhibitory strategy in AML.
Materials and methods
Cells and reagents
Human AML cell lines U937 (p53-null), MV-4-11 (p53-mutant, FLT3–internal tandem duplication [ITD]), MOLM-13 (wild-type [wt]-p53, FLT3-ITD), and OCI-AML-3 (wt-p53) were maintained as before.6 Experiments used logarithmically growing cells (3-6 × 105 cells per mL). Bone marrow (BM) or peripheral blood samples were obtained with informed consent from patients with histologically documented AML undergoing routine diagnostic procedures (Virginia Commonwealth University Institutional Review Board approval #HM 12517). Primary AML (blasts >70% and viability >95%) or MDS samples and normal human cord blood (CB) CD34+ cells were isolated as before.6 Clinical, molecular, and cytogenetic characteristics of 6 selected patients are described in supplemental Table 1 (available on the Blood Web site).
The selective NAE inhibitor MLN4924 and the pan-HDAC inhibitor belinostat (Beleodaq, formerly PXD-101) were provided by Millennium Pharmaceuticals, Inc (Cambridge, MA) and Spectrum Pharmaceuticals (Henderson, NV), respectively. Reagents were formulated in dimethylsulfoxide (DMSO) and stored at −20°C. Final DMSO concentrations were <0.1%.
Next-generation sequencing
Mononuclear cells isolated from primary AML samples were analyzed by next-generation sequencing [NGS] using the Ion AmpliSeq Cancer Hotspot Panel v2, which surveys hotspot regions, including ∼2800 COSMIC mutations of 50 oncogenes and tumor suppressor genes (average amplicon length, 154 bp; average depth of coverage >2000x; coverage uniformity, ≥98%; on-target reads, ≥96%; single nucleotide polymorphism detection sensitivity, 98% detection rate for 5% variant frequency). NGS analyses used Ion Torrent targeted sequencing technology (Life Technologies, Grand Island, NY).
In vitro procedures
Please see supplemental Materials and methods.
Animal studies
Two mouse (The Jackson Laboratory) models were used: subcutaneous (s.c.) inoculation in the flank with 5 × 106 luciferase-labeled U937 cells or IV tail vein injection with 1 × 106 luciferase-labeled MV-4-11 cells. MLN4924 (reconstituted with 20% (2-hydroxypropyl)-β-cyclodextrin [HPbCD]) and belinostat were administrated by s.c. and intraperitoneal (i.p.) injection, respectively. Controls received equal vehicle volumes. Mice were monitored every other day after luciferin i.p. injection using an IVIS 200 imaging system (Xenogen Corporation, Alameda, CA). For the flank model, tumor size was measured by caliper and volumes calculated using the formula (L × W2)/2, with L = length and W = width. Body weights were measured every other day. When tumor size reached 1700 mm3 or other humane end points (eg, abscessed or necrotic tumors) reached, mice were euthanized in accordance with institutional guidelines.
Statistical analysis
Values represent the means ± standard deviation (SD) for at least 3 independent experiments performed in triplicate. Significance of differences between experimental variables was determined using the Student t test or 1-way analysis of variance with Tukey-Kramer multiple comparisons test (2-sided). Significance values were *P < .05 and **P < .01 wherever indicated. Analysis of synergism was performed by median dose-effect analysis using Calcusyn software (Biosoft, Ferguson, MO). Kaplan-Meier survival analysis used IBM SPSS Statistics software.
Results
MLN4924 and belinostat interact synergistically in AML cell lines with various genetic backgrounds, whereas FLT3-ITD or p53 knockdown increase sensitivity
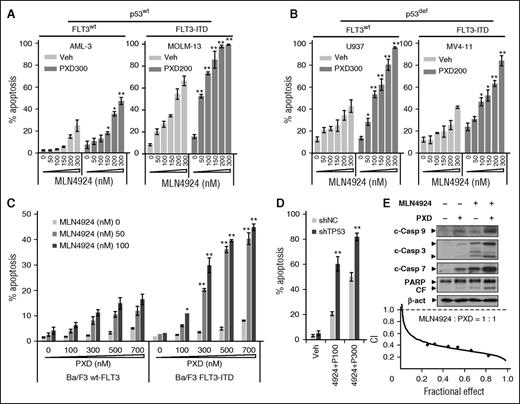
MLN4924 interactions with belinostat in leukemia cell lines expressing wt or mutant p53 and/or FLT3-ITD, adverse prognostic markers in AML,6 were first examined. Although sensitivity to MLN4924 alone varied between lines, MLN4924/belinostat coexposure significantly increased apoptosis in all AML lines (Figure 1A-B; supplemental Figure 1A), whereas p53-deficient cells (eg, U937; Figure 1B left panel) were more sensitive than wt p53 cells (eg, OCI-AML-3; Figure 1A left panel). Interestingly, wt-p53 cells carrying FLT3-ITD (eg, MOLM-13; Figure 1A right panel) were substantially more sensitive to MLN4924 ± belinostat compared with wt-p53 OCI-AML-3 cells without FLT3 mutation (Figure 1A left panel).
MLN4924 interacts synergistically with belinostat to induce apoptosis in both p53 wt and deficient leukemia cells. (A-B) p53wt OCI-AML-3 and MOLM-13 (FLT3-ITD), as well as U937 (p53-null) and MV-4-11 (p53mut, FLT-ITD), cells were exposed to the indicated concentrations of MLN4924 ± belinostat (PXD-101, nM) for 24 hours. (C) Murine Ba/F3 cells were stably transfected with constructs encoding either wt-FLT3 or FLT3-ITD. Cells were exposed to the indicated concentrations of PXD-101 ± MLN4924 for 24 hours. (D) wt-p53 OCI-AML-3 cells were stably transfected with shRNA constructs targeting human TP53 (shTP53) or scrambled sequence as a negative control (shNC), and then treated with 100 to 300 nM PXD-101 (P) ± 300 nM MLN4924 for 24 hours. For panels A-D, the percentage of Annexin V+ apoptotic cells was determined by FCM (*P < .05, **P < .01). (E) U937 cells were incubated with 300 nM PXD-101 ± 100 nM MLN4924 for 24 hours, after which cleavage of caspase 3, 7, 9, and PARP was monitored by western blot analysis (top panels). Alternatively, U937 cells were exposed (24 hours) to varying concentrations of MLN4924 ± PXD-101 at a fixed ratio (1:1), after which the percentage of Annexin V+ cells was determined, and median dose-effect analysis was then used to characterize the nature of the interaction between these agents. Combination index (CI) values <1.0 denote a synergistic interaction. The results are representative of 3 separate experiments. β-act, β-actin; c-Casp, cleaved caspase; CF, cleaved fragment; PXD, PXD-101; Veh, vehicle.
MLN4924 interacts synergistically with belinostat to induce apoptosis in both p53 wt and deficient leukemia cells. (A-B) p53wt OCI-AML-3 and MOLM-13 (FLT3-ITD), as well as U937 (p53-null) and MV-4-11 (p53mut, FLT-ITD), cells were exposed to the indicated concentrations of MLN4924 ± belinostat (PXD-101, nM) for 24 hours. (C) Murine Ba/F3 cells were stably transfected with constructs encoding either wt-FLT3 or FLT3-ITD. Cells were exposed to the indicated concentrations of PXD-101 ± MLN4924 for 24 hours. (D) wt-p53 OCI-AML-3 cells were stably transfected with shRNA constructs targeting human TP53 (shTP53) or scrambled sequence as a negative control (shNC), and then treated with 100 to 300 nM PXD-101 (P) ± 300 nM MLN4924 for 24 hours. For panels A-D, the percentage of Annexin V+ apoptotic cells was determined by FCM (*P < .05, **P < .01). (E) U937 cells were incubated with 300 nM PXD-101 ± 100 nM MLN4924 for 24 hours, after which cleavage of caspase 3, 7, 9, and PARP was monitored by western blot analysis (top panels). Alternatively, U937 cells were exposed (24 hours) to varying concentrations of MLN4924 ± PXD-101 at a fixed ratio (1:1), after which the percentage of Annexin V+ cells was determined, and median dose-effect analysis was then used to characterize the nature of the interaction between these agents. Combination index (CI) values <1.0 denote a synergistic interaction. The results are representative of 3 separate experiments. β-act, β-actin; c-Casp, cleaved caspase; CF, cleaved fragment; PXD, PXD-101; Veh, vehicle.
To explore the role of FLT3-ITD in MLN4924/belinostat sensitivity, murine Ba/F3 cells were transfected with constructs encoding either wt-FLT3 or FLT-ITD. Notably, FLT3-ITD expression significantly sensitized Ba/F3 cells to MLN4924 with belinostat (Figure 1C, 24 hours) associated with markedly increased γH2A.X induction (supplemental Figure 1B), a DNA double-stranded break (DSB) marker,6 or MLN4924 alone following longer exposures (eg, 48-72 hours; data not shown). Interestingly, MLN4924 alone modestly increased Y591 phosphorylation of FLT3 in MV-4-11 cells with FLT3-ITD, which was sharply blocked by belinostat in association with FLT3 protein downregulation (supplemental Figure 1C). Moreover, the selective FLT3 inhibitor AC220 (quizartinib), at very low subtoxic concentrations (eg, 1-3 nM), significantly enhanced MLN4924/belinostat lethality in these cells (supplemental Figure 1D). To evaluate p53 functional significance, p53 was stably knocked down using short hairpin RNA (shRNA) in wt-p53 OCI-AML-3 cells, which did not disrupt cell cycle checkpoints (eg, phosphorylated [p] Wee1 and Chk1 [p-Wee1 and p-Chk1]; supplemental Figure 1E). Isogenic p53 knockdown clearly increased MLN4924/belinostat lethality compared with controls (shRNA scrambled sequence as negative control [shNC]; Figure 1D). Consistently, the regimen was highly active in p53-deficient AML cells, for example, U937 and MV-4-11 cells (Figure 1B), as well as HL-60, and KG-1 cells (data not shown).
Combined MLN4924/belinostat treatment triggered caspase cascade activation and poly ADP ribose polymerase (PARP) cleavage (Figure 1E; supplemental Figure 1F), whereas median dose-effect analysis yielded CI values <1.0, denoting synergism,33 in U937 cells (Figure 1E) and other lines, or when other HDACIs (eg, suberic bishydroxamic acid) were used (supplemental Figure 1G). These findings indicate that MLN4924 and belinostat interact synergistically in AML cells, and also suggest that cells with either p53 deficiency or FLT3-ITD may be particularly sensitive to this strategy.
MLN4924 prevents HDACI-induced NF-κB activation whereas Bim knockdown attenuates apoptosis induced by cotreatment with MLN4924 and belinostat
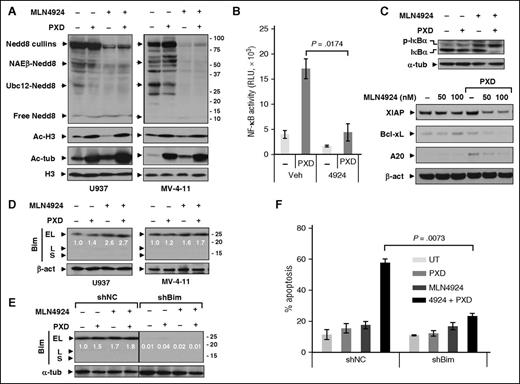
Consistent with previous reports,34 whereas belinostat had modest effects, MLN4924 (± belinostat) inhibited protein neddylation (Figure 2A), reflected by diminished expression of Nedd8-cullins, NAEβ-Nedd8, and Ubc12-Nedd8. Belinostat increased both histone H3 and tubulin acetylation, indicating class I and class IIb HDAC inhibition, respectively (Figure 2A).
Coadministration of MLN4924 with belinostat disrupts NF-κB activation whereas Bim knockdown markedly attenuates apoptosis induced by this regimen. (A) U937 and MV-4-11 cells were exposed to PXD-101 (U937, 300 nM; MV-4-11, 200 nM) ± 100 nM MLN4924 for 24 hours, after which western blot analysis was performed using antibodies that specifically recognize neddylated (Nedd8) or acetylated (Ac) proteins. (B) U937 cells were incubated with 300 nM PXD-101 ± 100 nM MLN4924 for 16 hours, after which nuclear extracts were prepared and subjected to the TransAM NF-κB p65–DNA-binding enzyme-linked immunosorbent assay (ELISA) as per the manufacturer’s instruction. (C) Alternatively, total and S32/34 phosphorylated IκBα (top panels) as well as expression of NF-κB–dependent genes (bottom panels) were monitored by western blot analysis in U937 cells. (D) U937 and MV-4-11 cells were treated for 24 hours as described in panel A, after which western blot analysis was performed to monitor expression of the BH3-only proapoptotic protein Bim (including EL, L, and S isoforms). (E-F) U937 cells stably transfected with constructs encoding shRNA targeting human BCL2L11/Bim (shBim) or scrambled sequence as negative control (shNC) were treated with 300 nM PXD-101 ± 100 nM MLN4924 for 24 hours, after which western blot analysis and FCM were carried out to monitor Bim expression (E) and apoptosis (Annexin V, determined by flow cytometry) (F), respectively. For panels D and E, blots for BimEL were quantified using ImageJ software. Values indicate fold-increase of BimEL vs untreated control (arbitrarily set as 1.0), after normalization to β-actin or α-tubulin (α-tub). RLU, relative light unit; UT, untreated control.
Coadministration of MLN4924 with belinostat disrupts NF-κB activation whereas Bim knockdown markedly attenuates apoptosis induced by this regimen. (A) U937 and MV-4-11 cells were exposed to PXD-101 (U937, 300 nM; MV-4-11, 200 nM) ± 100 nM MLN4924 for 24 hours, after which western blot analysis was performed using antibodies that specifically recognize neddylated (Nedd8) or acetylated (Ac) proteins. (B) U937 cells were incubated with 300 nM PXD-101 ± 100 nM MLN4924 for 16 hours, after which nuclear extracts were prepared and subjected to the TransAM NF-κB p65–DNA-binding enzyme-linked immunosorbent assay (ELISA) as per the manufacturer’s instruction. (C) Alternatively, total and S32/34 phosphorylated IκBα (top panels) as well as expression of NF-κB–dependent genes (bottom panels) were monitored by western blot analysis in U937 cells. (D) U937 and MV-4-11 cells were treated for 24 hours as described in panel A, after which western blot analysis was performed to monitor expression of the BH3-only proapoptotic protein Bim (including EL, L, and S isoforms). (E-F) U937 cells stably transfected with constructs encoding shRNA targeting human BCL2L11/Bim (shBim) or scrambled sequence as negative control (shNC) were treated with 300 nM PXD-101 ± 100 nM MLN4924 for 24 hours, after which western blot analysis and FCM were carried out to monitor Bim expression (E) and apoptosis (Annexin V, determined by flow cytometry) (F), respectively. For panels D and E, blots for BimEL were quantified using ImageJ software. Values indicate fold-increase of BimEL vs untreated control (arbitrarily set as 1.0), after normalization to β-actin or α-tubulin (α-tub). RLU, relative light unit; UT, untreated control.
MLN4924 inhibits NF-κB by blocking IκBα ubiquitination and degradation.24 To determine whether MLN4924 inhibited HDACI-mediated NF-κB activation,35 a RelA/p65–DNA-binding assay was performed. Whereas belinostat markedly induced NF-κB activation (more than threefold), this was blocked by MLN4924 (Figure 2B) in association with phosphorylated IκBα accumulation (Figure 2C), reflecting diminished degradation. Additionally, MLN4924 downregulated multiple NF-κB–dependent antiapoptotic proteins (eg, X-linked inhibitor of apoptosis protein [XIAP], Bcl-xL, and A20) in belinostat-treated cells (Figure 2C). Combined treatment also inhibited RelA/p65 phosphorylation (S536) (supplemental Figure 2A) and NF-κB2/p100 precursor accumulation (supplemental Figure 2B), reflecting failure of proteasome-mediated processing to p52, essential for noncanonical NF-κB pathway activation.13
Given evidence that HDACIs upregulate proapoptotic Bim,36 the contribution of this BH3-only protein to MLN4924/belinostat interactions was then examined. Although each agent (particularly MLN4924) alone upregulated Bim in U937 (Figure 2D) and HL-60 cells (supplemental Figure 2C), and to a lesser extent in MV-4-11 cells (Figure 2D), combined treatment did not further increase Bim levels in these cells. Notably, other BH3-only proteins remained either unchanged or in some cases downregulated after drug treatment, whereas the multidomain protein Bak, but not Bax, was upregulated (supplemental Figure 2D). Moreover, shRNA prevention of Bim upregulation (Figure 2E) dramatically diminished MLN4924/belinostat-induced apoptosis in both U937 (24 hours, Figure 2F, and 48 hours, supplemental Figure 2E) and MV-4-11 cells (supplemental Figure 2F), but interestingly, did not affect attenuation of colony formation by this regimen (supplemental Figure 2G) or cell cycle checkpoints (supplemental Figure 2H). Together, these findings suggest roles for NF-κB inactivation and Bim in MLN4924/belinostat lethality.
Belinostat blocks ATR/Chk1/Wee1-mediated checkpoint activation by MLN4924, accompanied by robust DNA damage
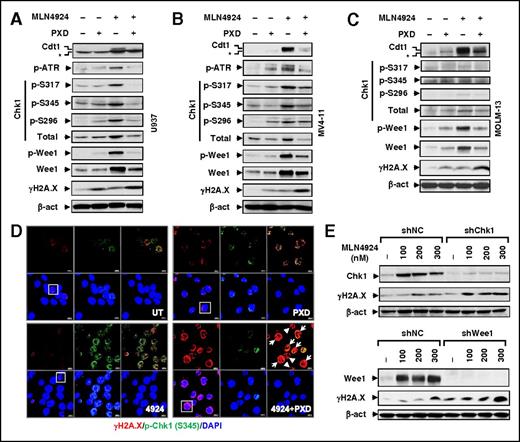
DDR disruption is involved in both HDACI and MLN4924 mechanisms of action (MOAs). An RT2 Profiler polymerase chain reaction array was performed to profile expression of DDR-related genes. Combined MLN4924/belinostat treatment yielded a profile distinct from each agent alone (supplemental Figure 3A), indicating disparate effects on the DDR. Consistent with previous reports,37 MLN4924 upregulated the DNA replication factor Cdt1 (Figure 3A-C), a substrate of cullins 1 and 4, an event related to Chk1 activation.38 Indeed, MLN4924 induced phosphorylations of Chk1 (S317, S345, and S296), ataxia-telangiectasia and Rad3-related (ATR) (supplemental Figure 3B, not detected in MOLM-13 cells), and Wee1, in association with increased protein levels of Chk1, Wee1 (Figure 3A-C), and to a lesser extent ATR (supplemental Figure 3C), but not phosphorylation of Chk2 (T68, supplemental Figure 3D). Notably, these actions were sharply attenuated by belinostat, associated with robust DNA damage induction, including both DSB (eg, γH2A.X, Figure 3A-C) and single-stranded breaks as detected by Alkaline Comet Assay (supplemental Figure 3E). Confocal microscopy revealed that although belinostat had modest effects, MLN4924 robustly triggered p-Chk1 (S345) foci formation, many of which colocalized with γH2A.X foci (Figure 3D), suggesting Chk1 activation at DSB sites. Notably, belinostat dramatically reduced MLN4924-induced p-Chk1 foci formation while markedly increasing γH2A.X foci (Figure 3D and supplemental Figure 3F-G, P < .05 for each case). Significantly, attenuation of MLN4924-induced Chk1 or Wee1 expression by shRNA sharply increased DNA damage (eg, γH2A.X, Figure 3E), suggesting that belinostat-mediated checkpoint blockade contributes to MLN4924 interactions.
Belinostat attenuates activation of the ATR/Chk1/Wee1-mediated DNA damage checkpoint pathway by MLN4924. (A-C) Cells were treated (24 hours) with PXD-101 (U937, 300 nM; MV-4-11 and MOLM-13, 200 nM) ± 100 nM MLN4924, after which western blot analysis was performed to monitor expression of Cdt1 (top band, *nonspecific bands), total and/or phosphorylated ATR (S428), Chk1, Wee1 (S642), H2A.X (S139). (D) MV-4-11 cells were treated with 200 nM PXD-101 ± 100 nM MLN4924 for 16 hours, after which cytospin slides were prepared and stained with the primary antibodies against p-Chk1 (S345) and γH2A.X, followed by Alexa Fluor 488– or Alexa Fluor 594–conjugated secondary antibody. Images (×64) were captured using a Zeiss LSM 700 confocal laser-scanning microscope. Arrow, Cells containing γH2A.X foci with no or few p-Chk1 colocalization foci; arrowhead, apoptotic cells. (Top panels) Left, γH2A.X (red); middle, p-Chk1 (green); right, γH2A.X and p-Chk1 merged. (Bottom panels) Left, γH2A.X and DAPI merged; middle, p-Chk1 and DAPI merged; right, DAPI (blue). Scale bars, 10 μm. Squares indicate areas corresponding to images with higher magnification as shown in supplemental Figure 3F. (E) U937 cells were stably transfected with constructs encoding shRNA targeting human CHEK1/Chk1 (shChk1), Wee1 (shWee1), or scrambled sequence (shNC). Cells were then treated with the indicated concentrations of MLN4924 for 24 hours, after which western blot analysis was performed to monitor expression of Chk1, Wee1, and γH2A.X.
Belinostat attenuates activation of the ATR/Chk1/Wee1-mediated DNA damage checkpoint pathway by MLN4924. (A-C) Cells were treated (24 hours) with PXD-101 (U937, 300 nM; MV-4-11 and MOLM-13, 200 nM) ± 100 nM MLN4924, after which western blot analysis was performed to monitor expression of Cdt1 (top band, *nonspecific bands), total and/or phosphorylated ATR (S428), Chk1, Wee1 (S642), H2A.X (S139). (D) MV-4-11 cells were treated with 200 nM PXD-101 ± 100 nM MLN4924 for 16 hours, after which cytospin slides were prepared and stained with the primary antibodies against p-Chk1 (S345) and γH2A.X, followed by Alexa Fluor 488– or Alexa Fluor 594–conjugated secondary antibody. Images (×64) were captured using a Zeiss LSM 700 confocal laser-scanning microscope. Arrow, Cells containing γH2A.X foci with no or few p-Chk1 colocalization foci; arrowhead, apoptotic cells. (Top panels) Left, γH2A.X (red); middle, p-Chk1 (green); right, γH2A.X and p-Chk1 merged. (Bottom panels) Left, γH2A.X and DAPI merged; middle, p-Chk1 and DAPI merged; right, DAPI (blue). Scale bars, 10 μm. Squares indicate areas corresponding to images with higher magnification as shown in supplemental Figure 3F. (E) U937 cells were stably transfected with constructs encoding shRNA targeting human CHEK1/Chk1 (shChk1), Wee1 (shWee1), or scrambled sequence (shNC). Cells were then treated with the indicated concentrations of MLN4924 for 24 hours, after which western blot analysis was performed to monitor expression of Chk1, Wee1, and γH2A.X.
Belinostat abrogates MLN4924-induced intra-S checkpoint and attenuates HR and NHEJ DNA repair pathway activation
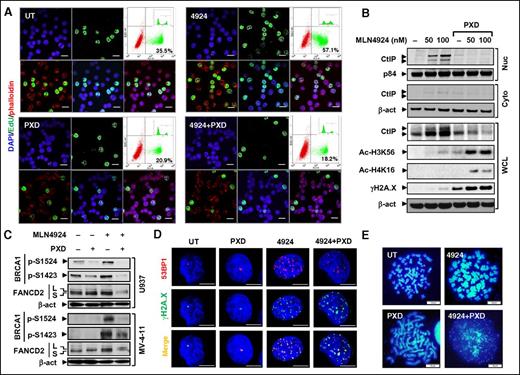
MLN4924 treatment markedly increased 5-ethynyl-2′-deoxyuridine (EdU) incorporation into DNA (Figure 4A and supplemental Figure 4A, P < .01) and S-phase cells (data not shown), consistent with DNA re-replication through replication licensing factor Cdt1 stabilization and the intra-S checkpoint.38 Significantly, belinostat abrogated these events (P < .01).
Cotreatment with MLN4924 and belinostat disrupts the intra-S-phase checkpoint and HR or NHEJ repairs. (A) MV-4-11 cells were treated with 200 nM PXD-101 ± 100 nM MLN4924 for 16 hours, after which cells were pulse-labeled with EdU for 30 minutes, followed by double staining for EdU and F-actin (Alexa Fluor phalloidin) with DAPI counterstaining. (Top panels) Left, DAPI (blue); middle, EdU (green); right, dot plot of FCM determining the percentage of EdU-positive cells (inset, histogram). (Bottom panels) Left, EdU and phalloidin (red) merged; middle, EdU and DAPI merged; right, EdU, phalloidin, and DAPI merged. Scale bars, 20 μm. (B) After exposure to 300 nM PXD-101 ± 50 to 100 nM MLN4924 for 16 hours, whole-cell lysates (WCL) and nuclear (Nuc) or cytosolic (Cyto) fractions of U937 cells were prepared and subjected to western blot analysis using the indicated antibodies. (C) MV-4-11 and U937 cells were treated as described in panels A and B, respectively, after which western blot analysis was performed to assess BRCA1 phosphorylation (S1524, S1423) and FANCD2 ubiquitination (S, an inactive unubiquitinated isoform; L, an active monoubiquitinated isoform). (D) U937 cells were incubated with 300 nM MLN4924 ± 100 nM PXD-101 for 16 hours, followed by double staining for 53BP1 (red) and γH2A.X (green) with DAPI counterstaining. Scale bars, 10 μm. (E) U937 cells were treated with 300 nM PXD-101 ± 100 nM for 16 hours, after which chromosome-spreading analysis (DAPI staining) was carried out to monitor chromosome morphology and pulverization, characterized by fragments of DAPI staining material that were clustered into a discrete location on the spread. Scale bars, 10 μm.
Cotreatment with MLN4924 and belinostat disrupts the intra-S-phase checkpoint and HR or NHEJ repairs. (A) MV-4-11 cells were treated with 200 nM PXD-101 ± 100 nM MLN4924 for 16 hours, after which cells were pulse-labeled with EdU for 30 minutes, followed by double staining for EdU and F-actin (Alexa Fluor phalloidin) with DAPI counterstaining. (Top panels) Left, DAPI (blue); middle, EdU (green); right, dot plot of FCM determining the percentage of EdU-positive cells (inset, histogram). (Bottom panels) Left, EdU and phalloidin (red) merged; middle, EdU and DAPI merged; right, EdU, phalloidin, and DAPI merged. Scale bars, 20 μm. (B) After exposure to 300 nM PXD-101 ± 50 to 100 nM MLN4924 for 16 hours, whole-cell lysates (WCL) and nuclear (Nuc) or cytosolic (Cyto) fractions of U937 cells were prepared and subjected to western blot analysis using the indicated antibodies. (C) MV-4-11 and U937 cells were treated as described in panels A and B, respectively, after which western blot analysis was performed to assess BRCA1 phosphorylation (S1524, S1423) and FANCD2 ubiquitination (S, an inactive unubiquitinated isoform; L, an active monoubiquitinated isoform). (D) U937 cells were incubated with 300 nM MLN4924 ± 100 nM PXD-101 for 16 hours, followed by double staining for 53BP1 (red) and γH2A.X (green) with DAPI counterstaining. Scale bars, 10 μm. (E) U937 cells were treated with 300 nM PXD-101 ± 100 nM for 16 hours, after which chromosome-spreading analysis (DAPI staining) was carried out to monitor chromosome morphology and pulverization, characterized by fragments of DAPI staining material that were clustered into a discrete location on the spread. Scale bars, 10 μm.
Whether MLN4924 ± belinostat might also affect HR and NHEJ repair, 2 major forms of DNA repair,9 was then examined. Interestingly, MLN4924 activated multiple HR-related pathways, including (1) marked nuclear accumulation of the DNA-end resection protein C-terminal binding protein (CtBP)-interacting protein (CtIP) (Figure 4B); (2) BRCA1 phosphorylation (S1524 and S1423; Figure 4C), reflecting HR pathway activation,39 associated with increased protein levels (supplemental Figure 3C); and (3) increased FANCD2-L (top band, Figure 4C), an active monoubiquitinated form of FANCD2, indicating Fanconi anemia pathway activation.40 Significantly, these HR-related events were largely blocked by belinostat (Figure 4B-C), consistent with earlier reports.41 In addition, MLN4924 and belinostat coadministration also downregulated other DNA repair proteins (eg, 53BP1 and RAD51) (supplemental Figures 3C and 4B) and attenuated their foci formation (Figure 4D; supplemental Figure 4C). Moreover, MLN4924/belinostat cotreatment promoted histone 3 K56 (H3K56) and histone 4 K16 (H4K16) acetylation (Figure 4B), reflecting defective HDAC1/2-mediated NHEJ repair.9 Inhibition of HDACI-induced NF-κB activation (eg, by the IKK inhibitor Bay 11-7082) sharply increased H3K56 and H4K16 acetylation, accompanied by robust DNA damage (supplemental Figure 4D), suggesting that impaired NHEJ repair might be associated with NF-κB inactivation.
Metaphase spread analysis revealed chromosome aberrations, including chromosome thickening or elongation following MLN4924 or belinostat exposure, respectively (Figure 4E), the latter presumably reflecting chromosome decondensation following histone hyperacetylation. Significantly, combined treatment induced chromosome pulverization in U937 (Figure 4E, in 35.7% ± 2.5% cells vs none in control or single-agent treatment) and MV-4-11 cells (supplemental Figure 4E), characterized by 4,6 diamidino-2-phenylindole (DAPI)-positive fragments clustered into discrete locations on the spread.42 Together, these findings indicate that MLN4924/belinostat coadministration disrupts the intra-S checkpoint as well as both HR and NHEJ repair pathways, triggering pronounced DNA damage and apoptosis.
The MLN4924/belinostat regimen targets AML blasts carrying NGS-defined cancer hotspot mutations, as well as populations enriched for leukemia-initiating cells, but not normal hematopoietic cells
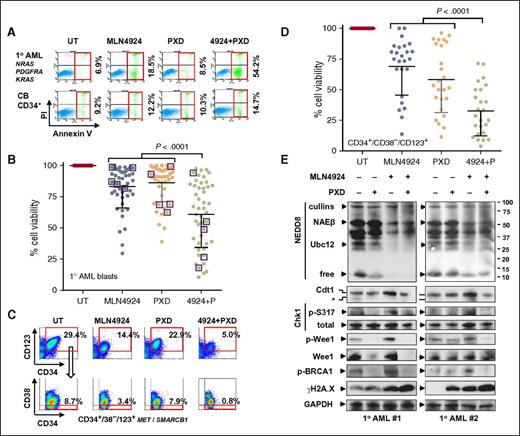
To characterize the activity and selectivity of the MLN4924/belinostat regimen, studies were performed in patient-derived leukemic cells vs normal hematopoietic cells.43 In representative AML specimens carrying NGS-defined cancer hotspot mutations of the indicated genes, MLN4924/belinostat coadministration sharply induced apoptosis (Annexin V+) of AML (Figure 5A top panels; supplemental Figure 5A-C) and MDS cells (supplemental Figure 6A-C; similar results were obtained in embryonic stem cells of NUP98-HOXD13 transgenic mice that recapitulate key human MDS features,44 and counterparts with activating NRAS mutations,45 supplemental Figure 6D-E). MDS cells are identified as CD34+ BM mononuclear cells of MDS patients.46 In contrast, identical regimens did not significantly increase cell death in normal CB CD34+ cells (Figure 5A bottom panels; supplemental Figure 5D). Parallel studies performed in a total of 47 specimens revealed a significant reduction in AML or MDS cell viability with combined treatment (Figure 5B, P < .0001 vs each single agent). Moreover, human HS-5 stromal cell coculture failed to protect AML blasts from MLN4924/belinostat lethality (supplemental Figure 5E).
The MLN4924/belinostat regimen is active against primary AML/MDS cells and diminishes the primitive CD34+/CD38−/CD123+ population enriched for LICs, while displaying minimal toxicity toward normal CD34+ cells. (A) A representative primary sample from a patient with AML carrying the indicated cancer hotspot mutations (top panels) and normal CB CD34+ cells (bottom panels) was exposed to 300 nM PXD-101 ± 200 nM MLN4924 for 24 hours, after which the percentage of Annexin V+ cells was determined by FCM. (B) Parallel experiments were carried out with 47 primary samples (AML = 42; MDS = 5, indicated by squares, treatment with 500 nM PXD-101 ± 500 nM MLN4924). Cell viability was analyzed by 7-aminoactinomycin D (7AAD) staining and FCM. Lines indicate means and SD. (C) After exposure to 300 nM PXD-101 ± 200 nM MLN4924 for 24 hours, the percentage of the primitive CD34–phycoerythrin-positive (PE+)/CD38–peridinin chlorophyll protein-negative (PerCP−)/CD123–allophycocyanin-positive (APC+) population, which is enriched for LICs, was determined by multicolor FCM in BM mononuclear cells from a primary AML sample carrying the indicated cancer hotspot mutations. (D) Parallel experiments were carried out with 25 primary AML samples. Viability of CD34+/CD38−/CD123+ cells was analyzed by 7AAD staining and FCM. (E) Alternatively, western blot analysis was performed to monitor the selected markers for inhibition of neddylation, disruption of the DDR, and induction of DNA damage in 2 representative primary samples that responded to the MLN4924/belinostat regimen. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PI, propidium iodide.
The MLN4924/belinostat regimen is active against primary AML/MDS cells and diminishes the primitive CD34+/CD38−/CD123+ population enriched for LICs, while displaying minimal toxicity toward normal CD34+ cells. (A) A representative primary sample from a patient with AML carrying the indicated cancer hotspot mutations (top panels) and normal CB CD34+ cells (bottom panels) was exposed to 300 nM PXD-101 ± 200 nM MLN4924 for 24 hours, after which the percentage of Annexin V+ cells was determined by FCM. (B) Parallel experiments were carried out with 47 primary samples (AML = 42; MDS = 5, indicated by squares, treatment with 500 nM PXD-101 ± 500 nM MLN4924). Cell viability was analyzed by 7-aminoactinomycin D (7AAD) staining and FCM. Lines indicate means and SD. (C) After exposure to 300 nM PXD-101 ± 200 nM MLN4924 for 24 hours, the percentage of the primitive CD34–phycoerythrin-positive (PE+)/CD38–peridinin chlorophyll protein-negative (PerCP−)/CD123–allophycocyanin-positive (APC+) population, which is enriched for LICs, was determined by multicolor FCM in BM mononuclear cells from a primary AML sample carrying the indicated cancer hotspot mutations. (D) Parallel experiments were carried out with 25 primary AML samples. Viability of CD34+/CD38−/CD123+ cells was analyzed by 7AAD staining and FCM. (E) Alternatively, western blot analysis was performed to monitor the selected markers for inhibition of neddylation, disruption of the DDR, and induction of DNA damage in 2 representative primary samples that responded to the MLN4924/belinostat regimen. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PI, propidium iodide.
The MLN4924/belinostat combination was also evaluated in populations enriched for leukemia-initiating cells (LICs), postulated to contribute to drug resistance and disease recurrence.14 Multicolor flow cytometry (FCM) revealed that combined treatment sharply reduced CD34+/CD38−/CD123+ cells within the bulk population (Figure 5C), in association with pronounced apoptosis in this primitive population (supplemental Figure 7A-D; genetic mutations indicated), enriched for LICs.6 Parallel studies were performed in 25 AML samples in which the CD34+/CD38−/CD123+ population was detectable. Combined treatment strikingly decreased primitive AML cell viability (Figure 5D, P < .0001 vs each agent alone).
To assess relationships between response and genetic mutations in primary samples, NGS using the Cancer Hotspot Panel v2, which surveys hotspot regions of 50 oncogenes and tumor suppressor genes, was performed in 16 AML specimens (Table 1). Interestingly, bulk blast populations carrying FLT3 mutations responded well to the MLN4924/belinostat regimen, whereas those bearing IDH2 mutations were less responsive. Interestingly, although leukemic blast responses varied with different genetic backgrounds, CD34+/CD38−/CD123+ cells responded to the combination regimen in virtually all samples, regardless of genetic aberrations including IDH2 mutations (patients 14 and 15).
Relation between NGS-defined cancer hotspot mutations and response to MLN4924/belinostat in primary AML samples (n = 16)
| Patient . | Gene . | CDS_mut . | AA_mut . | Frequency, % . | Viability, %* . | |
|---|---|---|---|---|---|---|
| Blast . | LIC . | |||||
| 1 | ATM | c.998C>T | p.S333F | 46 | 13.7 | 8.8 |
| FLT3 | FLT3-ITD | FLT3-ITD | 27 | |||
| NPM1 | c.863_864insTCTG | p.W288fs*12 | 48 | |||
| SMARCB1 | c.1119-41G>A | (Unknown) | 51 | |||
| 2 | FLT3 | FLT3-ITD | FLT3-ITD | 8 | 15.1 | 14.9 |
| HRAS | c.81T>C | p.H27H | 50 | |||
| NPM1 | c.863_864insTCTG | p.W288fs*12 | 43 | |||
| 3 | KDR | c.1416A>T | p.Q472H | 100 | 18.0 | 70.8 |
| KIT | c.1621A>C | p.M541L | 49 | |||
| NRAS | c.34G>A | p.G12S | 64 | |||
| PDGFRA | c.2472C>T | p.V824V | 47 | |||
| 4 | HRAS | c.81T>C | p.H27H | 49 | 32.5 | — |
| KRAS | c.35G>A | p.G12D | 8.2 | |||
| NRAS | c.181C>A | p.Q61K | 8.5 | |||
| 5 | HRAS | c.81T>C | p.H27H | 48 | 39.1 | 59.7 |
| KIT | c.2447A>T | p.D816V | 48 | |||
| 6 | MET | c.1124A>G | p.N375S | 6 | 43.9 | 40.1 |
| KRAS | c.35G>T | p.G12V | 2 | |||
| FLT3 | c.2504A>T | p.D835V | 3 | |||
| STK11 | c.1062C>G | p.F354L | 42 | |||
| 7 | TP53 | c.722C>T | p.S241F | 97 | 52.9 | 53.8 |
| 8 | NRAS | c.35G>A | p.G12D | 4 | 55.1 | 43.6 |
| PDGFRA | c.2472C>T | p.V824V | 49 | |||
| KRAS | c.182A>C | p.Q61P | 13 | |||
| KRAS | c.38G>A | p.G13D | 15 | |||
| 9 | BRAF | c.1799T>A | p.V600E | 29 | 59.3 | — |
| SMARCB1 | c.1119-41G>A | (Unknown) | 50 | |||
| 10 | MET | c.3029C>T | p.T1010I | 50 | 60.8 | 9.2 |
| SMARCB1 | c.1119-41G>A | (Unknown) | 52 | |||
| 11 | FLT3 | c.2503G>G | p.D835H | 57 | 69.6 | — |
| HRAS | c.81T>C | p.H27H | 51 | |||
| IDH1 | c.395G>A | p.R132H | 48 | |||
| IDH1 | c.315C>T | p.G105G | 51 | |||
| KDR | c.1416A>T | p.Q472H | 51 | |||
| NPM1 | c.863_864insTCTG | p.W288fs*12 | 44 | |||
| 12 | APC | c.4326T>A | p.P1442P | 43 | 78.8 | — |
| 13 | SMARCB1 | c.1119-41G>A | (Unknown) | 51 | 85.1 | 21.4 |
| 14 | HRAS | c.81T>C | p.H27H | 50 | 88.1 | — |
| IDH2 | c.419G>A | p.R140Q | 47 | |||
| STK11 | c.1062C>G | p.F354L | 49 | |||
| 15 | IDH2 | c.419G>A | p.R140Q | 49 | 92.3 | 11.1 |
| NRAS | c.38G>A | p.G13D | 42 | |||
| PDGFRA | c.2472C>T | p.V824V | 47 | |||
| STK11 | c.816C>T | p.Y272Y | 100 | |||
| TP53 | c.216_217delCCinsGT | p.P72R | 52 | |||
| 16 | NPM1 | c.863_864TCTG | p.W288fs*12 | 49 | 94.4 | — |
| Patient . | Gene . | CDS_mut . | AA_mut . | Frequency, % . | Viability, %* . | |
|---|---|---|---|---|---|---|
| Blast . | LIC . | |||||
| 1 | ATM | c.998C>T | p.S333F | 46 | 13.7 | 8.8 |
| FLT3 | FLT3-ITD | FLT3-ITD | 27 | |||
| NPM1 | c.863_864insTCTG | p.W288fs*12 | 48 | |||
| SMARCB1 | c.1119-41G>A | (Unknown) | 51 | |||
| 2 | FLT3 | FLT3-ITD | FLT3-ITD | 8 | 15.1 | 14.9 |
| HRAS | c.81T>C | p.H27H | 50 | |||
| NPM1 | c.863_864insTCTG | p.W288fs*12 | 43 | |||
| 3 | KDR | c.1416A>T | p.Q472H | 100 | 18.0 | 70.8 |
| KIT | c.1621A>C | p.M541L | 49 | |||
| NRAS | c.34G>A | p.G12S | 64 | |||
| PDGFRA | c.2472C>T | p.V824V | 47 | |||
| 4 | HRAS | c.81T>C | p.H27H | 49 | 32.5 | — |
| KRAS | c.35G>A | p.G12D | 8.2 | |||
| NRAS | c.181C>A | p.Q61K | 8.5 | |||
| 5 | HRAS | c.81T>C | p.H27H | 48 | 39.1 | 59.7 |
| KIT | c.2447A>T | p.D816V | 48 | |||
| 6 | MET | c.1124A>G | p.N375S | 6 | 43.9 | 40.1 |
| KRAS | c.35G>T | p.G12V | 2 | |||
| FLT3 | c.2504A>T | p.D835V | 3 | |||
| STK11 | c.1062C>G | p.F354L | 42 | |||
| 7 | TP53 | c.722C>T | p.S241F | 97 | 52.9 | 53.8 |
| 8 | NRAS | c.35G>A | p.G12D | 4 | 55.1 | 43.6 |
| PDGFRA | c.2472C>T | p.V824V | 49 | |||
| KRAS | c.182A>C | p.Q61P | 13 | |||
| KRAS | c.38G>A | p.G13D | 15 | |||
| 9 | BRAF | c.1799T>A | p.V600E | 29 | 59.3 | — |
| SMARCB1 | c.1119-41G>A | (Unknown) | 50 | |||
| 10 | MET | c.3029C>T | p.T1010I | 50 | 60.8 | 9.2 |
| SMARCB1 | c.1119-41G>A | (Unknown) | 52 | |||
| 11 | FLT3 | c.2503G>G | p.D835H | 57 | 69.6 | — |
| HRAS | c.81T>C | p.H27H | 51 | |||
| IDH1 | c.395G>A | p.R132H | 48 | |||
| IDH1 | c.315C>T | p.G105G | 51 | |||
| KDR | c.1416A>T | p.Q472H | 51 | |||
| NPM1 | c.863_864insTCTG | p.W288fs*12 | 44 | |||
| 12 | APC | c.4326T>A | p.P1442P | 43 | 78.8 | — |
| 13 | SMARCB1 | c.1119-41G>A | (Unknown) | 51 | 85.1 | 21.4 |
| 14 | HRAS | c.81T>C | p.H27H | 50 | 88.1 | — |
| IDH2 | c.419G>A | p.R140Q | 47 | |||
| STK11 | c.1062C>G | p.F354L | 49 | |||
| 15 | IDH2 | c.419G>A | p.R140Q | 49 | 92.3 | 11.1 |
| NRAS | c.38G>A | p.G13D | 42 | |||
| PDGFRA | c.2472C>T | p.V824V | 47 | |||
| STK11 | c.816C>T | p.Y272Y | 100 | |||
| TP53 | c.216_217delCCinsGT | p.P72R | 52 | |||
| 16 | NPM1 | c.863_864TCTG | p.W288fs*12 | 49 | 94.4 | — |
— represents CD34+/CD38−/CD123+ cells undetectable.
AA_mut, amino acid mutation; CDS_mut, coding DNA sequence mutation; LIC, CD34+/CD38−/CD123+ cells enriched for leukemia-initiating cells.
Cell viability after combined treatment with MLN4924 and belinostat, compared with untreated control, for which value is arbitrarily set as 100%.
Finally, western blot analysis was conducted in 2 representative AML specimens for which sufficient blasts were available. Whereas MLN4924 ± belinostat attenuated NAEβ and Ubc12 neddylation, belinostat sharply diminished MLN4924-mediated expression of Cdt1, p-Chk1 (S317), p-Wee1, and p-BRCA1 (S1524; S1423 phosphorylation not detected in these primary samples) (Figure 5E). Moreover, combined treatment induced robust DNA damage, reflected by increased γH2A.X expression. These findings argue that a regimen combining MLN4924 and belinostat is active against both primary AML/MDS cells and primitive cell populations enriched for LICs, including those harboring unfavorable genetic mutations, while displaying limited toxicity toward normal hematopoietic cells. They also suggest that such interactions may operate through similar mechanisms observed in leukemia cell lines.
Coadministration of MLN4924/belinostat suppresses tumor growth and improves animal survival in both subcutaneous and orthotopic AML xenograft models
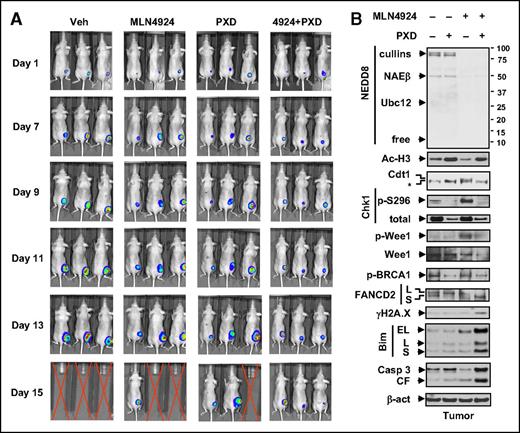
Finally, AML xenograft models were used to validate the in vivo activity of the regimen. Nude mice were inoculated s.c. with luciferase-labeled U937 cells. Combined administration clearly reduced bioluminescent signal intensity compared with each agent alone (Figure 6A). To identify potential pharmacodynamic (PD) markers, western blot analysis revealed that multiple molecular alterations observed in vitro were recapitulated in vivo (Figure 6B), particularly those involving cell cycle checkpoints (eg, Chk1 and Wee1 dephosphorylation), DNA repair (eg, p-BRCA1 and FANCD2-L downregulation), and apoptosis (eg, Bim upregulation), accompanied by increased DNA damage (γH2A.X) and apoptosis (eg, caspase 3 cleavage).
Coadministration of MLN4924 and belinostat suppresses tumor growth in a murine xenograft flank model. Nude mice were s.c. inoculated in the right rear flank with 5 × 106 luciferase-expressing U937 cells. Treatment was initiated after luciferase signal was detected. MLN4924 was reconstituted weekly with 20% HPbCD (HY-100; ONBIO) and administrated at a dose of 30 mg/kg via s.c. injection twice daily for 3 continuous days followed by 2 days off every 5 days. Stock solution of 50 mg/mL belinostat in 100 mg/mL l-arginine (Sigma A8094; belinostat: l-arginine = 1:2) was formulated and stored at −20°C. Belinostat stock solution was freshly diluted in isotonic saline and administrated at a dose of 40 mg/kg via i.p. injection daily 5 days a week. Control animals were administered equal volumes of vehicle. (A) Tumor growth was monitored every other day after i.p. injection with 150 mg/kg luciferin using the IVIS 200 imaging system. Representative images of 3 mice are shown. Red crossed lines indicate that mice were euthanized when tumor size reached 1700 mm3 or other humane end points (eg, abscessed or necrotic tumors) were reached. (B) Western blot analysis was performed to monitor the indicated candidate PD markers, identified from in vitro experiments, in tumors excised from representative mice.
Coadministration of MLN4924 and belinostat suppresses tumor growth in a murine xenograft flank model. Nude mice were s.c. inoculated in the right rear flank with 5 × 106 luciferase-expressing U937 cells. Treatment was initiated after luciferase signal was detected. MLN4924 was reconstituted weekly with 20% HPbCD (HY-100; ONBIO) and administrated at a dose of 30 mg/kg via s.c. injection twice daily for 3 continuous days followed by 2 days off every 5 days. Stock solution of 50 mg/mL belinostat in 100 mg/mL l-arginine (Sigma A8094; belinostat: l-arginine = 1:2) was formulated and stored at −20°C. Belinostat stock solution was freshly diluted in isotonic saline and administrated at a dose of 40 mg/kg via i.p. injection daily 5 days a week. Control animals were administered equal volumes of vehicle. (A) Tumor growth was monitored every other day after i.p. injection with 150 mg/kg luciferin using the IVIS 200 imaging system. Representative images of 3 mice are shown. Red crossed lines indicate that mice were euthanized when tumor size reached 1700 mm3 or other humane end points (eg, abscessed or necrotic tumors) were reached. (B) Western blot analysis was performed to monitor the indicated candidate PD markers, identified from in vitro experiments, in tumors excised from representative mice.
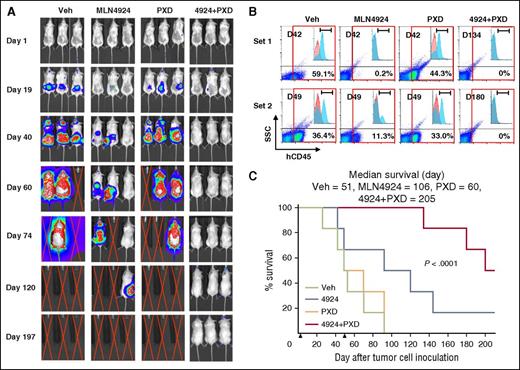
Although the subcutaneous xenograft model described in the previous paragraph facilitates in vivo PD studies, it may not be representative of leukemia. To address this issue, parallel studies were performed in an orthotopic model using luciferase-labeled MV-4-11 cells carrying FLT3-ITD. Bioluminescent imaging demonstrated that whereas MLN4924, but not belinostat, displayed moderate single-agent activity, tumor signals in mice receiving combined treatment were either significantly delayed (eg, >120 days; data not shown) in half of the mice or not observed in the remaining animals on study termination at day 200 post-tumor cell inoculation (Figure 7A), despite treatment discontinuation at day 50. These results were concordant with analysis of cells expressing the human leukemia marker CD45 in mouse BMs. FCM indicated that although belinostat alone had minimal effects, single-agent MLN4924 clearly reduced the percentage of CD45+ cells. However, marrows obtained from mice cotreated with MLN4924/belinostat revealed <0.3% in half of the mice or no detectable CD45+ cells in the remaining mice (Figure 7B; supplemental Figure 8A). Significantly, mice receiving both agents experienced a substantial increase in survival (Figure 7C, P < .0001 vs single agent). In both mouse models, MLN4924/belinostat treatment was well tolerated, with essentially no weight loss (supplemental Figure 8B) or signs of hematologic (supplemental Table 2) or other toxicities (eg, lethargy, fur loss, behavioral changes, etc). These findings argue that a regimen combining MLN4924/belinostat is highly active in both in vitro and in vivo AML models.
The combination of MLN4924 and belinostat reduces tumor burden and significantly prolongs animal survival in a murine IV AML xenograft model carrying FLT3-ITD. NOD/SCID-γ (NSG) mice were inoculated via tail vein with 1 × 106 luciferase-labeled MV-4-11 cells harboring FLT3-ITD. Mice were randomized to 4 groups (n = 6 per group), and treatment was initiated 2 days after injection of tumor cells. MLN4924 (30 mg/kg) and belinostat (40 mg/kg) were administrated as described in Figure 6. Control animals were administered equal volumes of vehicle. (A) Tumor growth was monitored every other day after i.p. injection with 150 mg/kg luciferin using the IVIS 200 imaging system. Representative images of mice are shown. Red crossed lines indicate that mice were euthanized when humane end points were reached. (B) When mice were sacrificed, BM was harvested and subjected to FCM to determine the percentage of human CD45+ tumor cells in BM mononuclear cells. Two sets of representative data are shown. (C) Kaplan-Meier analysis was performed to analyze survival of animals (n = 6 per group). Inset, Median survival. Arrows indicate time when treatment began (day 2) and discontinued (day 50). D, Day when bone barrow was harvested after tumor cell inoculation; h, human; SSC, side scatter.
The combination of MLN4924 and belinostat reduces tumor burden and significantly prolongs animal survival in a murine IV AML xenograft model carrying FLT3-ITD. NOD/SCID-γ (NSG) mice were inoculated via tail vein with 1 × 106 luciferase-labeled MV-4-11 cells harboring FLT3-ITD. Mice were randomized to 4 groups (n = 6 per group), and treatment was initiated 2 days after injection of tumor cells. MLN4924 (30 mg/kg) and belinostat (40 mg/kg) were administrated as described in Figure 6. Control animals were administered equal volumes of vehicle. (A) Tumor growth was monitored every other day after i.p. injection with 150 mg/kg luciferin using the IVIS 200 imaging system. Representative images of mice are shown. Red crossed lines indicate that mice were euthanized when humane end points were reached. (B) When mice were sacrificed, BM was harvested and subjected to FCM to determine the percentage of human CD45+ tumor cells in BM mononuclear cells. Two sets of representative data are shown. (C) Kaplan-Meier analysis was performed to analyze survival of animals (n = 6 per group). Inset, Median survival. Arrows indicate time when treatment began (day 2) and discontinued (day 50). D, Day when bone barrow was harvested after tumor cell inoculation; h, human; SSC, side scatter.
Discussion
The first-in-human NAE inhibitor MLN4924 acts proximally in the UPS to prevent CRL activation, resulting in accumulation of multiple protein substrates,26,27 including those involved in NF-κB, DNA re-replication, DNA damage checkpoint, and DNA repair.37,38,47,48 Recent evidence has also highlighted the role of NF-κB, DNA damage, cell cycle checkpoint, and DNA repair in HDACI lethality.49-52 It therefore seemed plausible that complementary HDACI and MLN4924 MOAs might result in enhanced antileukemia activity. Indeed, the present results indicate that these agents interact synergistically against AML cell lines, primary AML/MDS cells, and LIC-enriched primitive cells, including those carrying diverse genetic mutations. Loss/mutation of p53 or FLT3 (ie, FLT3-ITD) is associated with poor prognoses in AML.6 With checkpoint inhibitors (eg, Chk1 or Wee1 inhibitors), p53 deficiency may be required for susceptibility,53 although this has not been a universal finding.54 Notably, the MLN4924/belinostat regimen was active against cells expressing either wt or mutant p53 or FLT3. Moreover, p53 shRNA knockdown sensitized wt-p53 OCI-AML-3 cells to this regimen, probably because G1/S checkpoint loss due to p53 deficiency cooperates with MLN4924/belinostat-mediated intra-S checkpoint disruption to promote cell death. Similarly, FLT3-ITD expression sensitized murine Ba/F3 cells to the regimen. This finding is supported by the observation that primary AML blasts carrying FLT3-ITD were most sensitive to this regimen. In this context, leukemia cells expressing FLT3 mutations exhibit impaired HR repair.55 Indeed, MLN4924/belinostat triggered more robust DSBs and apoptosis in FLT3-ITD Ba/F3 cells, compared with their wt-FLT3 counterparts, suggesting that certain AML subsets carrying specific genetic abnormalities might be particularly vulnerable to this combination regimen.
Previous studies demonstrated that HDACIs trigger NF-κB activation, through a DNA damage– and ATM/NEMO-dependent mechanism in leukemia cells,16 which limits HDACI lethality.15 The present findings argue that accumulation of the phosphorylated form of IκBα and NF-κB inactivation by MLN492424 may promote HDACI lethality in leukemia cells.56 Moreover, NAE inhibition also prevents proteasomal degradation of multiple proapoptotic proteins, including BimEL.57 HDACIs have also been shown to upregulate Bim.58 Notably, Bim knockdown essentially abrogated apoptosis induced by MLN4924/belinostat, suggesting a role for Bim in apoptosis induced by this regimen, presumably by lowering the death threshold. However, the possibility that other proapoptotic Bcl-2 family proteins (eg, Bak) and their interactions with antiapoptotic proteins might be also involved in this setting cannot be excluded.
MLN4924 triggers the intra-S checkpoint38 and DNA re-replication59 in neoplastic cells, while HDACIs disrupt the G2/M checkpoint51,60 and induce aberrant firing of dormant replication origins.61 Consistently, MLN4924 triggered the intra-S checkpoint, accompanied by marked upregulation/activation of multiple checkpoint kinases, including ATR, Chk1, and Wee1, due in part to increased protein levels, although the possible involvement of other checkpoints (eg, G2/M checkpoint due to Chk2 activation by belinostat) cannot presently be ruled out. Significantly, these events were substantially attenuated by belinostat. The observations that either Chk1 or Wee1 shRNA knockdown significantly increased MLN4924 lethality argues that inhibition/downregulation of Chk1 and Wee1 contributes functionally to interactions between these agents. Interestingly, several recent studies have demonstrated that epithelial neoplasms62 and AML cells63 are highly vulnerable to concomitant Chk1 and Wee1 inhibition. Finally, belinostat also diminished MLN4924-mediated upregulation of Cdt1, a DNA-licensing protein. Further studies will be required to define the functional role of Cdt1 downregulation in belinostat/MLN4924 interactions. Of note, MLN4924 triggered activation of the HR pathway for DSB repair, including (1) upregulation of CtIP, which converts DSB ends into 3′-single-stranded DNA overhangs for HR initiation64 ; (2) activation of BRCA1, a major in vivo partner of CtIP64 ; and (3) an increase in the monoubiquitinated (active) form of FANCD2 (FANCD2-L), implicated in 3 classic DNA repair pathways including HR, nucleotide excise repair, and mutagenic translesion synthesis.65 Significantly, whereas events associated with HR pathway activation by MLN4924 were largely blocked by belinostat, coexposure to these agents also increased acetylation of histone H3 (K56) and H4 (K16), indicating impaired NHEJ repair.9,66 Interestingly, the latter may also be related to NF-κB inactivation. Finally, combined treatment sharply increased DSBs (eg, γH2A.X expression/foci formation) and chromosome pulverization due to premature chromosome condensation in S-phase nuclei.67 Collectively, these findings suggest that both HR and NHEJ disruption contribute to MLN4924/belinostat lethality. However, further studies (eg, using approaches that distinguish between NHEJ and HR)68 will be required to determine more definitively the functional contributions of either or both of these repair mechanisms to HDAC/NAE inhibitor interactions. In addition, the possibility that alterations in these DNA repair proteins may be secondary to other MOAs of this combination regimen (eg, DNA damage, cell cycle disruption, etc) cannot presently be excluded.
The MLN4924/belinostat regimen effectively induced apoptosis of primary AML cells carrying various genetic aberrations, including those associated with poor prognoses, but was relatively sparing to normal hematopoietic progenitors. The mechanism underlying ex vivo selectivity is uncertain but may reflect intrinsic DDR (particularly DNA repair) deficiencies of neoplastic cells, for example, in the case of HDACIs.69 Although the basis for MLN4924 selectivity is unclear, other UPS inhibitors (eg, proteasome inhibitors) may, however, be selectively lethal to neoplastic cells.70 The regimen was also active against primary primitive AML populations enriched for LICs. However, further studies using more definitive approaches (eg, long-term culture of primary AML LICs and serial transplant animal studies)71 will be necessary to validate whether HDAC/NAE inhibition targets true LICs. Such studies are currently in progress. Finally, this regimen was highly effective in reducing tumor burden and prolonging survival in AML xenograft models, while exerting minimal toxicity. Moreover, evidence for several PD events (eg, DDR disruption, DSB induction) suggest that similar MOAs operate both in vitro and in vivo. Given emerging evidence of MLN4924 single-agent activity in AML,32 the present findings suggest that the NAE/HDAC inhibitory strategy warrants consideration in this disease, particularly in certain genetic/molecular subsets.
The microarray data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE77560).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Peter D. Aplan (National Institutes of Health [NIH], National Cancer Institute [NCI], Center for Cancer Research) for providing 188G3 and 188NRG-2D cells.
This work was supported by the NIH (NCI awards CA93738, CA167708, P50 CA142509, and NCATS UH2 TR001373) and the Leukemia & Lymphoma Society of America (grant R6472). Human tissues, patient consents, clinical data (or other appropriate service) were provided by the Virginia Commonwealth University (VCU) Tissue and Data Acquisition and Analysis Core Facility, supported, in part, with the funding from NIH–NCI Cancer Center Core Support grant P30 CA016059, as well as through the Department of Pathology, School of Medicine, and Massey Cancer Center of VCU. Plasmid preparation was performed at the VCU Macromolecule Core Facility, supported, in part, with funding from NIH–NCI Cancer Center Core grant 5P30CA016059-29. Confocal microscopies were performed at the VCU Department of Anatomy and Neurobiology Microscopy Facility, supported, in part, with funding from NIH–National Institute of Neurological Disorders and Stroke Center Core grant 5P30NS047463.
Authorship
Contribution: Y.D., L.Z., S. Chen, and S.G. designed research and analyzed data; L.Z., S. Chen, Y.D., Y.Z., Y.L., L.L., and H.L. performed research and collected data; M.K. performed flow cytometry analysis on patient samples; M.R. generated Ba/F3 wt-FLT3, Ba/F3 FLT3-ITD, and MV-4-11/luciferase cells; L.P. and S. Chalasani assisted with DNA repair analyses; K.A.R., C.I.D., and A.F.-G. provided patient samples and performed NGS; Y.D. and S.G. wrote the manuscript; and A.J.B. helped write the manuscript.
Conflict-of-interest disclosure: A.J.B. is an employee of Millennium Pharmaceuticals, Inc, a wholly owned subsidiary of Takeda Pharmaceutical Company Ltd. The remaining authors declare no competing financial interests.
Correspondence: Steven Grant, Division of Hematology/Oncology, Virginia Commonwealth University, PO Box 980035, Richmond, VA 23298; e-mail: stgrant@vcu.edu; and Yun Dai, Virginia Commonwealth University and the Massey Cancer Center, Room 234, Goodwin Research Building, 401 College St, Richmond, VA 23298; e-mail: ydai@vcu.edu.
References
Author notes
L.Z. and S. Chen contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal