Key Points
HIF-1α critically regulates the interaction of neoplastic CLL cells with the leukemic microenvironment.
HIF-1α is regulated at the transcriptional level in CLL patients and correlates with CXCR4 expression.
Abstract
Hypoxia-inducible transcription factors (HIFs) regulate a wide array of adaptive responses to hypoxia and are often activated in solid tumors and hematologic malignancies due to intratumoral hypoxia and emerging new layers of regulation. We found that in chronic lymphocytic leukemia (CLL), HIF-1α is a novel regulator of the interaction of CLL cells with protective leukemia microenvironments and, in turn, is regulated by this interaction in a positive feedback loop that promotes leukemia survival and propagation. Through unbiased microarray analysis, we found that in CLL cells, HIF-1α regulates the expression of important chemokine receptors and cell adhesion molecules that control the interaction of leukemic cells with bone marrow and spleen microenvironments. Inactivation of HIF-1α impairs chemotaxis and cell adhesion to stroma, reduces bone marrow and spleen colonization in xenograft and allograft CLL mouse models, and prolongs survival in mice. Of interest, we found that in CLL cells, HIF-1α is transcriptionally regulated after coculture with stromal cells. Furthermore, HIF-1α messenger RNA levels vary significantly within CLL patients and correlate with the expression of HIF-1α target genes, including CXCR4, thus further emphasizing the relevance of HIF-1α expression to CLL pathogenesis.
Introduction
Hypoxia-inducible transcription factor (HIF)-1α is an essential regulator of cell adaptation to hypoxia and is often upregulated in tumors due to intratumoral hypoxia or activation of oncogenic pathways.1 In solid tumors, HIF-1α fosters different tumor-promoting mechanisms, including metabolic adaptation, neoangiogenesis, and metastasis.1,2 Recent evidence indicates that HIF-1α is also implicated in the development of hematologic malignancies such as chronic lymphocytic leukemia (CLL).3
CLL is the most common leukemia in adults and is characterized by the accumulation of mature CD5+ B cells in peripheral blood (PB), bone marrow (BM), and secondary lymphoid organs.4 CLL is clinically and biologically heterogeneous: patients may suffer from an indolent disease with long life expectancy or an aggressive malignancy with dismal prognosis. Gene expression and genetic profiling have uncovered a number of markers and genetic lesions that are implicated in the pathogenesis of CLL and predict predisposition to clinical progression.5 From a therapeutic standpoint, introduction of chemoimmunotherapy such as combined fludarabine, cyclophosphamide, and rituximab and treatment with B-cell receptor signaling pathway inhibitors such as ibrutinib have significantly prolonged disease-free survival for low- and high-risk CLL patients; current therapeutic efforts aim to eliminate minimal residual disease toward reaching a cure for patients with CLL.6,7
However, the biology and drug responsiveness of CLL is complicated by the evidence that CLL cells establish crucial connections with leukemia microenvironments in BM and secondary lymphoid organs, where they receive protective signals from a number of accessory cells.8,9 For this reason, dissecting the role of the microenvironment in the pathogenesis of CLL may provide new strategies for improved treatment.
In this study, we identify a novel mechanism that drives the interaction of CLL cells with the microenvironment. We find that in CLL, HIF-1α regulates the expression of genes that promote the interaction of neoplastic B cells with leukemia microenvironments. As a consequence, inhibiting HIF-1α impairs BM chemotaxis and colonization of BM and spleen, in addition to regulating neoangiogenesis, and prolongs survival in mice. Remarkably, HIF-1α messenger (m)RNA levels vary significantly within CLL patients, and HIF-1α is transcriptionally upregulated in neoplastic CLL cells upon contact with stromal cells in a positive feedback loop that may foster CLL expansion and protection from apoptosis. In summary, our data indicate that HIF-1α plays important tumor-promoting functions in CLL and suggest that targeting this pathway may have clinical implications.
Materials and methods
Cell culture and reagents
MEC-1 (German Collection of Microorganisms and Cell Cultures) and HEK-293T and Hs5 cells (American Type Culture Collection) were maintained in RPMI 1640, Iscove modified Dulbecco medium, and Dulbecco’s modified Eagle medium with 10% fetal bovine serum (FBS) and antibiotics (Lonza), at 37°C, 5% carbon dioxide. EZN-2208, control locked nucleic acid (LNA)-oligonucleotide (EZN-3088), and HIF-1α LNA-oligonucleotide (EZN-2968) were provided by Belrose Pharma.10,11 In vitro treatment with EZN-2208 (24 hours) was performed at the indicated concentrations. Cobalt chloride (CoCl2), AMD3100 (CXCR4 inhibitor), and puromycin were from Sigma, 5-chloromethylfluorescein diacetate (CMFDA) was from Life Technologies, and stromal cell–derived factor (SDF)-1α (CXCL12) was from PeproTech. GIPZ HIF-1α short hairpin RNA or control short hairpin RNA plasmids were from Open Biosystems. Lentiviral infections were performed as previously described.12 MEC-1 cells were selected with puromycin (1 μg/mL).
Animals
Rag2−/−γc−/− and C57BL/6 EμTCL1 mice13 were maintained in a specific pathogen-free animal facility and treated in accordance with European Union and Institutional Animal Care and Use Committee guidelines. For homing experiments, Rag2−/−γc−/− mice were injected IV with 20 × 106 MEC-1 cells and euthanized after 16 hours. BM and spleen cells were incubated with anti-human CD19 (PC-7; Beckman Coulter). CD19+ MEC-1 cells from the BM or spleen were counted in 2 × 106 events. For survival experiments, mice were injected with 10 × 106 MEC-1 cells and euthanized when terminally sick. For EZN-2208 treatment, mice were injected with 10 × 106 cells and treated IV with 5 mg/kg every other day for 5 administrations (every 2 days × 5 schedule).
For leukemia propagation, EμTCL1 mice were euthanized when CD19+CD5+ cells reached 90% in PB. A total of 10 × 106 splenic cells were injected intraperitoneally into syngeneic mice for leukemia expansion, and experiments were performed after the second transplant. Leukemic mice were treated with 5 mg/kg EZN-2208 on an every 2 days × 5 schedule. For ex vivo organ cultures, animals were euthanized when terminally sick, and their spleens were divided into 20-mg fragments and cultured in a bioreactor-based cell culture system. Immunophenotypic analysis was performed with the following antibodies: anti-mouse CD5 (APC) and anti-mouse CD19 (PECY-7), from BD Biosciences.13 Annexin V staining was performed using the PE Annexin V Apoptosis Detection Kit I (BD Pharmingen).
Human primary samples
CLL patients were diagnosed according to the updated National Cancer Institute Working Group guidelines.14 All patients (supplemental Table 1, available on the Blood Web site) were either untreated or off therapy for at least 6 months before the beginning of the study. Leukemic CD19+ cells were selected as previously reported.13 CLL lymph node stromal cells (LNS1) were derived by trypsin digestion of a surgical lymph node fragment of a CLL patient with intermediate Rai stage and stable disease, who underwent lymphadenectomy to confirm diagnosis in lymph node. Primary adherent cells were cultured in Dulbecco’s modified Eagle medium supplemented with 20% FBS and standard antibiotics. Cell cultures with a constant growth rate were established after 15 to 20 passages and were characterized as CD45−CD31−CD14−CD33−. All tissue samples were obtained after informed consent as approved by the institutional ethics committee of San Raffaele Hospital.
mRNA expression of CXCR4 and SLC2A1 (calculated by the 2−ΔCt method relative to 18S expression and multiplied by 105) was compared with HIF-1α expression by Spearman rank-order correlation after conversion of normalized mRNA levels to log10. To obtain HIF-1α silencing, primary CLL cells were electroporated with 10 μM LNA-oligonucleotides by Amaxa Nucleofector Technology (Lonza) with the Ingenio Electroporation Kit (MIR50115; Mirus Bio). All subsequent assays were performed 48 hours after electroporation. Cell viability was measured by trypan blue exclusion assay.
Coculture assays
A total of 5 × 106 MEC-1 cells and primary CLL cells from patients were plated on 80% confluent Hs5 cells or LNS1 cells. Nonadherent cells were collected after 24 hours for RNA extraction and analysis of CXCR4 expression (anti-hCXCR4 PE; R&D Systems) by flow cytometry. Mouse CLL B cells were purified from PB of leukemic EμTCL1 mice with EasySep Mouse Pan-B Cell Isolation Kit (STEMCELL Technologies), maintained in RPMI 1640 supplemented with 20% FBS and standard antibiotics and plated for 24 hours on Hs5 cells. For evaluation of cell viability, primary CLL cells were cocultured with LNS1 cells for 72 hours and analyzed with the PE Annexin V Apoptosis Detection Kit I (BD Pharmingen).
Adhesion and migration assays
MEC-1 cell adhesion to stroma was assessed as previously reported.13 Briefly, Hs5 stromal cells were seeded on 48-well plates. After 24 hours, MEC-1 cells were labeled with 1 μM CMFDA and added to the stroma monolayer. After overnight culture, nonadherent cells were collected from cell supernatants and 3 washes in phosphate-buffered saline, whereas adherent cells were recovered after trypsinization. MEC-1 CMFDA+ cells were counted by flow cytometer for 1 minute in both fractions. The percentage of MEC-1 adhesion was evaluated as the number of adherent CMFDA+ cells divided by the total number of CMFDA+ cells.
For migrations assays, 1 × 106 MEC-1 cells were seeded in the upper chamber of 6.5-mm diameter, 8-μm pore Costar Transwell inserts with 100 ng/mL SDF-1α in the lower chamber. Cells in the lower chamber were counted as the number of cells acquired per minute (High Throughput Sampler FACSCanto; Becton Dickinson) 4 hours after seeding. Where indicated, cells were preincubated with 50 μM AMD3100. For migration experiments of patients’ CLL cells, 5 × 105 PB leukemic cells were seeded on 5-μm pores 24 hours after electroporation with EZN-2968 or EZN-3088 oligonucleotides, and migration was assessed after 4 hours.
Immunoblot
MEC-1 cells were lysed with radioimmunoprecipitation assay buffer (Sigma) supplemented with protease inhibitor cocktail (Roche). Fifty micrograms of total proteins were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and immunoblot analysis was performed with rabbit polyclonal anti-HIF-1α (Cayman Chemicals) and mouse anti-β-actin (Sigma) as loading control. Where indicated, MEC-1 cells were treated with 200 nM CoCl2 for 5 hours, and primary CLL B cells were incubated at 1% oxygen for 8 hours.
Real-time PCR
RNA was isolated with the RNeasy Mini Kit (QIAGEN), retrotranscribed with the Advantage RT-for-PCR Kit (Clontech), and analyzed by real-time polymerase chain reaction (PCR) in a 7900HT Fast Real-Time PCR System (Applied Biosystems). Probes for TaqMan assays were purchased from Applied Biosystems. 18S was used as internal control. The relative expression of different complementary DNAs was calculated using the 2−ΔΔCt method.
Gene expression analysis
Gene expression profiling was performed on MEC-1 cells using an Illumina HumanHT-12 v3 Expression BeadChip Kit. RNA was extracted using the RNeasy Mini Kit (QIAGEN). Five hundred nanograms of RNA was reverse transcribed (Roche Transcriptor First Strand cDNA Synthesis Kit) and used to synthesize complementary RNA labeled with biotinylated uridine triphosphate. Complementary RNA was hybridized to the chips, and data were collected with an Illumina BeadArray Reader. Analysis of gene expression data was performed as previously described15 using gene set enrichment analysis.16
Histopathology and immunohistochemistry
Mice tissues were fixed in 4% formalin, paraffin embedded, cut into 5-μm thick sections, and stained with hematoxylin and eosin according to standard protocols. Immunohistochemistry was performed using CD31 antibody (Thermo Scientific), and a certified pathologist evaluated histologic sections. Images were taken with a Zeiss Axioskop 40 microscope equipped with a Zeiss AxioCam MRc digital camera.
Statistics
Unless otherwise stated, a 2-sided Student t test was used to measure statistical significance. For survival experiments, Kaplan-Meier curves were analyzed with the Mantel-Cox test.
Results
HIF-1α regulates chemotaxis and adhesion to stroma in CLL
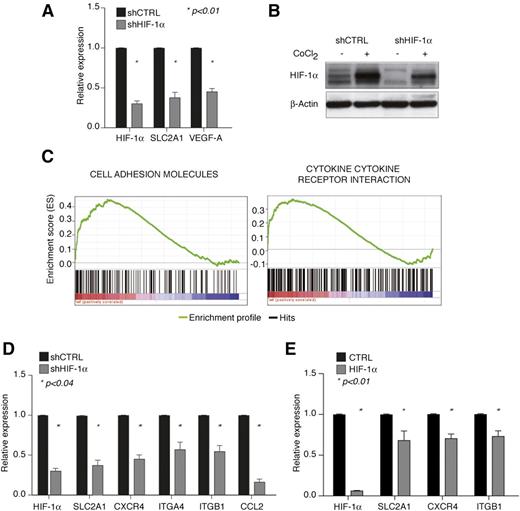
It was recently observed that HIF-1α is constitutively expressed in CLL cells compared with normal B cells due to microRNA-mediated downregulation of the HIF-1α ubiquitin ligase, von Hippel–Lindau protein.17,18 Constitutive HIF-1α expression was suggested to induce vascular endothelial growth factor (VEGF) upregulation and neoangiogenesis.19-21 However, because HIF-1α regulates a variety of other functions in solid tumors and hematologic malignancies,1,22,23 we asked whether HIF-1α also controlled other aspects of CLL pathogenesis. Microarray analysis was performed upon stable HIF-1α silencing in the human CLL cell line MEC-1 (Figure 1A-B), where HIF-1α is constitutively expressed in normoxia and further stabilized by the hypoxia-mimetic agent CoCl2 (Figure 1B). HIF-1α silencing led to reduced expression of HIF-1α in normoxic and hypoxia-mimetic conditions (Figure 1B) and to significant downregulation of the classical HIF-1α target genes SLC2A1 (Glut-1) and VEGF-A (Figure 1A). Of interest, gene set enrichment analysis revealed that reduced expression of HIF-1α caused deregulation of a number of genes mediating cell adhesion and cytokine-cytokine receptor interactions, such as genes belonging to the “Cell Adhesion Molecules” and “Cytokine-Cytokine Receptor Interaction” gene sets of the Kyoto Encyclopedia of Genes and Genomes pathway database (Figure 1C). These gene sets contain HIF-1α target genes importantly implicated in regulating CLL cell trafficking, interaction with protective microenvironments, and cell survival, such as CXCR4 and VLA-4 (ITGA4 and ITGB1), which regulate homing and retention of CLL cells in microenvironments rich in CXCL12,24 and CCL2, a prosurvival CLL chemokine produced by CLL cells in an autocrine manner when cocultured with accessory cells.25 Because of their role in CLL, we focused on validating the expression of these genes upon modulation of HIF-1α activity.
HIF-1α regulates genes involved in chemotaxis and cell adhesion in CLL. (A) Relative expression of HIF-1α and common HIF-1α target genes in MEC-1 cells upon HIF-1α stable silencing (shHIF-1α) with respect to control cells (shCTRL). Data represent mean values ± SEM of 3 independent experiments. (B) Immunoblot of HIF-1α in shHIF-1α and shCTRL MEC-1 cells shows constitutive expression of HIF-1α and downregulation by shRNA both at basal conditions and upon further protein stabilization with CoCl2. Representative experiment of 2 with similar results. (C) Expression of Cell Adhesion Molecules (left panel) and Cytokine-Cytokine Receptor Interaction (right panel) gene sets from the Kyoto Encyclopedia of Genes and Genomes in microarray data from MEC-1 shHIF-1α cells compared with shCTRL cells (respective normalized enrichment score and false discovery rate of 1.98 and 0.008 for shHIF-1α cells and 1.74 and 0.078 for shCTRL cells). The bar-code plot indicates the position of the genes belonging to each gene set on the expression data rank sorted by its association with HIF-1α knockdown, with red and blue colors indicating overexpression and underexpression, respectively, in the HIF-1α-silenced group. (D-E) Downregulation of HIF-1α target genes involved in cell adhesion and cytokine interaction in MEC-1 cells upon independent shHIF-1α-mediated silencing (D) and in primary CLL B cells (n = 10) after electroporation with a control (CTRL) or HIF-1α-targeting oligonucleotide (HIF-1α) (E). Data represent mean values ± SEM of 3 (D) and 10 (E) independent experiments. SEM, standard error of the mean.
HIF-1α regulates genes involved in chemotaxis and cell adhesion in CLL. (A) Relative expression of HIF-1α and common HIF-1α target genes in MEC-1 cells upon HIF-1α stable silencing (shHIF-1α) with respect to control cells (shCTRL). Data represent mean values ± SEM of 3 independent experiments. (B) Immunoblot of HIF-1α in shHIF-1α and shCTRL MEC-1 cells shows constitutive expression of HIF-1α and downregulation by shRNA both at basal conditions and upon further protein stabilization with CoCl2. Representative experiment of 2 with similar results. (C) Expression of Cell Adhesion Molecules (left panel) and Cytokine-Cytokine Receptor Interaction (right panel) gene sets from the Kyoto Encyclopedia of Genes and Genomes in microarray data from MEC-1 shHIF-1α cells compared with shCTRL cells (respective normalized enrichment score and false discovery rate of 1.98 and 0.008 for shHIF-1α cells and 1.74 and 0.078 for shCTRL cells). The bar-code plot indicates the position of the genes belonging to each gene set on the expression data rank sorted by its association with HIF-1α knockdown, with red and blue colors indicating overexpression and underexpression, respectively, in the HIF-1α-silenced group. (D-E) Downregulation of HIF-1α target genes involved in cell adhesion and cytokine interaction in MEC-1 cells upon independent shHIF-1α-mediated silencing (D) and in primary CLL B cells (n = 10) after electroporation with a control (CTRL) or HIF-1α-targeting oligonucleotide (HIF-1α) (E). Data represent mean values ± SEM of 3 (D) and 10 (E) independent experiments. SEM, standard error of the mean.
Real-time PCR analysis after independent HIF-1α silencing confirmed CXCR4, ITGA4, ITGB1, and CCL2 downregulation in MEC-1 cells (Figure 1D), and acute HIF-1α silencing in primary PB leukemic cells from CLL patients confirmed downregulation of SCL2A1, CXCR4, and ITGB1 (Figure 1E), whereas basal expression of ITGA4 and CCL2 was undetectable after in vitro manipulation (data not shown).
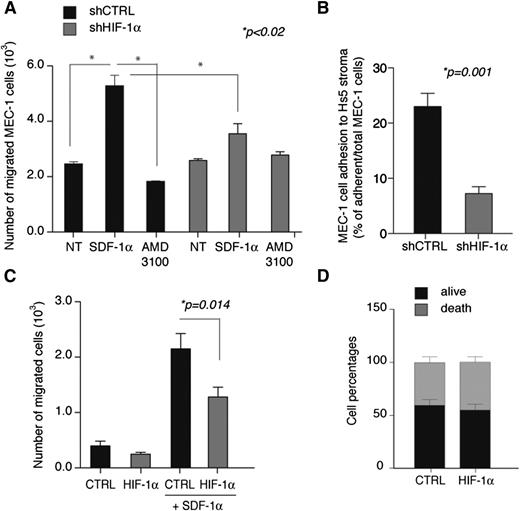
Based on these data and on the function of HIF factors in promoting homing and adhesion to chemokine-rich microenvironments in solid tumors,26,27 we asked whether HIF-1α regulated chemotaxis and cell adhesion to stroma in CLL. HIF-1α silencing did not affect proliferation or apoptosis of MEC-1 cells (data not shown) but impaired CXCL12 (SDF-1α)-mediated chemotaxis (specifically blocked by a CXCR4 inhibitor) and adhesion to the stromal BM cell line Hs528 (Figure 2A-B). In addition, acute HIF-1α silencing in primary cells from CLL patients reduced CXCL12-mediated chemotaxis without inducing further apoptosis (Figure 2C-D).
HIF-1α silencing inhibits SDF-1-mediated chemotaxis and adhesion to stroma in CLL. (A) SDF-1α-mediated migration of MEC-1 cells is inhibited upon chronic silencing of HIF-1α cells (shHIF-1α) compared with control cells (shCTRL). Where indicated, cells were pretreated with the CXCR4 inhibitor AMD3100 as a control. Data are represented as mean values ± SEM of 3 independent experiments. (B) Cell adhesion of MEC-1 cells to Hs5 stromal cells is inhibited upon chronic silencing of HIF-1α cells (shHIF-1α) compared with control cells (shCTRL). MEC-1 cell adhesion was calculated as the percentage of MEC-1 cells adhering to Hs5 cells over total MEC-1 cells in coculture. Data are represented as mean values ± SEM of triplicates from 1 of 3 experiments with similar results. (C) SDF-1α-mediated migration of primary CLL B cells is inhibited after electroporation with a HIF-1α-targeting oligonucleotide (HIF-1α) compared with control oligonucleotide (CTRL). Data represent mean values ± SEM (n = 10). (D) Primary CLL B cells viability after electroporation with control oligonucleotide (CTRL) or HIF-1α-targeting oligonucleotide (HIF-1α). Data are expressed as percentages of total population and represent mean values ± SEM (n = 14). NT, untreated.
HIF-1α silencing inhibits SDF-1-mediated chemotaxis and adhesion to stroma in CLL. (A) SDF-1α-mediated migration of MEC-1 cells is inhibited upon chronic silencing of HIF-1α cells (shHIF-1α) compared with control cells (shCTRL). Where indicated, cells were pretreated with the CXCR4 inhibitor AMD3100 as a control. Data are represented as mean values ± SEM of 3 independent experiments. (B) Cell adhesion of MEC-1 cells to Hs5 stromal cells is inhibited upon chronic silencing of HIF-1α cells (shHIF-1α) compared with control cells (shCTRL). MEC-1 cell adhesion was calculated as the percentage of MEC-1 cells adhering to Hs5 cells over total MEC-1 cells in coculture. Data are represented as mean values ± SEM of triplicates from 1 of 3 experiments with similar results. (C) SDF-1α-mediated migration of primary CLL B cells is inhibited after electroporation with a HIF-1α-targeting oligonucleotide (HIF-1α) compared with control oligonucleotide (CTRL). Data represent mean values ± SEM (n = 10). (D) Primary CLL B cells viability after electroporation with control oligonucleotide (CTRL) or HIF-1α-targeting oligonucleotide (HIF-1α). Data are expressed as percentages of total population and represent mean values ± SEM (n = 14). NT, untreated.
Taken together, these data suggest that in CLL, HIF-1α may be an important regulator of the interaction of neoplastic B cells with the leukemia microenvironment.
HIF-1α inhibition impairs BM homing and colonization of BM and spleen in CLL
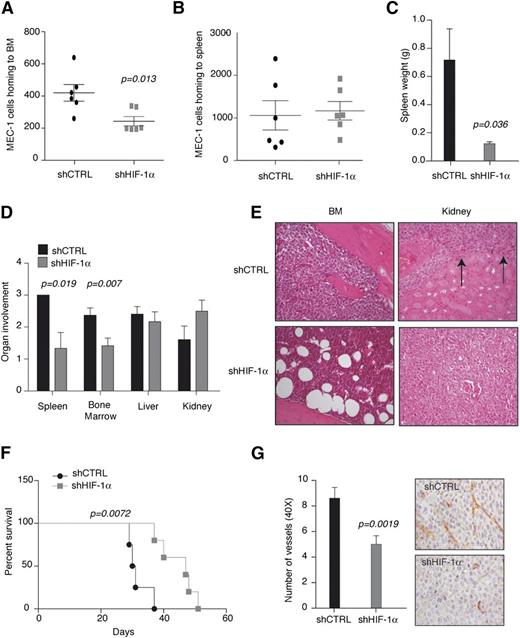
To evaluate the role of HIF-1α in regulating CLL homing and adhesion in vivo, Rag2−/−γc−/− mice were transplanted with MEC-1 cells upon HIF-1α silencing.29
Reduced expression of HIF-1α significantly impaired homing of MEC-1 cells to BM, but not to the spleen (Figure 3A-B). Of interest, however, macroscopic and histopathological examination revealed that HIF-1α downregulation significantly impaired long-term colonization of both BM and spleen, 30 to 50 days after injection, when animals were euthanized because they were terminally sick (Figure 3C-F). In detail, spleen weight was remarkably reduced (Figure 3C), and spleen and BM colonization was specifically affected by HIF-1α downregulation, whereas liver colonization was not impaired and kidney colonization was increased (Figure 3D-E). Although impaired BM colonization is consistent with reduced homing at shorter time points, the defect in spleen colonization does not appear to be mediated by the inability of MEC-1 cells to reach the spleen upon HIF-1α downregulation (Figure 3B-C). In addition, HIF-1α silencing did not induce MEC-1 apoptosis in the spleen (data not shown), thus suggesting that HIF-1α downregulation may impair the retention of MEC-1 cells in the spleen, which may explain increased tumor burden in the kidney (Figure 3D-E).
HIF-1α inhibition impairs BM homing and BM and spleen colonization. HIF-1α stable silencing (shHIF-1α) in MEC-1 cells impairs homing to BM (A), but not to the spleen (B) in Rag2−/−γc−/− mice 16 hours after tail vein injection (n = 6). (C) Spleen weight of Rag2−/−γc−/− mice injected with shCTRL and shHIF-1α MEC-1 cells and euthanized when terminally sick (panel F). Data represent mean values ± SEM (n = 6). (D) Histopathological evaluation of organs colonization in mice euthanized when terminally sick. Data represent organ involvement as evaluated by a certified pathologist expressed as mean values ± SEM (n = 6). (E) Representative images (original magnification ×20; hematoxylin and eosin stain) of BM and kidney from Rag2−/−γc−/− mice injected with the indicated cells and euthanized when terminally sick. shCTRL BM is filled with leukemic cells, whereas shHIF-1α BM shows normal architecture. In shCTRL kidney, arrows indicate leukemia involvement, whereas shHIF-1α kidney is filled with leukemic cells. (F) Kaplan-Meier survival curve of Rag2−/−γc−/− mice injected with MEC-1 shCTRL or shHIF-1α cells and euthanized when terminally sick (n = 6). Representative experiment of 2 with similar results. (G) CD31 immunostaining of spleens colonized by MEC-1 shCTRL or shHIF-1α. The graph (left) shows the average number of microvessels per field (original magnification ×40). Data are represented as mean values ± SEM (n = 3). Representative images are shown (right).
HIF-1α inhibition impairs BM homing and BM and spleen colonization. HIF-1α stable silencing (shHIF-1α) in MEC-1 cells impairs homing to BM (A), but not to the spleen (B) in Rag2−/−γc−/− mice 16 hours after tail vein injection (n = 6). (C) Spleen weight of Rag2−/−γc−/− mice injected with shCTRL and shHIF-1α MEC-1 cells and euthanized when terminally sick (panel F). Data represent mean values ± SEM (n = 6). (D) Histopathological evaluation of organs colonization in mice euthanized when terminally sick. Data represent organ involvement as evaluated by a certified pathologist expressed as mean values ± SEM (n = 6). (E) Representative images (original magnification ×20; hematoxylin and eosin stain) of BM and kidney from Rag2−/−γc−/− mice injected with the indicated cells and euthanized when terminally sick. shCTRL BM is filled with leukemic cells, whereas shHIF-1α BM shows normal architecture. In shCTRL kidney, arrows indicate leukemia involvement, whereas shHIF-1α kidney is filled with leukemic cells. (F) Kaplan-Meier survival curve of Rag2−/−γc−/− mice injected with MEC-1 shCTRL or shHIF-1α cells and euthanized when terminally sick (n = 6). Representative experiment of 2 with similar results. (G) CD31 immunostaining of spleens colonized by MEC-1 shCTRL or shHIF-1α. The graph (left) shows the average number of microvessels per field (original magnification ×40). Data are represented as mean values ± SEM (n = 3). Representative images are shown (right).
Survival analysis revealed that HIF-1α downregulation delayed leukemia progression (Figure 3F). Lastly, consistent with VEGF downregulation (Figure 1A) and previous data correlating HIF-1α levels with VEGF in CLL cells,18 neoangiogenesis was impaired in MEC-1-colonized spleens upon HIF-1α downregulation (Figure 3G).
Taken together, these data indicate that HIF-1α plays multiple protumorigenic functions in CLL, which include regulating chemotaxis and retention within BM and spleen and promoting neoangiogenesis in colonized organs. As a consequence, inhibiting HIF-1α reduces CLL aggressiveness.
A compound with HIF-1α-inhibiting activity recapitulates HIF-1α silencing
To corroborate the results obtained with chronic HIF-1α silencing, we aimed to test the consequences of acute HIF-1α inhibition.30-32 Camptothecin-11 is a cytotoxic topoisomerase I inhibitor that also inhibits HIF-1α at both cytotoxic and noncytotoxic concentrations.30 EZN-2208 is a water-soluble, polyethylene glycol conjugate of the camptothecin-11 analog SN38 with improved delivery and pharmacokinetics in xenograft models of solid tumors.31,32
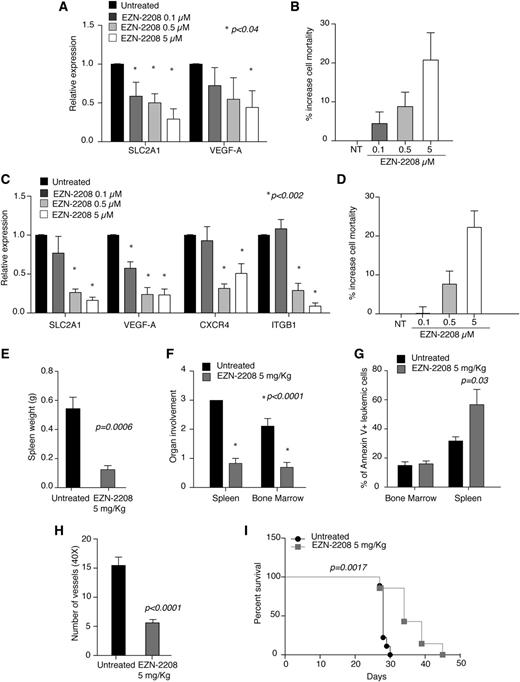
Treatment of MEC-1 cells and primary CLL cells with EZN-2208 reduced mRNA levels of HIF-1α target genes, even at concentrations that did not exert overt toxicity (Figure 4A-D, albeit surface expression of CXCR4 did not decrease accordingly [supplemental Figure 1]), thus confirming that this compound inhibits HIF-1α function in CLL cells.
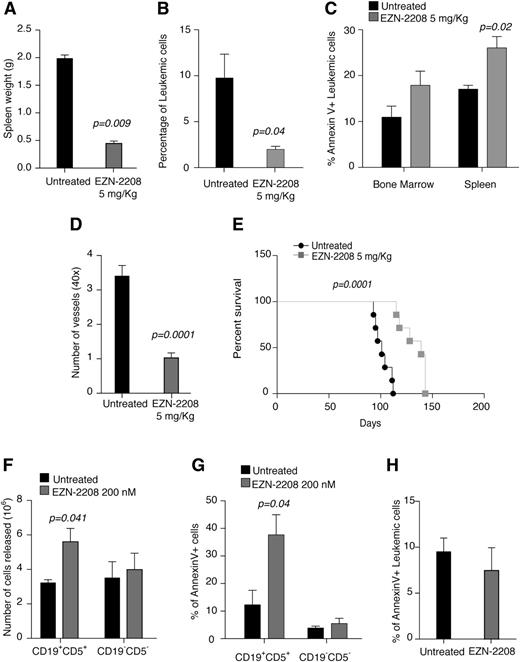
EZN-2208 treatment recapitulates HIF-1α silencing. (A) Relative expression of the indicated genes in MEC-1 cells treated with the indicated doses of EZN-2208 as compared with untreated cells. Data represent mean values ± SEM of 3 independent experiments. (B) Increase of cell mortality of MEC-1 cells treated with the indicated doses of EZN-2208 with respect to basal cell death in untreated cells. Data are expressed as percentages and represent mean values ± SEM of 3 independent experiments. (C) Relative expression of the indicated genes in primary CLL B cells from 8 patients treated with the indicated doses of EZN-2208 as compared with untreated cells. Data represent mean values ± SEM (n = 8). (D) Increase of cell mortality of primary CLL B cells treated with the indicated doses of EZN-2208 with respect to basal cell death in NT cells. Data are expressed as percentages and represent mean values ± SEM (n = 8). (E) Spleen weight of Rag2−/−γc−/− mice injected with MEC-1 cells, treated with EZN-2208 at day 18 after leukemia challenge, and euthanized at the end of treatment (day 27). Data are expressed as mean values ± SEM (n = 6). (F) Histopathological evaluation of BM and spleen colonization of Rag2−/−γc−/− mice injected with MEC-1 cells, treated and euthanized as in panel E. Data represent organ involvement as evaluated by a certified pathologist and are expressed as mean values ± SEM (n = 6). (G) Analysis of annexin V+ MEC-1 cells in the BM and spleen of leukemic mice treated and euthanized as in panel E. Data are expressed as percentage of annexin V+ cells within CD19+ cells in the indicated organs (n = 6). (H) Graph of average number of microvessels per field (original magnification ×40) in spleen colonized by MEC-1 cells at the end of EZN-2208 treatment as in panel E. Data are represented as mean values ± SEM (n = 3). (I) Kaplan-Meier survival curve of Rag2−/−γc−/− mice injected with MEC-1 cells, treated with EZN-2208 at day 18 after leukemia challenge, and euthanized when terminally sick (untreated, n = 9; EZN-2208-treated animals, n = 7). Data are representative of 1 of 2 experiments with similar results.
EZN-2208 treatment recapitulates HIF-1α silencing. (A) Relative expression of the indicated genes in MEC-1 cells treated with the indicated doses of EZN-2208 as compared with untreated cells. Data represent mean values ± SEM of 3 independent experiments. (B) Increase of cell mortality of MEC-1 cells treated with the indicated doses of EZN-2208 with respect to basal cell death in untreated cells. Data are expressed as percentages and represent mean values ± SEM of 3 independent experiments. (C) Relative expression of the indicated genes in primary CLL B cells from 8 patients treated with the indicated doses of EZN-2208 as compared with untreated cells. Data represent mean values ± SEM (n = 8). (D) Increase of cell mortality of primary CLL B cells treated with the indicated doses of EZN-2208 with respect to basal cell death in NT cells. Data are expressed as percentages and represent mean values ± SEM (n = 8). (E) Spleen weight of Rag2−/−γc−/− mice injected with MEC-1 cells, treated with EZN-2208 at day 18 after leukemia challenge, and euthanized at the end of treatment (day 27). Data are expressed as mean values ± SEM (n = 6). (F) Histopathological evaluation of BM and spleen colonization of Rag2−/−γc−/− mice injected with MEC-1 cells, treated and euthanized as in panel E. Data represent organ involvement as evaluated by a certified pathologist and are expressed as mean values ± SEM (n = 6). (G) Analysis of annexin V+ MEC-1 cells in the BM and spleen of leukemic mice treated and euthanized as in panel E. Data are expressed as percentage of annexin V+ cells within CD19+ cells in the indicated organs (n = 6). (H) Graph of average number of microvessels per field (original magnification ×40) in spleen colonized by MEC-1 cells at the end of EZN-2208 treatment as in panel E. Data are represented as mean values ± SEM (n = 3). (I) Kaplan-Meier survival curve of Rag2−/−γc−/− mice injected with MEC-1 cells, treated with EZN-2208 at day 18 after leukemia challenge, and euthanized when terminally sick (untreated, n = 9; EZN-2208-treated animals, n = 7). Data are representative of 1 of 2 experiments with similar results.
In vivo, short-term treatment of leukemic Rag2−/−γc−/− mice with EZN-2208 (5 mg/kg; every 2 days × 5 schedule)33 reduced spleen weight (Figure 4E) and colonization of spleen and BM (Figure 4F), similarly to chronic HIF-1α silencing (Figure 3). Of interest, although EZN-2208 did not induce cell death in BM, it increased MEC-1 cells apoptosis in the spleen (Figure 4G). Because chronic HIF-1α silencing did not cause apoptosis in vitro or in vivo, the proapoptotic effect of EZN-2208 toward splenic MEC-1 cells may be caused by mechanisms that do not depend on HIF-1α inhibition. Similar to HIF-1α silencing, and consistent with VEGF downregulation (Figure 4A), EZN-2208 treatment also reduced microvessel density in MEC-1-colonized spleens (Figure 4H) and increased survival in mice (Figure 4I).
In conclusion, these data show that a pharmacological agent that inhibits HIF-1α recapitulates chronic HIF-1α silencing by affecting colonization of hematopoietic and lymphoid organs, neoangiogenesis, and CLL progression, even though some consequences of EZN-2208 treatment may be HIF-1α independent.
EZN-2208 affects BM and spleen colonization in mouse CLL and promotes ex vivo cell mobilization
We extended these results to an immunocompetent CLL transplantable model derived from EμTCL1 transgenic mice34-36 that is characterized by higher involvement of leukemia in lymphoid organs (Figure 5A). Transplanted CLL was used instead of EμTCL1 transgenic mice to avoid long-term leukemia development34 and long treatment with EZN-2208, which may lead to the accumulation of nonspecific effects.
HIF-1α inhibition affects BM and spleen colonization in mouse CLL and promotes ex vivo cell mobilization. (A) Spleen weight at the end of treatment with EZN-2208 (day 63) of mice transplanted with EµTCL1-derived leukemia and treated with EZN-2208 at day 54. Data are presented as mean values ± SEM (n = 3). (B) Percentage of leukemic cells (CD5+CD19+) in the BM of mice transplanted with EµTCL1-derived leukemia, treated with EZN-2208, and euthanized as in panel A (n = 3). (C) Percentage of annexin V+ leukemic cells (CD5+CD19+) in the BM and spleen of mice transplanted with EµTCL1-derived leukemia, treated with EZN-2208, and euthanized as in panel A (n = 3). (D) Number of microvessels per field (original magnification ×40) in spleen colonized by leukemic cells of mice treated and euthanized as in panel A. Data represent mean values ± SEM (n = 2). (E) Kaplan-Meier survival curve of mice transplanted with EµTCL1-derived leukemia, treated with EZN-2208, and euthanized when terminally sick (untreated, n = 7; EZN-2208-treated animals, n = 5). (F) Number of leukemic (CD5+CD19+) and nonleukemic (CD5−CD19−) cells released in the culture supernatant from spleen fragments of EµTCL1-derived leukemic mice after 48 hours ex vivo culture in bioreactor. Data are represented as mean values ± SEM of 3 independent experiments. (G) Percentage of annexin V+ leukemic (CD5+CD19+) and nonleukemic (CD5−CD19−) cells released from spleen fragment after ex vivo culture in bioreactor for 48 hours. Data are represented as mean values ± SEM of 3 independent experiments. (H) Percentage of annexin V+ leukemic (CD5+CD19+) cells within spleen fragments after ex vivo culture in bioreactor for 48 hours. Data represent mean values ± SEM of 3 independent experiments.
HIF-1α inhibition affects BM and spleen colonization in mouse CLL and promotes ex vivo cell mobilization. (A) Spleen weight at the end of treatment with EZN-2208 (day 63) of mice transplanted with EµTCL1-derived leukemia and treated with EZN-2208 at day 54. Data are presented as mean values ± SEM (n = 3). (B) Percentage of leukemic cells (CD5+CD19+) in the BM of mice transplanted with EµTCL1-derived leukemia, treated with EZN-2208, and euthanized as in panel A (n = 3). (C) Percentage of annexin V+ leukemic cells (CD5+CD19+) in the BM and spleen of mice transplanted with EµTCL1-derived leukemia, treated with EZN-2208, and euthanized as in panel A (n = 3). (D) Number of microvessels per field (original magnification ×40) in spleen colonized by leukemic cells of mice treated and euthanized as in panel A. Data represent mean values ± SEM (n = 2). (E) Kaplan-Meier survival curve of mice transplanted with EµTCL1-derived leukemia, treated with EZN-2208, and euthanized when terminally sick (untreated, n = 7; EZN-2208-treated animals, n = 5). (F) Number of leukemic (CD5+CD19+) and nonleukemic (CD5−CD19−) cells released in the culture supernatant from spleen fragments of EµTCL1-derived leukemic mice after 48 hours ex vivo culture in bioreactor. Data are represented as mean values ± SEM of 3 independent experiments. (G) Percentage of annexin V+ leukemic (CD5+CD19+) and nonleukemic (CD5−CD19−) cells released from spleen fragment after ex vivo culture in bioreactor for 48 hours. Data are represented as mean values ± SEM of 3 independent experiments. (H) Percentage of annexin V+ leukemic (CD5+CD19+) cells within spleen fragments after ex vivo culture in bioreactor for 48 hours. Data represent mean values ± SEM of 3 independent experiments.
Mice transplanted with murine CLL cells were treated with EZN-2208 when they had 50% of leukemic cells (CD5+CD19+) in PB. As observed in the xenograft model, treatment with EZN-2208 in allotransplanted mice also caused marked reduction of spleen weight (Figure 5A) and BM colonization (Figure 5B), although in this model, EZN-2208 was more toxic to BM cells (Figure 5C). Short-term treatment with EZN-2208 also resulted in decreased microvessel density (Figure 5D) and increased survival in mice (Figure 5E).
Because leukemia cell death induced by EZN-2208 in BM and spleen of xeno- and allotransplanted mice could not account for the extent of spleen weight reduction and decreased BM infiltration (Figures 4 and 5), and because HIF-1α regulates genes involved in cell adhesion in CLL cells (Figure 1), we asked whether EZN-2208 induced release of CLL cells from protective niches, thus causing their further elimination by cell death upon mobilization.37 To measure cell mobilization in a confined ex vivo system, small fragments of leukemic spleens were maintained for 48 hours in bioreactor-based rotary cell cultures, which preserve organ architecture and ensure long-term cell viability.38 Ex vivo treatment with EZN-2208 induced specific mobilization of CLL cells from leukemic spleens into the culture supernatant without affecting the release of other cell types (Figure 5F). In addition, CLL cells mobilized to the culture medium specifically underwent apoptosis as compared with mobilized non-CLL cells (Figure 5G), as well as to CLL cells that remained embedded in the spleen (Figure 5H), thus demonstrating that EZN-2208 induced CLL mobilization, followed by cell death.
In conclusion, experiments with allotransplanted CLL cells confirmed previous results obtained in xenograft experiments and further indicated that HIF-1α controls homing and retention of CLL cells in protective microenvironments.
HIF-1α is transcriptionally regulated in CLL cells upon coculture with stromal cells
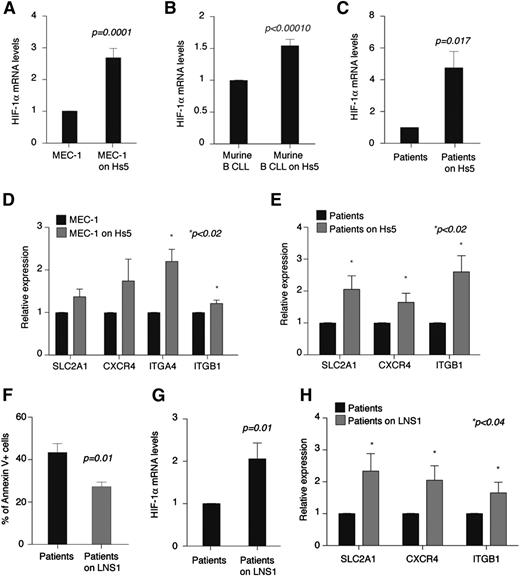
While performing coculture experiments, we observed that HIF-1α is transcriptionally upregulated in CLL cells in culture with stromal cells. As shown in Figure 6A-C, upon normoxic coculture with Hs5 cells, BM stromal cells HIF-1α mRNA levels increased in MEC-1 cells, CLL cells from EμTCL1 leukemic mice, and primary CLL cells from patients. HIF-1α upregulation was accompanied by increased expression of HIF-1α target genes SLC2A1, CXCR4, ITGB1, and ITGA4 when expressed (Figure 6D-E), albeit surface CXCR4 did not vary considerably overall (CXCR4 surface levels did not change, whereas the number of MEC-1 cells expressing surface CXCR4 slightly increased, leading to increased CXCL12-mediated chemotaxis; supplemental Figure 2A,F).
HIF-1α is transcriptionally upregulated in CLL cells upon coculture with stromal cells. (A-C) Relative expression of HIF-1α after 24 hours of coculture with Hs5 cells as compared with cells cultured without stroma. (A) MEC-1 cells. Data represent mean values ± SEM of 3 independent experiments. (B) Leukemic murine cells (CD5+CD19+) from PB of EµTCL1-transplanted mice. Data represent mean values ± SEM of 2 independent experiments. (C) Primary CLL B cells. Data represent mean values ± SEM (n = 10). (D-E) Relative expression of the indicated genes in MEC-1 cells (D) and primary CLL B cells (E) after 24 hours of coculture with Hs5 cells as compared with cells cultured without stroma. Data represent mean values ± SEM of 3 independent experiments (D) and mean values ± SEM (n = 10) (E). (F) Percentage of annexin V+ primary CLL B cells (CD19+) after 72 hours of culture or coculture with CLL lymph node stromal cells (LNS1) (n = 5). (G) Relative expression of HIF-1α in primary CLL B cells after 24 hours of coculture with LNS1 as compared with cells cultured without stroma (n = 9). (H) Relative expression of the indicated genes in primary CLL B cells after 24 hours of coculture with LNS1 cells as compared with cells cultured without stroma (n = 9).
HIF-1α is transcriptionally upregulated in CLL cells upon coculture with stromal cells. (A-C) Relative expression of HIF-1α after 24 hours of coculture with Hs5 cells as compared with cells cultured without stroma. (A) MEC-1 cells. Data represent mean values ± SEM of 3 independent experiments. (B) Leukemic murine cells (CD5+CD19+) from PB of EµTCL1-transplanted mice. Data represent mean values ± SEM of 2 independent experiments. (C) Primary CLL B cells. Data represent mean values ± SEM (n = 10). (D-E) Relative expression of the indicated genes in MEC-1 cells (D) and primary CLL B cells (E) after 24 hours of coculture with Hs5 cells as compared with cells cultured without stroma. Data represent mean values ± SEM of 3 independent experiments (D) and mean values ± SEM (n = 10) (E). (F) Percentage of annexin V+ primary CLL B cells (CD19+) after 72 hours of culture or coculture with CLL lymph node stromal cells (LNS1) (n = 5). (G) Relative expression of HIF-1α in primary CLL B cells after 24 hours of coculture with LNS1 as compared with cells cultured without stroma (n = 9). (H) Relative expression of the indicated genes in primary CLL B cells after 24 hours of coculture with LNS1 cells as compared with cells cultured without stroma (n = 9).
Because Hs5 cells do not originate from a CLL patient and do not epitomize the microenvironment of secondary lymphoid organs, we also analyzed HIF-1α regulation upon coculture with stromal cells derived from the lymph node of a CLL patient (LNS1 cells). In cocultures, LNS1 protected primary CLL B cells from apoptosis (Figure 6F) but did not induce CLL proliferation (data not shown). Transcriptional upregulation of HIF-1α and HIF-1α target genes was confirmed upon coculture of primary CLL cells with LNS1 cells (Figure 6G-H). However, surface CXCR4 was downregulated in LNS1 cocultures (supplemental Figure 2G-H). This finding is consistent with the observations that compared with Hs5 cells, LNS1 cells express high levels of CXCL12 (supplemental Figure 2I), as previously reported for stromal cells in CLL lymph nodes,39 and surface CXCR4 is downregulated via receptor endocytosis in CLL cells in the presence of high CXCL12.8
In summary, these experiments indicate that in CLL cells, HIF-1α levels are controlled not only through protein stabilization18 but also transcriptionally, as a consequence of the interaction of CLL cells with stromal cells. Although the specific signal or signals inducing HIF-1α expression remain to be identified, this observation reinforces the concept that in CLL, HIF-1α is an important regulator of the interaction of leukemic cells with the microenvironment.
Variable expression of HIF-1α and HIF target genes in CLL patients
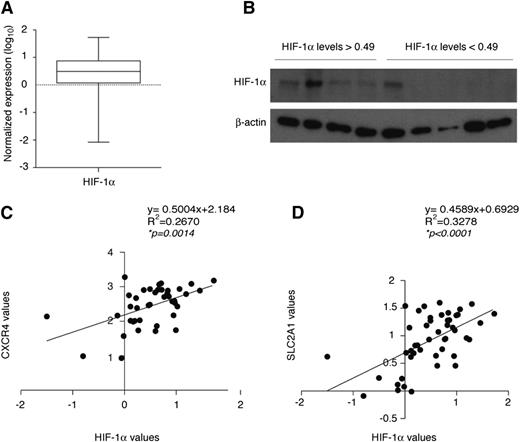
Because HIF-1α is transcriptionally regulated in CLL cells, we asked whether HIF-1α mRNA levels differ across CLL patients. PB leukemic cells from 50 CLL patients showed considerable variation in HIF-1α mRNA expression, which largely correlated with protein expression upon hypoxic stabilization (Figure 7A-B). Correlation curves with the HIF-1α target genes CXCR4 and SLC2A1 indicated that HIF-1α mRNA expression correlates with CXCR4 and SLC2A1 and is therefore indicative of differential transcriptional activity in CLL patients (Figure 7C-D).
HIF-1α expression is variable and correlates with HIF-1α target genes in CLL patients. (A) Box plot representing mRNA expression levels of HIF-1α in primary CLL B cells (n = 50). Expression was calculated using the 2−ΔCt method relative to 18S expression and expressed as log10. The median value was 0.49. (B) Western blot analysis of HIF-1α in primary CLL B cells expressing HIF-1α mRNA levels greater than or less than the median value of 0.49 calculated in panel A and exposed to 1% oxygen for 8 hours to stabilize HIF-1α. Spearman correlation analysis of HIF-1α mRNA levels and mRNA levels of CXCR4 (n = 41) (C) and SLC2A1 (n = 48) (D) in CLL B cells from patients, calculated by the 2−ΔCt method relative to 18S expression and expressed as log10.
HIF-1α expression is variable and correlates with HIF-1α target genes in CLL patients. (A) Box plot representing mRNA expression levels of HIF-1α in primary CLL B cells (n = 50). Expression was calculated using the 2−ΔCt method relative to 18S expression and expressed as log10. The median value was 0.49. (B) Western blot analysis of HIF-1α in primary CLL B cells expressing HIF-1α mRNA levels greater than or less than the median value of 0.49 calculated in panel A and exposed to 1% oxygen for 8 hours to stabilize HIF-1α. Spearman correlation analysis of HIF-1α mRNA levels and mRNA levels of CXCR4 (n = 41) (C) and SLC2A1 (n = 48) (D) in CLL B cells from patients, calculated by the 2−ΔCt method relative to 18S expression and expressed as log10.
In conclusion, these data suggest that there are 2 levels of transcriptional regulation of HIF-1α in CLL: in addition to intrinsic variability of HIF-1α mRNA levels in PB CLL cells, HIF-1α is also transcriptionally regulated upon interaction with the microenvironment, independently of basal expression (supplemental Table 2). Although further studies in larger patients cohorts will be necessary to elucidate the significance of different baseline expression of HIF-1α, our work suggests that regulation of HIF-1α within the leukemia microenvironment may contribute to the regulation of CLL pathogenesis.
Discussion
CLL is a chronic malignancy of mature B cells that exemplifies the importance of tumor-microenvironment cross talks. CLL B cells localize within protective niches in BM and secondary lymphoid organs, where they receive protective signals in the form of secreted factors and cell-cell interaction proteins and acquire resistance to spontaneous and chemotherapy-induced apoptosis.8,9 Here, we find that the transcription factor HIF-1α is a novel regulator of the interaction of CLL cells with leukemia microenvironments and, in turn, is regulated by this interaction through a mechanism involving transcriptional regulation.
Unbiased microarray analysis after HIF-1α silencing revealed that HIF-1α regulates a number of genes involved in mediating chemokine responses and cell adhesion in CLL, including crucial homing and retention factors such as CXCR4 and VLA-4.24 As a consequence, abating HIF-1α expression in CLL cells impaired CXCL12-directed chemotaxis, adhesion to stroma, BM homing, and BM and spleen colonization, and resulted in increased survival in a CLL mouse model. In confirmation of these results, short-term treatment with EZN-2208 (a topoisomerase I inhibitor and cytotoxic agent that also inhibits HIF-1α) impaired BM and spleen colonization and extended survival in mice, even after leukemia establishment and in the absence of overt cytotoxicity. Because HIF-1α silencing caused impaired spleen colonization in the absence of specific homing defects, we hypothesized that decreased HIF-1α function may lead to impaired spleen retention, followed by elimination of CLL cells in circulation. Ex vivo leukemic spleen cultures in a bioreactor-based system showed that EZN-2208 induced specific mobilization of CLL cells and restored their cell death proficiency once outside the organ, thus suggesting that also upon specific HIF-1α silencing, CLL cell mobilization may similarly cause cell death in PB.
Although our data indicate that in CLL, HIF-1α causes homing and retention in protective microenvironments, it is interesting to note that in other hematologic malignancies, including multiple myeloma and acute and chronic myeloid leukemia, hypoxia or HIF-1α activity instead induces cell egress and disease dissemination.3,22,40,41 This difference may be caused by the selectivity of HIF-1α-mediated responses in different cell types,42-44 such that adhesion factors may be regulated by HIF-1α specifically in CLL. Further studies should address in greater detail the specificity of HIF-1α activity in different tumor types, especially if HIF inhibitors will be considered for therapeutic intervention.
Although EZN-2208 was used in this study as a means to acutely inhibit HIF-1α, our data suggest that HIF-1α may be investigated in the future as a new therapeutic target in CLL, because HIF-1α inhibitors may not only exert toxic activities toward CLL cell but also synergize with chemotherapy through CLL mobilization. Because EZN-2208 is not a specific HIF-1α inhibitor, more specific compounds should be tested in the future.
Of interest, we found that HIF-1α responds to environmental stimuli in CLL B cells by being upregulated in coculture with stromal cells. This regulation occurs in normoxic conditions and at the transcriptional level, a way of regulating HIF-1α expression that is poorly characterized compared with posttranslational stabilization. Nonetheless, recent studies are beginning to implicate a number of paracrine factors in the transcriptional regulation of HIF-1α.45 Therefore, it will be important to understand whether HIF-1α expression in CLL is induced by direct cell interaction with stromal cell components or by secreted factors. Our data extend recent evidence whereby HIF-1α was found upregulated in in breast cancer cells upon coculture with stromal cells,28 and although in CLL (unlike breast cancer), HIF-1α upregulation occurs at the transcriptional level, our data further strengthen the concept that HIF-1α is an important regulator of the interaction of neoplastic cells with local microenvironments.
Lastly, in keeping with our observations on the transcriptional regulation of HIF-1α, we found that HIF-1α mRNA levels vary considerably among CLL patients. These differences appear to reflect differential HIF-1α activity, because HIF-1α levels correlate with expression of HIF-1α target genes. These data extend previous work in which the HIF-1α protein was found highly expressed in CLL cells compared with normal B cells through posttranslational accumulation.18 Therefore, in combination with posttranslational protein stabilization,18 transcriptional regulation may lead to differential expression of HIF-1α in different patients.
In summary, our results demonstrate that HIF-1α plays critical and pleiotropic roles in CLL pathogenesis and suggest that it may represent a new therapeutic target for CLL treatment to be evaluated by future preclinical experiments.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all current and previous members of the laboratory for valuable discussion and support, with special thanks to Silvia Cavalli; M. Rocchi for tissue preparation and immunohistochemistry; and L. Greenberger, Y. Zhang, and Belrose Pharma for providing EZN-2968, EZN-3088, and EZN-2208.
This work was supported by the Giovanni Armenise–Harvard Foundation with a Career Development Award (R.B.), by grants from the Italian Association for Cancer Research Special Program Molecular Clinical Oncology, 5 per mille #9965 and Investigator Grant #12769 (R.B.), and by the CLL Global Research Foundation (R.B.).
Authorship
Contribution: N.C. performed the cytofluorimetric analyses; D.B. performed the ex vivo organ cultures in bioreactor; M. Ponente performed the real-time PCR analyses; E.t.H. performed the in vitro adhesion assays; C.S. and L.S. performed the analysis of prognostic markers in CLL patients; M.T.S.B. performed the preparation and analysis of EμTCL1 mice; P.B. and F.M.B. performed the microarray analysis; E.L. and A.B. stabilized and studied the LNS1 cells; G.T. performed the analysis of microarray data; R.V. performed all other experiments; E.F. and M.F. designed the bioreactor experiments; P.G. designed the experiments with patients cells; M. Ponzoni performed the immunohistopathological evaluation of leukemic mice; F.C.-C. supervised the study and provided expertise with patient analysis; R.B. designed the study; and R.B. and R.V. analyzed the data and wrote the paper. All authors discussed the results and commented on the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for E.t.H. is Department of Leukemia, The University of Texas MD Anderson Cancer Center, Houston, TX.
Correspondence: Rosa Bernardi, Via Olgettina 60, 20132 Milan, Italy; e-mail: bernardi.rosa@hsr.it.