Key Points
Targeting of PAK1 inhibits primary AML and MDS patients' cells including leukemia stem cells but spares healthy stem and progenitor cells.
Inhibition of PAK1 induces differentiation and apoptosis of AML cells through downregulation of MYC and a core network of MYC target genes.
Abstract
Poor clinical outcome of acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS) has been attributed to failure of current chemotherapeutic regimens to target leukemic stem cells. We recently identified p21-activated kinase (PAK1) as a downstream effector molecule of H2.0-like homeobox (HLX), a gene functionally relevant for AML pathogenesis. In this study, we find that inhibition of PAK1 activity by small molecule inhibitors or by RNA interference leads to profound leukemia inhibitory effects both in vitro and in vivo. Inhibition of PAK1 induces differentiation and apoptosis of AML cells through downregulation of the MYC oncogene and a core network of MYC target genes. Importantly, we find that inhibition of PAK1 inhibits primary human leukemic cells including immature leukemic stem cell-enriched populations. Moreover, we find that PAK1 upregulation occurs during disease progression and is relevant for patient survival in MDS. Our studies highlight PAK1 as a novel target in AML and MDS and support the use of PAK1 inhibitors as a therapeutic strategy in these diseases.
Introduction
Acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS) are heterogeneous neoplastic blood diseases with poor clinical outcome. AML has a 5-year overall survival of only 3% to 8% in patients ≥60 years of age.1,2 Although disease course and prognosis are highly variable in MDS, ∼30% to 40% of patients with MDS will progress to AML,1,2 and allogeneic stem cell transplantation remains the only curative option. Current chemotherapeutic approaches, which have remained unchanged for the last 40 years, target rapidly dividing cells, having limited effects on the preleukemic and leukemic stem cells (LSC) responsible for disease propagation and relapse. Targeting of (pre)leukemic stem cells is vital to maintaining remission and improving survival in AML and MDS.
We recently identified p21-activated kinase (PAK1) as a downstream effector molecule of H2.0-like homeobox (HLX), a gene upregulated in preleukemic stem cells and functionally relevant for AML pathogenesis.3 Using data from a large cohort of AML patients, we found that higher levels of PAK1 in AML are associated with significantly inferior overall patient survival, implicating PAK1 as a potential clinical target in this disease. PAK1 is a member of the PAK family of serine/threonine kinases that act downstream of the Rho family small GTPases Cdc42 and Rac1 in a variety of signaling pathways. PAKs in mammalian cells are classified into group I (PAK1, PAK2, and PAK3) and group II (PAK4, PAK5, and PAK6) on the basis of structural similarities, although group members exhibit significant functional differences.4 PAK family members have been shown to play important roles in cell proliferation and motility, as well as in cell transformation and tumor progression.5 PAK1 has been implicated in oncogenic transformation in several malignancies including breast, ovarian, melanoma, T-cell lymphoma, liver, and bladder cancers.6-14
In this study, we assessed the importance of PAK1 in the myeloid malignancies AML and MDS. We find that inhibition of PAK1 both chemically and using short-hairpin RNA (shRNA) targeting has profound antileukemic effects both in vitro and in vivo. Inhibition or reduction of PAK1 leads to induction of monocytic differentiation and apoptosis via repression of the oncoprotein c-MYC (MYC) and a MYC transcriptional network. Moreover, we show that leukemia cells from human patients with AML are reliant on PAK1 function and that chemical inhibition of PAK1 preferentially inhibits leukemic cells over healthy cells, including in immature LSC-enriched populations. Our studies highlight PAK1 as a novel target in AML and MDS and support the use of PAK1 inhibitors as a therapeutic strategy in these diseases.
Methods
Cell culture
THP-1, MOLM-13, HL-60, and KG1a cells were purchased from American Type Culture Collection (ATCC) and cultured according to ATCC recommendations. All cell lines were maintained in an incubator at 37°C and 5% CO2. Primary mononuclear samples were obtained from patients with AML or from healthy donors. Patient characteristics are shown in supplemental Figure 5A available on the Blood Web site. The studies were approved by the institutional review board (#2008-942).
Knockdown of PAK1 and PAK2 by shRNA and chemical inhibition of PAK1
To silence PAK1 and PAK2 by RNAi, we transduced cells with plko.1-based lentiviruses (Open Biosystems) by spinfection at 1800 rpm for 90 minutes at 37°C in 8 μg/mL polybrene. Stably transduced cell lines were selected with 1.2 μg/mL puromycin for 6 days. PAK1-specific sequences used were TRCN0000002225 (CGATGAGAAATACCAGCACTA) and TRCN0000002226 (GCGATCCTAAGAAGAAATATA). PAK2 specific sequences used were TRCN0000002115 (CTCTAGGAACCAAAGTGATTT) and TRCN0000002118 (TGGGAATGGAAGGATCTGTTA). P21-activated kinase inhibitor III (IPA-3; 1,1′-dithiodi-2-naphthtol; Tocris Bioscience), negative control of IPA-3 (PIR 3.5; 6,6′-dithiodi-2-naphthtol; Tocris Bioscience), and the small molecule pyridopyrimidinone FRAX-597 (Genentech) were suspended in dimethyl sulfoxide (DMSO) and assayed at the indicated concentrations.
Proliferation, apoptosis, differentiation, and clonogenic assays
Manual cell counts were performed by culturing 2 × 105 cells/mL in 1 mL culture medium in 24-well plates. Viable cells were counted daily using trypan blue exclusion. Cell differentiation was assessed by morphologic observation after Diff Quik (IMEB) staining, as well as fluorescence-activated cell sorter (FACS) analysis using antibodies directed against human CD11b (CR3; BD Biosciences) and CD15 (VIMC6; Life Technologies) on a BD FACSAria II machine. Cell line and stem cell apoptosis was measured with Annexin-V FLUOS Staining Kit (Roche) and 4′,6-diamidino-2-phenylindole (DAPI; 1:1000), followed by flow cytometric analysis. Viable cells were defined as Annexin-V−/DAPI−. For AML cell line colony assays, 3000 cells were plated in HSC002SF methylcellulose (R&D Systems) with 5% fetal bovine serum. Colonies were scored at day 10. For primary cell colony assays, 1 to 3 × 105 mononuclear cells were plated in 1 mL HSC003 (R&D Systems) methylcellulose media with 40 μg/mL human low-density lipoprotein (hLDL) (Calbiochem), 10 ng/mL human interleukin-6, and 25 ng/mL human FMS-like tyrosine kinase 3 ligand (hFLT3L). For serial replating assays, colonies were resuspended, and all cells were replated as above with indicated doses of IPA-3 and FRAX-597. Colonies were scored at day 10.
Western blotting
Proteins were resolved on 10% to 12% polyacrylamide gels after lysis in denaturing buffer (R&D Systems) and transferred to Nitrobind nitrocellulose membranes (Maine Manufacturing). Membranes were blocked for 1 hour in 2% milk in Tris-buffered saline containing 0.1% Tween-20 (TBS-T). For detection of protein phosphorylation, membranes were blocked in TBS-T with 2.5% bovine serum albumin (Roche). Primary antibody incubations were carried out at 4°C overnight or at room temperature for 1 hour in TBS-T containing 2.5% bovine serum albumin. The following primary antibodies were used: AKTS473 (1:1000, #9271S; Cell Signaling), AKT (1:1000, C67E7 #4691S; Cell Signaling), actin (1:1000, C-11 #sc-1615; Santa Cruz Biotech), MYC (1:1000, D84C12 #5605; Cell Signaling), and PAK1 (1:1000, #2602; Cell Signaling). Secondary antibody incubations (LI-COR 926-32213 IRDye 800CW donkey anti-rabbit immunoglobulin IgG [H+L] 1:5000 and LI-COR 926-68074 IRDye 680RD donkey anti-Goat IgG [H+L] 1:5000) were performed at room temperature for 1 hour. Blots were imaged on a LI-COR Odyssey Imaging System.
Microarray experiments, analysis, and quantitative reverse transcriptase-polymerase chain reaction
Microarray data are available in the Gene Expression Omnibus, accession no. GSE70791. For microarray, RNA was isolated from THP-1 cells after 5 hours of treatment with IPA-3 (6 μg/mL), FRAX-597 (2 μg/mL), or an equal volume of DMSO using Trizol Reagent (Invitrogen). After evaluation of the quality of RNA with an Agilent2100 Bioanalyzer, RNA was labeled with the GeneChip WT terminal labeling kit (Affymetrix), and labeled cRNA of each individual sample was hybridized to Affymetrix Human Gene 2.0ST microarrays (Affymetrix) and scanned by GeneChip Scanner 3000 7G system (Affymetrix) according to standard protocols. For evaluation of individual genes, reverse transcriptase (RT) reaction was performed using iScript cDNA Synthesis Kit (Bio-Rad) and quantitative polymerase chain reaction (qPCR) was performed using Power SYBR Green Master Mix (ThermoFisher) on a ViiA 7 Real Time PCR System. qRT-PCR primers were as follows: hPAK1—fwd, GATGCTGGAACCCTAAACCA; rev, GGCCGCTCTTTCTCTTTCTT; hPAK2—fwd, ACAGAAGCACCCGCAGTAGT; rev, AAAGACTTGGCAGCACCATC; hDUSP7—fwd, GCGAGTTCACCTACAAGCAG; rev, TCTGCATCAGATAGGCCACA; hMYC—fwd, CCTGGTGCTCCATGAGGAGAC; rev, CAGACTCTGACCTTTTGCCAGG; hAMD1—fwd, CTCACCAAGGGTACCCACAC; rev, CGTCCCATACAATATGCTGCT.
In vivo leukemia models
All animal experiments were performed in compliance with institutional guidelines and approved by the Animal Institute Committee of the Albert Einstein College of Medicine (#20130102). NSG mice were sublethally irradiated (200 rad) and were intravenously injected (tail vein) with 3 × 105 THP-1 cells. At 46 days, tissues were fixed in neutral buffered formalin fixative and embedded in paraffin for hematoxylin and eosin (H&E) staining. Tissues were imaged using EVOS Cell Imaging station (Life Technologies; 4 animals per construct per experiment; n = 2). For xenografting of AML patient samples, mononuclear cells were isolated and treated for 12 hours with either DMSO or IPA-3; 1 × 106 cells were transplanted into each animal of a cohort of 2 to 5 NSG mice per condition. Six weeks after transplantation, murine bone marrows were assessed via flow cytometry for the presence of CD45+CD33+ human myeloid cells. The experiment was performed twice, with 2 independent primary AML patient samples.
Stem cell-enriched apoptosis assays
Viability of stem/progenitor cells was assessed based on a previously published assay.15 AML or healthy cells (AllCells) were cultured in Myelocult H5100 media (Stem Cell Technologies) supplemented with Primocin 100 μg/mL, human interleukin 3 (hIL-3) 10 ng/mL, human stem cell factor (hSCF)100 ng/mL, human thrombopoietin (hTPO) 50 ng/mL, human interleukin 6 (hIL-6) 10 ng/mL, hFLT3L 20 ng/mL, and hLDL 40 μg/mL for 24 hours with IPA-3, FRAX-597, or DMSO. All drug treatments were performed in triplicate. After 24 hours in culture, cells were isolated, washed twice with phosphate-buffered saline, and labeled with anti-CD38-phycoerythrin-Cy7 (PE-Cy7; HIT2) and anti-CD34-allophycocyanin (APC) for 20 minutes.
Data sets and statistical analysis
For MDS analyses, we used the publicly available data set GSE19429 consisting of gene expression data of CD34+ cells isolated from the bone marrow of 183 MDS patients. For survival analysis, patients were grouped into low- and high-expression categories by use of median expression of PAK1 or PAK2, and survival was assessed by log-rank test. Survival package in R was used to plot Kaplan-Meier curves. Correlation of PAK1 expression and survival in individual patients was assessed by correlation coefficient R and regression P value. Multivariate analysis using Cox regression model was performed with SAS9.3 using the clinical information available for patients from the GSE19429 data set (including sex, French-American-British (FAB) classification, hemoglobin, bone marrow blasts, white blood cell, PAK1 expression status [low/high], and International Prognostic Scoring System (IPSS) category). For AML analyses, we used the publicly available data set GSE14468 consisting of 598 patients with AML. Patients were grouped into low- and high-expression categories by the 33rd percentile of PAK1 expression. Fisher’s exact test of independence was used to assess correlations of patient characteristics (FAB subtype/karyotype/mutation) and PAK1 expression. Error bars represent mean ± standard deviation unless specifically indicated. P values are by 2-tailed Student t test unless specifically indicated.
Results
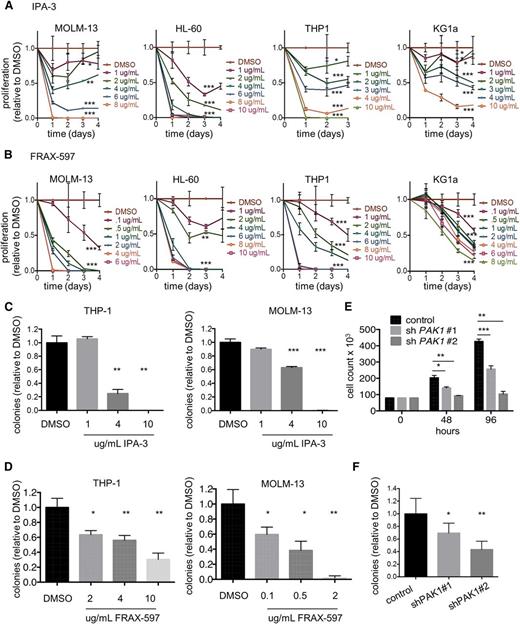
Targeting PAK1 signaling slows growth and reduces leukemic colony formation capacity of human acute myeloid leukemia cells
To determine whether PAK1 is functionally important in AML, we first examined the effects of PAK1 inhibition on the growth of a panel of human AML cell lines (MOLM-13, HL-60, THP-1, and KG1a). We selected 2 small molecule inhibitors that target PAK1 directly: IPA-3, an allosteric inhibitor of PAK1-3 activation, and FRAX-597, an ATP-competitive PAK1-3 inhibitor currently under advanced preclinical investigation for solid tumor malignancies.16,17 Treatment with IPA-3 or FRAX-597 inhibited the proliferation of all 4 cell lines in a dose-dependent manner with 24-hour IC50s in the range of 0.45 to 5.3 μg/mL (6-15 μM for IPA-3; 800 nM to 8 μM for FRAX-597; Figure 1A-B; supplemental Figure 1A-B). Leukemic colony formation capacity of THP-1 and MOLM-13 AML cells was also inhibited on treatment with both PAK1 inhibitors (Figure 1C-D). Importantly, we observed only mild toxicity with PIR3.5, a structural isomer of IPA-3 that has no inhibitory activity toward PAK1 and serves as a negative control for PAK1 inhibition17 (supplemental Figure 1C). To confirm PAK1 inhibition by both IPA-3 and FRAX-597 in AML cell lines, we evaluated levels of phosphorylated AKT at Ser473, an established PAK1 target.16,18,19 Treatment with either drug resulted in decreased phosphorylation of AKT at Ser473 in a dose-dependent manner (supplemental Figure 1D-E).
Targeting PAK1 inhibits growth and reduces clonogenicity of human acute myeloid leukemia cells. (A) MOLM-13, HL-60, THP-1, and KG1a cells were treated with increasing concentrations of IPA-3. Cell viability was measured by cell counts with trypan blue exclusion every 24 hours. Experiment performed in triplicate (P values are shown for last time point indicated). (B) MOLM-13, HL-60, THP-1, and KG1a cells were treated with increasing concentrations of FRAX-597. Statistics are indicated as in A. A total of 3000 THP-1 or MOLM-13 cells were seeded in methylcellulose containing indicated concentrations of (C) IPA-3 or (D) FRAX-597. (E) Two unique shRNAs targeting human PAK1 were expressed in THP-1 cells. Cell growth was measured by cell counts with trypan blue exclusion. Average of 3 independent experiments. (F) Colony formation analysis of THP-1 cells treated with 2 independent PAK1 knockdown sequences. Average of 2 independent experiments. *P < .05, **P < .01, and ***P < .001 for all experiments.
Targeting PAK1 inhibits growth and reduces clonogenicity of human acute myeloid leukemia cells. (A) MOLM-13, HL-60, THP-1, and KG1a cells were treated with increasing concentrations of IPA-3. Cell viability was measured by cell counts with trypan blue exclusion every 24 hours. Experiment performed in triplicate (P values are shown for last time point indicated). (B) MOLM-13, HL-60, THP-1, and KG1a cells were treated with increasing concentrations of FRAX-597. Statistics are indicated as in A. A total of 3000 THP-1 or MOLM-13 cells were seeded in methylcellulose containing indicated concentrations of (C) IPA-3 or (D) FRAX-597. (E) Two unique shRNAs targeting human PAK1 were expressed in THP-1 cells. Cell growth was measured by cell counts with trypan blue exclusion. Average of 3 independent experiments. (F) Colony formation analysis of THP-1 cells treated with 2 independent PAK1 knockdown sequences. Average of 2 independent experiments. *P < .05, **P < .01, and ***P < .001 for all experiments.
To confirm these findings were due to specific inhibition of PAK1, we used RNA interference with 2 independent shRNA sequences targeting human PAK1. We achieved reductions of 89% (shPAK1#1) and 54% (shPAK1#2) in PAK1 transcript levels and a corresponding decrease in the amount of PAK1 protein (supplemental Figure 1F-G). PAK1 knockdown was confirmed by significant reduction in AKT Ser473 phosphorylation (supplemental Figure 1G). PAK1 knockdown inhibited both the proliferation and leukemic colony formation capacity of THP-1 cells (Figure 1E-F). Notably, PAK1 knockdown was specific for PAK1 and did not result in reducing levels of PAK2, nor did PAK1 knockdown result in a compensatory increase in PAK2 levels (supplemental Figure 1F). PAK3 was not expressed in the AML cell lines. Furthermore, to assess the importance of PAK2 in AML, we used 2 different shRNAs to reduce PAK2 expression in THP-1 and MOLM-13 cells. We found that PAK2 inhibition had no effect on proliferation of these AML cell lines (supplemental Figure 1H-I).
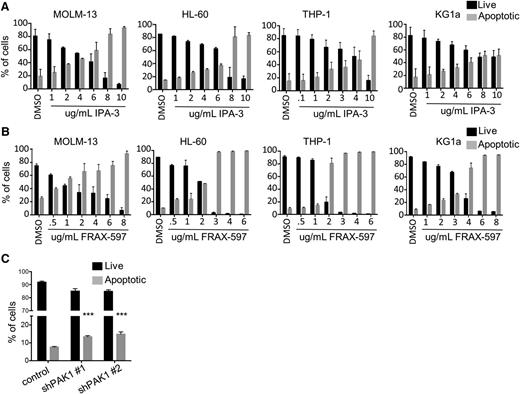
PAK1 inhibition induces apoptosis of AML cells
To investigate the cellular mechanism of PAK1-dependent inhibition of cell growth and clonogenic capacity, cell lines were treated with increasing concentrations of IPA-3 or FRAX-597 for 24 hours and assessed for apoptosis and cell cycle arrest. Treatment with either compound resulted in a dose-dependent induction of apoptotic cell death in all 4 of the malignant myeloid cell lines tested, with observable progression through an early apoptotic stage (Figure 2A-B; supplemental Figure 2A). Additionally, we observed a significant increase in apoptosis of AML cells with PAK1 knockdown compared with the nonsilencing control (Figure 2C). Treatment of AML cell lines with either IPA-3 or FRAX-597 for 24 hours resulted in no changes in the cell cycle, despite reported roles for PAK1 in the regulation of cell cycle progression in other cell types20 (supplemental Figure 2B).
PAK1 inhibition induces apoptosis of AML cells. (A) MOLM-13, HL-60, THP-1, and KG1a cells were treated with increasing concentrations of IPA-3. Apoptosis was measured at 24 hours by flow cytometry using Annexin V and DAPI staining. Average of 2 independent experiments. (B) MOLM-13, HL-60, THP-1, and KG1a cells were treated with increasing concentrations of FRAX-597. Average of 2 independent experiments. (C) Human THP-1 AML cells were infected with lentiviruses targeting PAK1 for knockdown or a nontargeting control. Average of 2 independent experiments (***P < .001).
PAK1 inhibition induces apoptosis of AML cells. (A) MOLM-13, HL-60, THP-1, and KG1a cells were treated with increasing concentrations of IPA-3. Apoptosis was measured at 24 hours by flow cytometry using Annexin V and DAPI staining. Average of 2 independent experiments. (B) MOLM-13, HL-60, THP-1, and KG1a cells were treated with increasing concentrations of FRAX-597. Average of 2 independent experiments. (C) Human THP-1 AML cells were infected with lentiviruses targeting PAK1 for knockdown or a nontargeting control. Average of 2 independent experiments (***P < .001).
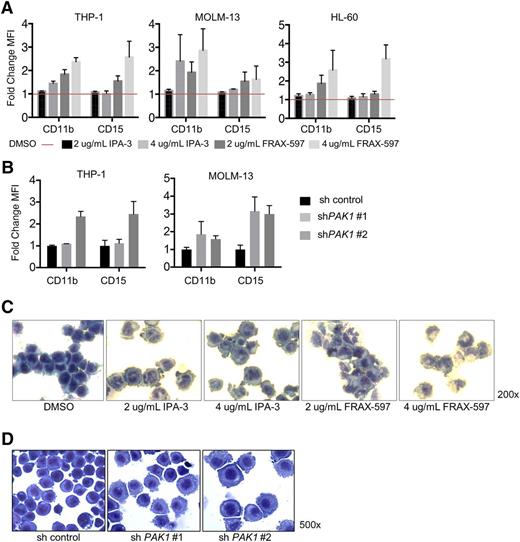
Inhibition of PAK1 induces myelomonocytic differentiation of AML cells
A block in terminal myeloid differentiation is a hallmark of myeloid leukemic cells. To determine whether modulating PAK1 activity could overcome the differentiation block in AML, we assessed the expression of myelomonocytic surface markers following incubation of AML cell lines with increasing concentrations of IPA-3 or FRAX-597 for 24 hours. Treatment with either drug induced upregulation of surface expression of mature myelomonocytic markers CD11b and CD15, indicating PAK1 inhibition differentiates AML cells toward the monocytic lineage (Figure 3A; supplemental Figure 3A). We confirmed a direct role for PAK1 in affecting myeloid differentiation, as shRNA knockdown of PAK1 similarly induced differentiation of AML cells toward the monocytic lineage (Figure 3B; supplemental Figure 3B). The appearance of cytomorphologic features of mature myeloid cells, including multilobed nuclei and an increase in the cytoplasm:nucleus ratio, correlated with the induction of myelomonocytic markers following drug treatment or PAK1 knockdown (Figure 3C-D; supplemental Figure 3C).
Inhibition of PAK1 induces myelomonocytic differentiation of AML cells. (A) THP-1, MOLM-13, and HL-60 cells were treated with indicated concentrations of IPA-3 or FRAX-597 for 24 hours and analyzed by flow cytometry for expression of CD11b and CD15 cell surface markers for myelomonocytic differentiation. Y-axis plots fold change mean fluorescence intensity. Average of 2 independent experiments; error bars represent standard error of the mean. (B) Expression of CD11b and CD15 cell surface markers on THP-1 infected with lentiviruses targeting PAK1 for knockdown. (C) HL-60 AML cells were treated with indicated concentrations of DMSO (0.2%), IPA-3, or FRAX-597 for 24 hours. Cells were then spun onto slides (cytospin) and stained with Wright-Giemsa stain. (D) THP-1 AML cells were infected with lentivirus targeting PAK1 for knockdown or a nontargeting control and selected with puromycin. Cells were spun onto slides and stained with Wright-Giemsa stain.
Inhibition of PAK1 induces myelomonocytic differentiation of AML cells. (A) THP-1, MOLM-13, and HL-60 cells were treated with indicated concentrations of IPA-3 or FRAX-597 for 24 hours and analyzed by flow cytometry for expression of CD11b and CD15 cell surface markers for myelomonocytic differentiation. Y-axis plots fold change mean fluorescence intensity. Average of 2 independent experiments; error bars represent standard error of the mean. (B) Expression of CD11b and CD15 cell surface markers on THP-1 infected with lentiviruses targeting PAK1 for knockdown. (C) HL-60 AML cells were treated with indicated concentrations of DMSO (0.2%), IPA-3, or FRAX-597 for 24 hours. Cells were then spun onto slides (cytospin) and stained with Wright-Giemsa stain. (D) THP-1 AML cells were infected with lentivirus targeting PAK1 for knockdown or a nontargeting control and selected with puromycin. Cells were spun onto slides and stained with Wright-Giemsa stain.
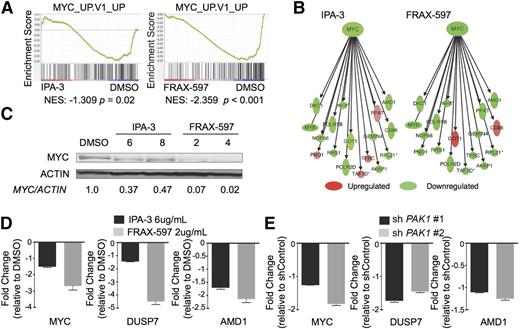
PAK1 inhibition leads to leukemic cell death by downregulation of a MYC transcriptional network
To obtain insight into the molecular mechanism for the induction of apoptosis and differentiation resulting from PAK1 inhibition in AML, we performed gene expression microarrays following treatment with either IPA-3 or FRAX-597. Consistent with our observed phenotypes, Ingenuity Pathway Analysis confirmed considerable enrichment for networks that regulate cancer, hematologic disease, cell growth and proliferation, and cell death and survival (supplemental Figure 4A). Gene Set Enrichment Analysis confirmed our results that PAK1 inhibition leads to an increase in differentiation and apoptosis (supplemental Figure 4B). Furthermore, we observed that treatment with either drug led to transcriptional changes inversely correlated with a MYC_UP.V1_UP gene set, comprising genes that are upregulated in primary breast cancer cells overexpressing MYC (NES IPA-3: −1.309, P = .02; NES FRAX-597: −2.359, P < .001; Figure 4A).21 Strikingly, both inhibitors of PAK1 led to a downregulation of MYC and a core set of genes directly targeted by MYC (Figure 4B). We validated reduction of MYC expression by western blot following treatment with both inhibitors (Figure 4C; supplemental Figure 4C). Furthermore, we confirmed MYC network downregulation by either inhibitor or PAK1 shRNA by validating a reduction in expression of several downstream targets of MYC including dual specificity phosphatase 7 (DUSP7) and adenosylmethionine decarboxylase 1 (AMD1) by qPCR (Figure 4D-E; supplemental Figure 4D).22-24
PAK1 inhibition leads to leukemic cell death by downregulation of MYC and a MYC transcriptional network. (A) Gene Set Enrichment Analysis of genes expressed differently in IPA-3 (6 μg/mL) or FRAX-597 (2 μg/mL) treated THP-1 cells compared with DMSO. (B) Ingenuity Pathway Analysis-generated image showing the downregulation of genes from the MYC network following treatment with either IPA-3 (6 μg/mL) or FRAX-597 (2 μg/mL). Intensities indicate degree of up- or downregulation. (C) Western blot shows reduction of MYC protein levels after 4 hours of treatment with IPA-3 or FRAX-597 in THP-1 cells. Concentrations shown are micrograms per milliliter. (D) qPCR validation of downregulated MYC network genes after treatment of THP-1 cells with either DMSO, IPA-3 (6 μg/mL) or FRAX-597 (2 μg/mL). (E) qPCR validation of downregulated MYC network genes after knockdown of PAK1 in THP-1 AML cells.
PAK1 inhibition leads to leukemic cell death by downregulation of MYC and a MYC transcriptional network. (A) Gene Set Enrichment Analysis of genes expressed differently in IPA-3 (6 μg/mL) or FRAX-597 (2 μg/mL) treated THP-1 cells compared with DMSO. (B) Ingenuity Pathway Analysis-generated image showing the downregulation of genes from the MYC network following treatment with either IPA-3 (6 μg/mL) or FRAX-597 (2 μg/mL). Intensities indicate degree of up- or downregulation. (C) Western blot shows reduction of MYC protein levels after 4 hours of treatment with IPA-3 or FRAX-597 in THP-1 cells. Concentrations shown are micrograms per milliliter. (D) qPCR validation of downregulated MYC network genes after treatment of THP-1 cells with either DMSO, IPA-3 (6 μg/mL) or FRAX-597 (2 μg/mL). (E) qPCR validation of downregulated MYC network genes after knockdown of PAK1 in THP-1 AML cells.
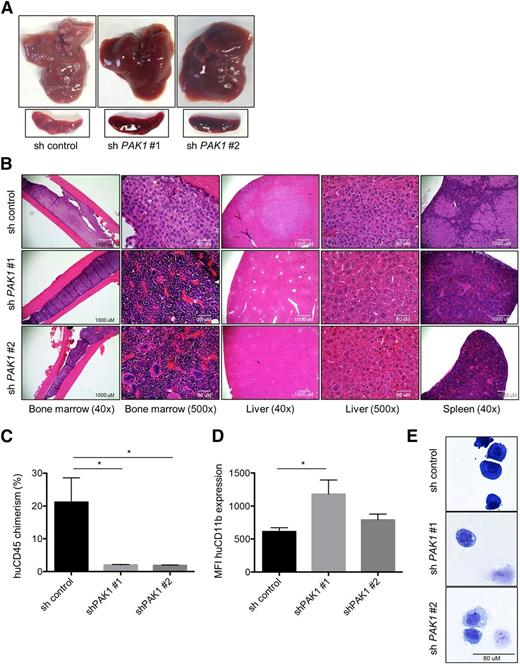
PAK1 reduction inhibits AML progression in vivo
Given the in vitro antileukemic effects of PAK1 inhibition, we examined the in vivo effects of loss of PAK1 using a murine xenotransplantation model of AML. THP-1 cells expressing a nonsilencing control or either of two PAK1 targeting constructs were injected into sublethally irradiated NSG mice. Gross organ analysis and histopathology at 6 weeks following transplantation revealed dramatically reduced infiltration of leukemic cells and preservation of normal NSG tissue architecture in the bone marrow, liver, and spleen of mice transplanted with PAK1 shRNAs compared with the nonsilencing control (Figure 5A-B). Mice injected with cells expressing either of the PAK1 shRNAs had a 91% reduction in engraftment of THP-1 cells in the liver compared with mice injected with the nonsilencing control cells (Figure 5C). Consistent with our in vitro studies, the few THP-1 cells that could be isolated from PAK1 knockdown experimental animals had increased CD11b cell surface expression and displayed a more differentiated morphology in cytospin preparations compared with nonsilencing control cells (Figure 5D-E).
PAK1 reduction inhibits AML progression in vivo. (A) Gross liver and spleen images 46 days after transplantation of 3 × 105 sh control, shPAK1#1, or shPAK1#2 THP-1 cells (4 animals per group per experiment; n = 2). (B) Hematoxylin and eosin (H&E) staining of femur, liver, and spleen sections of transplanted animals. (C) Chimerism of human THP-1 cells in the liver quantified by flow cytometry for cells expressing human CD45. Data represent the mean ± standard deviation. (D) CD11b expression levels on the surface of human CD45 expressing THP-1 cells isolated from the livers of animals transplanted with THP-1 cells expressing the control shRNA lentivirus or 1 of 2 PAK1 targeting sequences. (E) Cytospin preparations and Wright-Giemsa staining of THP-1 cells FACS sorted from the livers of transplanted animals. The few remaining cells that could be isolated from animals receiving PAK1 knockdown cells exhibit signs of myelomonocytic differentiation. *P < .05 for all experiments.
PAK1 reduction inhibits AML progression in vivo. (A) Gross liver and spleen images 46 days after transplantation of 3 × 105 sh control, shPAK1#1, or shPAK1#2 THP-1 cells (4 animals per group per experiment; n = 2). (B) Hematoxylin and eosin (H&E) staining of femur, liver, and spleen sections of transplanted animals. (C) Chimerism of human THP-1 cells in the liver quantified by flow cytometry for cells expressing human CD45. Data represent the mean ± standard deviation. (D) CD11b expression levels on the surface of human CD45 expressing THP-1 cells isolated from the livers of animals transplanted with THP-1 cells expressing the control shRNA lentivirus or 1 of 2 PAK1 targeting sequences. (E) Cytospin preparations and Wright-Giemsa staining of THP-1 cells FACS sorted from the livers of transplanted animals. The few remaining cells that could be isolated from animals receiving PAK1 knockdown cells exhibit signs of myelomonocytic differentiation. *P < .05 for all experiments.
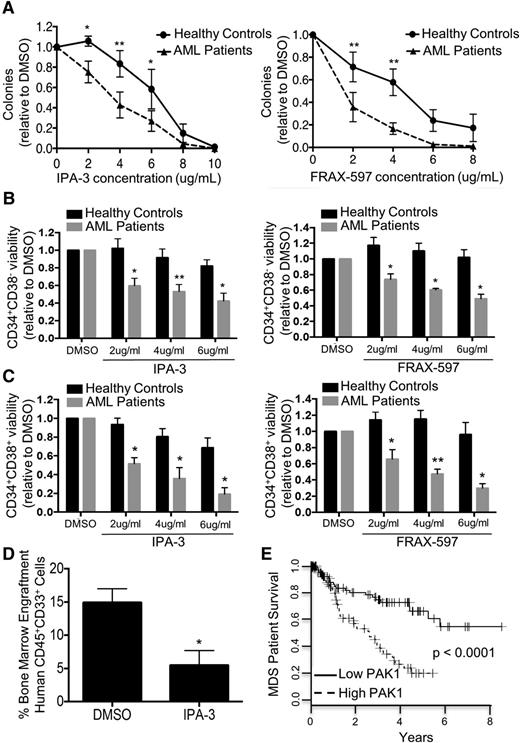
Targeting of PAK1 inhibits primary AML patients’ cells including immature, leukemia stem cell-enriched populations while sparing healthy stem and progenitor cells
Current chemotherapeutic approaches in AML and MDS target rapidly dividing cells, having minimal effects on the quiescent leukemic and preleukemic stem cells responsible for disease propagation and relapse. To investigate whether leukemic stem and progenitor cells are dependent on PAK1 function, we assessed the efficacy of PAK1 inhibition on leukemic colony formation of primary human mononuclear cells isolated from healthy controls or patients with active AML. PAK1 inhibition by either IPA-3 or FRAX-597 led to a dose-dependent reduction of leukemic colony formation capacity of AML patient cells, whereas colony formation capacity of healthy control cells was only modestly affected, indicating that primary leukemic cells exhibit significantly greater sensitivity to PAK1 inhibition than healthy cells (Figure 6A). Furthermore, we found that PAK1 inhibition with either IPA-3 or FRAX-597 inhibits the ability of AML primary patient cells to serially replate in a dose-dependent manner, in comparison to treatment with DMSO (supplemental Figure 5B).
Targeting of PAK1 inhibits AML patient cells including immature, leukemia stem cell-enriched populations while sparing healthy stem and progenitor cells. (A) Colony formation assay of primary AML patient MNC samples or healthy control MNC samples treated with IPA-3 (healthy, n = 4; AML, n = 5) or FRAX-597 (healthy, n = 4; AML, n = 3). (B) Cell viability (DAPI−Annexin V−) of CD34+CD38− cells from AML patients and healthy controls after 24 hours in vitro culture with IPA-3 or FRAX-597 (n = 3 for each drug). (C) Cell viability (DAPI−Annexin V−) of CD34+CD38+ cells from AML patients and healthy controls after 24 hours in vitro culture with IPA-3 or FRAX-597 (n = 3 for each drug). (D) Percentage bone marrow engraftment of human CD45+CD33+ cells 6 weeks following xenografting with primary AML patient samples treated with IPA-3. Experiment performed in duplicate with 2 independent primary AML patient samples. (E) Kaplan-Meier curve of overall survival of patients with high vs low PAK1 expression in CD34+ bone marrow cells isolated from MDS patients. Patients with high PAK1 expression are those with PAK1 expression levels higher than the median expression level of PAK1 in the dataset. The P value (log-rank test) is indicated. *P < .05 and **P < .01 for all experiments.
Targeting of PAK1 inhibits AML patient cells including immature, leukemia stem cell-enriched populations while sparing healthy stem and progenitor cells. (A) Colony formation assay of primary AML patient MNC samples or healthy control MNC samples treated with IPA-3 (healthy, n = 4; AML, n = 5) or FRAX-597 (healthy, n = 4; AML, n = 3). (B) Cell viability (DAPI−Annexin V−) of CD34+CD38− cells from AML patients and healthy controls after 24 hours in vitro culture with IPA-3 or FRAX-597 (n = 3 for each drug). (C) Cell viability (DAPI−Annexin V−) of CD34+CD38+ cells from AML patients and healthy controls after 24 hours in vitro culture with IPA-3 or FRAX-597 (n = 3 for each drug). (D) Percentage bone marrow engraftment of human CD45+CD33+ cells 6 weeks following xenografting with primary AML patient samples treated with IPA-3. Experiment performed in duplicate with 2 independent primary AML patient samples. (E) Kaplan-Meier curve of overall survival of patients with high vs low PAK1 expression in CD34+ bone marrow cells isolated from MDS patients. Patients with high PAK1 expression are those with PAK1 expression levels higher than the median expression level of PAK1 in the dataset. The P value (log-rank test) is indicated. *P < .05 and **P < .01 for all experiments.
It has previously been shown that LSC most often reside in the CD34+CD38−, but also in the CD34+CD38+ cell compartments in patients with AML.25,26 We sought to assess the effectiveness of PAK1 inhibition specifically on these LSC-enriched populations by culturing bone marrow mononuclear cells from human AML patients and healthy controls with increasing concentrations of IPA-3 or FRAX-597. Flow cytometry analysis revealed that PAK1 inhibition led to significantly decreased viability of CD34+CD38− cells from AML patients. In contrast, no significant decrease in cell viability was observed in healthy control CD34+CD38− cells with either PAK1 inhibitor (Figure 6B). Similarly, we found significant differential reduction in viability in the CD34+CD38+ cell compartment in human leukemic patient samples compared with healthy donor samples (Figure 6C). To assess the in vivo leukemia-initiating capacity of human AML cells in the presence of PAK1 inhibition, we performed xenografting studies in NSG mice using primary samples from 2 patients with AML. We detected significantly reduced CD45+CD33+ human AML cell engraftment in the bone marrow after 6 weeks for both patient samples treated with IPA-3 compared with DMSO (Figure 6D; supplemental Figure 5C).
To further assess the relevance of PAK1 in myeloid malignancies, we evaluated PAK1 expression in patients with MDS, a heterogeneous hematologic myeloid stem cell disorder that frequently progresses to AML. We found that MDS patients with high PAK1 expression in bone marrow CD34+ cells have significantly worse survival compared with PAK1 low expressing patients (P < .0001; Figure 6E; GSE19429)27 and that there is a linear correlation between PAK1 mRNA levels and worse overall survival in individual patients (P < .0001, R2 = 0.08607; supplemental Figure 6A). The hazard ratio (HR) of PAK1 high vs PAK1 low expressers was HR = 3.120 (95% confidence interval: 1.865-5.413). PAK2 expression showed no correlation with overall survival (supplemental Figure 6B). Of note, we found that elevation of PAK1 expression is significantly more frequent in patients with the MDS subtype refractory anemia with excess blasts (RAEB), the subtype of MDS with the highest risk for progression to AML (supplemental Figure 6C). To assess whether the impact of PAK1 expression on overall survival is independent of known prognostic factors for MDS, we performed multivariate analysis using a Cox regression model (taking into account sex, hemoglobin, FAB subtype, bone marrow blast percentage, and white blood cell count). In this analysis, high PAK1 expression remained an independent prognostic factor (P = .0334; HR = 1.884; supplemental Figure 6D). In addition, we performed multivariate analysis considering IPSS score as a covariable. In this analysis, high PAK1 expression also remained an independent prognostic factor (P = .0003; supplemental Figure 6E).
Discussion
Our studies highlight the importance of PAK1 as a novel target in AML and MDS. We show that leukemia cells are reliant on PAK1 function and that PAK1 inhibition has significant effects on leukemic tumor cells in vitro and in vivo. Inhibition of PAK1 with small molecule inhibitors IPA-3 and FRAX-597, or PAK1 shRNA-mediated knockdown, dramatically reduces the proliferation, as well as the leukemic colony formation capacity of AML cell lines and primary patient samples. Furthermore, reduction of PAK1 in an in vivo xenotransplantation AML model results in nearly complete loss of infiltration of leukemic cells and preservation of normal tissue architecture in the bone marrow, liver, and spleen.
MDS and AML are defined as clonal hematopoietic stem cell disorders arising de novo or as a result of previous chemo- or radiotherapy. Current treatment regimens in AML and MDS target rapidly dividing cells, having minimal effects on the quiescent leukemic and preleukemic stem cells responsible for disease progression and relapse. Novel therapies that are able to target preleukemic and leukemic stem cells are urgently needed to maintain remission and improve survival. MDS is considered a preleukemic disease as MDS patients frequently progress to AML. We find that PAK1 upregulation is more frequent in RAEB MDS, the subtype at highest risk of progression to AML, and that higher levels of PAK1 in MDS CD34+ cells correlate with inferior patient survival. PAK1 inhibition induces apoptosis of LSC-enriched cell populations and inhibits leukemic colony formation capacity and in vivo engraftment of primary AML cells in xenotransplantation experiments. Importantly, PAK1 inhibition targets primary human leukemic cells while sparing healthy control stem cells. These findings provide a promising therapeutic window that can be exploited by targeting of PAK1 in leukemia.
Several potent and specific small molecule inhibitors of group I PAKs are in the advanced stages of preclinical development, including a phase 1 trial in solid tumors. Our own group initially identified PAK1 as a downstream effector molecule of HLX, a gene upregulated in preleukemic stem cells and functionally relevant for AML pathogenesis.3 PAK1 was also recently shown to act downstream of mutations in the FLT3 and c-Kit (KIT) receptor proteins (FLT3-ITD, KITD816V) via focal adhesion kinase (FAK) signaling in myeloproliferative disease.28 Interestingly, 3 of the 4 AML cell lines and several patient samples we tested in this study do not contain either FLT3-ITD or KITD816V mutations yet were still sensitive to PAK1 inhibition. Furthermore, the AML patient samples we studied included FAB subtypes M4 and M5, which more frequently have elevated PAK1 expression from our assessments of a large cohort of AML patients (GSE14468; supplemental Figure 7A-B). Four patients from these experiments, however, had subtype M1 or M2 and also responded to PAK1 inhibition, despite these subtypes more frequently having low PAK1 expression (supplemental Figure 7A-B). Together, these results indicate PAK1 could be a broadly applicable target to combat the heterogeneity of AML, and PAK1 signaling may be downstream of multiple pathways important for AML growth and less dependent on mutation status (supplemental Figure 7A-E).
Despite reported roles for PAK1 in the regulation of cell cycle progression in other cell types,20 we find that in AML, PAK1 inhibition limits the growth of cells by inducing apoptosis and monocytic differentiation without affecting cell cycling. Using microarray studies, we identified downregulation of a large MYC oncogene regulated transcriptional network following PAK1 inhibition, including MYC target genes previously implicated in AML pathogenesis such as DUSP7. MYC oncoprotein has roles in regulating cell proliferation, apoptosis, and differentiation.29 Dysregulation of MYC is one of a series of oncogenic events implicated in myeloid leukemias.29,30 Retroviral overexpression of MYC has been shown to induce AML,31,32 and several studies have shown that fusion oncogenes in myeloid leukemias including AML1-ETO, PML-RARα, and PLZF-RARα induce leukemogenesis through activation of MYC.31 Exactly how PAK1 activates transcription of MYC in AML is still under investigation. For instance, in gastric cells, PAK1 modification of β-catenin was previously shown to lead to β-catenin nuclear localization and subsequent activation of MYC transcription.33 However, we did not find this effect in AML cells (data not shown). In FLT3-ITD–expressing myeloproliferative disease, PAK1 inhibition reduced the presence of activated nuclear signal transducer and activator of transcription 5 (STAT5), leading to reduced transcription of MYC.28 Additionally, other recent PAK1 studies have indicated PAK1 may itself activate transcription in the nucleus in addition to its kinase activities in the cytoplasm.34
In summary, our studies present PAK1 as a novel target in AML and MDS and show that inhibition of PAK1 has leukemia inhibitory effects not only on bulk leukemia blast cells but also in more immature LSC-enriched populations. These conclusions support the further development and clinical testing of PAK1 inhibitors as a therapeutic strategy in AML and MDS.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Swathi-Rao Narayanagari of the Stem Cell Isolation and Xenotransplantation Core Facility (funded by New York State Stem Cell Science (NYSTEM) grant C029154) of the Gottesman Institute for Stem Cell and Regenerative Medicine Research for expert technical support.
This work was supported by Albert Einstein Cancer Center Core Support grant P30CA013330, National Institutes of Health, National Cancer Institute (NCI) grants R01CA166429 (to U.S.) and R01CA142928 (to J.C.), Medical Scientist Training Program (MSTP) training grant T32GM007288 (to A.P. and R.F.S.), National Heart, Lung, and Blood Institute (NHLBI) fellowships F30HL117545 (to A.P.) and F30HL122085 (to R.F.S.), and Cellular and Molecular Biology and Genetics (CMBG) Training Grant T32GM007491 (to R.F.S.). J.B. acknowledges support from Leukaemia and Lymphoma Research of the United Kingdom. U.S. is a Research Scholar of the Leukemia & Lymphoma Society and the Diane and Arthur B. Belfer Faculty Scholar in Cancer Research of the Albert Einstein College of Medicine.
Authorship
Contribution: A.P. and R.F.S. designed and performed research, analyzed data, made figures, and wrote the manuscript; Y.Y. performed statistical analyses and made figures; B.B. performed statistical analyses and made figures; G.P. assisted in sample collection; J.B. contributed vital clinical data; K.G., J.C., and A.V. designed research and analyzed and interpreted data; and U.S. designed and analyzed research and contributed to writing of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ulrich Steidl, Department of Cell Biology, Department of Medicine (Oncology), Division of Hemato-Oncology, Albert Einstein College of Medicine/Montefiore Medical Center, Chanin Bldg, Room 601-605, 1300 Morris Park Ave, Bronx, NY 10461; e-mail: ulrich.steidl@einstein.yu.edu.
References
Author notes
A.P. and R.F.S. contributed equally to this study.