Key Points
Dicer1-mediated miRNA biogenesis is essential for induction and maintenance of Notch1-driven T-ALL.
miR-21 regulates survival of T-ALL, in part through the repression of the tumor suppressor gene Pdcd4.
Abstract
The modulatory function of individual microRNAs (miRNAs) in Notch-driven T-cell acute lymphoblastic leukemias (T-ALLs) has recently been established. Although protumorigenic and tumor-suppressive miRNAs are implicated in disease onset in murine models of Notch-driven T-cell leukemia, whether Dicer1-processed miRNAs are essential for Notch-driven T-ALL is currently unknown. Here we used conditional and inducible genetic loss-of-function approaches to test whether the development and maintenance of Notch-driven T-ALL was dependent on Dicer1 function. Mice with specific inactivation of both Dicer1 alleles in the T-cell lineage did not develop Notch-driven T-ALL. In contrast, loss of 1 functional Dicer1 allele did not significantly perturb T-ALL onset and tumor progression. Inducible inactivation of Dicer1 in early stage polyclonal T-ALL cells was sufficient to abrogate T-ALL progression in leukemic mice, whereas late-stage monoclonal T-ALL cells were counterselected against loss of Dicer1. Lineage-tracing experiments revealed that Dicer1 deficiency led to the induction of apoptosis in T-ALL cells, whereas cell cycle progression remained unaltered. Through microarray-based miRNA profiling, we identified miR-21 as a previously unrecognized miRNA deregulated in both mouse and human T-ALL. Herein, we demonstrate that miR-21 regulates T-ALL cell survival via repression of the tumor suppressor Pdcd4.
Introduction
MicroRNAs (miRNAs) are small noncoding RNA molecules, which posttranscriptionally regulate gene expression.1,2 Multiple miRNAs have been implicated in the regulation of hematopoiesis and leukemia.3,4 A global assessment for miRNA requirement within the hematopoietic system using tissue and stage-specific Cre-recombinase-mediated inactivation of Dicer1 has been performed.5 These studies revealed that the absolute requirement for Dicer1-processed miRNAs differs between hematopoietic lineages and/or developmental stages. Dicer1 inactivation in hematopoietic stem cells (HSCs) results in the induction of apoptosis and loss of HSC self-renewal and reconstitutive capacity, suggesting that Dicer1 is essential for HSC survival.5 Whereas, the requirement of Dicer1 for other blood lineages such as T cells seems to be less stringent. Loss of Dicer1 during T-cell development results in rather mild phenotypes.6,7
Aberrant expression of miRNAs has been observed in several cancers including hematologic malignancies. Using Dicer1 loss-of-function (LOF) studies, the global requirement for miRNA biogenesis in B-cell acute lymphoblastic leukemia (B-ALL), myelodysplastic syndrome, and acute myeloid leukemia (AML) was recently assessed.7-9 It became clear that miRNA dependency varies widely between different types of leukemia.7-9 Whereas the global requirement of miRNAs in T-cell acute lymphoblastic leukemia (T-ALL) remains elusive, various studies have been carried out identifying several miRNA candidates relevant to T-ALL onset.10-14 A comprehensive miRNA expression study using human T-ALL samples revealed that 5 highly abundantly expressed miRNAs function in a protumorigenic manner in a mouse model of Notch1-driven T-ALL. Adoptive transfer experiments using bone marrow (BM) cells that express individual oncomiRs together with the dominant active Notch1-intracellular domain (N1ICD) resulted in accelerated leukemia development, demonstrating cooperation with Notch to drive disease development.10 These miRNAs exhibited overlapping and cooperative effects on various known tumor suppressor genes. Downregulation of individual miRNAs had therefore only limited effects.10 Other studies identified miR-181a-1/b-1 as a modulator of oncogenic Notch signaling12 and showed that repression of miR-451, miR-709, and miR-193b-3p cooperates with N1ICD-driven T-cell leukemia.11,13 These studies clearly demonstrate that miRNAs can influence disease onset in Notch-driven T-ALL. However, it is unclear whether these effects reflect a critical role of miRNAs during the development/progression of Notch-driven T-ALL, or if they merely represent a potentiating effect following disease onset.
We demonstrate that development, progression, and maintenance of Notch1-driven T-ALL is entirely dependent on Dicer1-mediated miRNA biogenesis. Cell fate–tracing experiments show that Dicer1 is essential for survival of T-ALL cells but does not affect cell cycle. Moreover, microarray-based expression analysis on mouse and human T-ALL samples identified miR-21 as an important, previously unrecognized miRNA in T-ALL that regulates survival, in part, via repression of the tumor suppressor Pdcd4.
Methods
Mice
Dicer1lox/lox,15 CD4-Cre,16 and Mx1-Cre17 mice were previously described. Mice were crossed to generate Dicer1lox/lox CD4-Cre, Dicer1wt/lox CD4-Cre, and Dicer1lox/lox Mx1-Cre mice and backcrossed into C57BL/6 for at least 6 generations. C57BL/6 CD45.1 (Jackson Laboratories, ME) and Rag2γc−/− (Rag2−/−γc−/−; Taconic Europe, Denmark) were initially purchased, then bred and maintained at the Ecole Polytechnique Fédérale de Lausanne animal facility. Mx1-Cre induced gene inactivation was achieved by intraperitoneal injections of 2 μg/g body weight polyI:polyC (p-I:C; Invivogen) at 2-day intervals. Genomic deletion of Dicer1 was assessed by polymerase chain reaction (PCR; primer sequences are listed in supplemental Methods; see the Blood Web site).
Analysis of lymphoid and leukemic cells
Single cell suspensions were prepared from spleen, thymus, BM, or peripheral blood lymphocyte (PBL) of corresponding mice. T-cell subsets were analyzed as previously described.18 Expansion of T-ALL cells was monitored by flow-cytometric analysis of nerve growth factor receptor + (NGFR+) cells in lymphoid organs. For clonality studies and miRNA expression assays, NGFR+ T-ALL cells were sorted using the FACSAria II cytometer (Becton Dickinson). The purity of all sorted subsets was >97%. Flow-cytometric data were acquired on a Cyan flow cytometer (Beckman Coulter). Analysis of flow-cytometric data were carried out using the FlowJo software (version 9.2, Tree Star). A detailed list of primary and secondary antibody conjugates is listed in the supplemental Methods section.
Generation of stable murine T-ALL cell lines
Late-stage primary Notch1-driven T-ALL tumor cells were isolated from spleens of T-ALL-bearing mice and expanded in vitro (RPMI 1640–Glutamax supplemented with 10% fetal bovine serum, 25 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, 50 μM β-mercaptoethanol, and 50 μg/mL gentamicin) to generate 2 independent T-ALL cell lines, M295 and R112 (both of Dicer1lox/lox hN1ICD-IRES-NGFR genotype), for use in in vitro studies.
Lineage tracing of Dicer1 LOF mouse T-ALL cells
A lentiviral lineage-tracing system based on the conditional expression of enhanced green fluorescent protein (eGFP) flanked by loxP sites was generated to monitor the fate of Dicer1-deficient T-ALL cells. eGFP is expressed in T-ALL cells in which CreERT is inactive (Dicer1-proficient cells). Upon Cre activation, eGFP is deleted concomitantly with Dicer1. Fluorescence-activated cell sorter (FACS) sorting of GFPhi vs GFPlo cells allows for differential assessment of Dicer1lox/lox vs Dicer1−/− cell fate.
Transfection procedure for locked nucleic acids (LNAs) and short interfering RNAs (siRNAs)
LNA transfection solution was prepared mixing 100 μL OptiMEM medium (Gibco) with 40 pg Fluorescein amidite (FAM)-LNA and 9 µL HiPerFect (Qiagen). Following incubation at room temperature for 10 minutes, 100 µL transfection solution (100 nM final LNA concentration) was added dropwise to 250 μL supplemented RPMI 1640 medium containing 50 000 T-ALL cells. Following incubation for 72 hours at 37°C, effects of miRNA inhibition were assessed by comparing anti-miR LNAs with control. Typically, T-ALL cell transfection yielded 20% to 60% of FAM+ cells 3 days posttransfection. For double knockdown (KD) experiments of miR-21 and Pdcd4, T-ALL cells (0.2 × 106) were transfected with FAM-LNA-Ctr or FAM-LNA-21 and either Cy5-siRNA-Ctr or a mixture of 2 siRNAs (100 nM final) targeting human or mouse programmed cell-death protein 4 (Pdcd4), respectively (Cy5-siRNA-Pdcd4-1 and Cy5-siRNA-Pdcd4-2; see supplemental Methods). FAM+Cy5+ cells were assessed 3 days posttransplantation by flow cytometry using Annexin-V for detection of apoptotic cells. Pdcd4 KD efficiency was assessed by quantitative reverse-transcription (qRT) PCR on FACS-sorted FAM+Cy5+ cells. B-cell lymphoma–extra large (BCL-xL) expression was assessed using fixed cells (BD Cytofix) and followed by incubation with the primary anti-BCL-xL antibody (rabbit monoclonal α-BCL-xL) or rabbit immunoglobulin G isotype control. For detection, cells were incubated with the secondary anti-rabbit immunoglobulin G antibody (AlexaFluor405 coupled). Antibody details are listed in supplemental Methods.
T-cell receptor β rearrangement and miRNA expression studies
T-cell clonality of NGFR+ primary T-ALL cells or T-ALL cell lines was assessed by T-cell receptor β rearrangement PCR.19 miRNA expression studies were performed on total RNA from primary T-ALL cells or T-ALL cell lines prepared using the miRNeasy extraction kit (Qiagen). Isolated RNA was polyadenylated and reverse transcribed according to manufacturer’s instructions using the miRNA first-strand cDNA Synthesis Kit (Agilent Technologies). All primer sequences used are listed in supplemental Methods.
Ethics statement
All animal work was carried out in accordance with Swiss national guidelines. This study was reviewed and approved by the cantonal veterinary service (Service vétérinaire cantonal de Vaud).
Results
Dicer1 is essential for the induction of Notch1-driven T-ALL
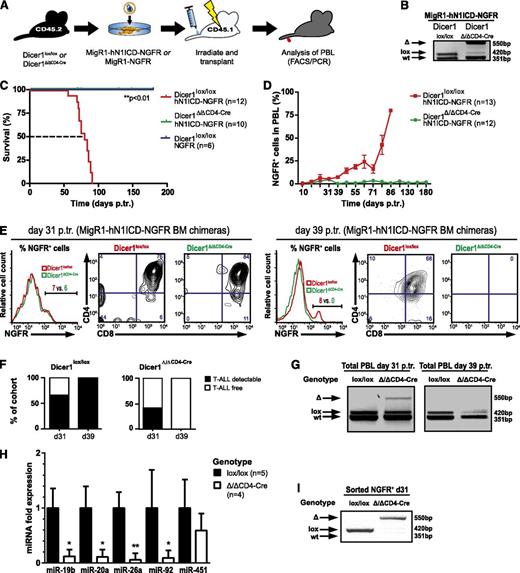
To investigate whether Dicer1-mediated miRNA processing is essential for the development of Notch1-driven T-ALL, we used a genetic LOF approach and crossed conditional Dicer1 mice with CD4-Cre transgenic mice15,16 (Dicer1Δ/ΔCD4-Cre). In this model, Dicer1 is inactivated as thymocytes or leukemogenic cells transit from the double-negative (DN) to the double-positive (DP) stage. Lineage negative (Lin−) CD45.2+ BM cells from Dicer1lox/lox and Dicer1Δ/ΔCD4-Cre mice were infected with a bicistronic retroviral (RV) vector expressing hN1ICD-NGFR or NGFR alone and subsequently transplanted into lethally irradiated CD45.1+ congenic hosts (Figure 1A). As expected, analysis of thymocytes 8 weeks posttransplantation revealed efficient recombination of the floxed Dicer1 allele in chimeric animals reconstituted with transduced Dicer1Δ/ΔCD4-Cre cells (Figure 1B). Interestingly, CD4-Cre-mediated inactivation of Dicer1 prevented tumor development (Figure 1C) but did not prevent the generation of Dicer1 mutant NGFR− peripheral T cells or other lymphoid lineages (supplemental Figure 1).20,21 In contrast, Dicer1-proficient chimeras developed Notch-driven T-ALL, with all animals dying of disease by 91 days posttransplantation with an MST of 80 days (Figure 1C and supplemental Figure 1A). NGFR+ CD4+CD8+ (DP) cells accumulated over time in the peripheral blood cells (PBLs) of Dicer1lox/lox chimeras, whereas these cells were only transiently present in Dicer1Δ/ΔCD4-Cre chimeras (Figure 1D-G). This suggests that loss of Dicer1 does not prevent the development of NGFR+ preleukemic DP cells, but does impair their tumorigenic potential and/or survival. To ensure that the transient presence of preleukemic NGFR+ DP cells in Dicer1 mutant chimeras was not because of the persistence of Dicer1-proficient escaper cells, Dicer1 deletion status and the expression levels of Dicer1-dependent miRNAs were confirmed on sorted early stage NGFR+ polyclonal (pre)leukemic cells (Figure 1H-I). The expression levels of Dicer1-dependent miRNAs were significantly reduced in cells sorted from Dicer1-mutant chimeras (Figure 1H), concomitant with efficient Dicer1 deletion (Figure 1I), whereas expression of miR-451, a Dicer1-independent miRNA, was unaffected (Figure 1H). Dicer1-dependent miR-19b miR-20a, miR-92a, and miR-26a have been previously shown to function as oncomiRs in Notch1-driven T-ALL.10,22
Dicer1 is essential for Notch-mediated T-ALL induction. (A) Lin− BM cells from CD45.2+ donors (Dicer1lox/lox or Dicer1Δ/Δ CD4-Cre) were transduced with MigR1-hN1ICD-NGFR RV or MigR1-NGFR RV and transplanted into CD45.1+ congenic recipients, and hosts were monitored for T-ALL development by PBL analysis. (B) Dicer1 deletion PCR performed on thymocyte DNA from MIGR1-hN1ICD-NGFR transduced chimeric Dicer1lox/lox or Dicer1Δ/Δ CD4-Cre mice 8 weeks posttransplantation (p.tr.). wt, wild-type Dicer1, 351 bp; lox, loxP Dicer1 allele, 420 bp; Δ, recombined Dicer1 allele, 550 bp. (C) Kaplan-Meier survival plot of BM chimeras as outlined in panel A. Depicted are 3 experimental cohorts pooled from 2 independent experiments: (1) Dicer1lox/lox transduced with NGFR control RV particles, Dicer1lox/loxNGFR; (2) Dicer1lox/lox transduced with hN1ICD-NGFR RV particles, Dicer1lox/lox hN1ICD-NGFR; and (iii) Dicer1Δ/ΔCD4-Cre transduced with hN1ICD-NGFR RV particles, Dicer1Δ/ΔCD4-Cre hN1ICD-NGFR. The dashed line indicates median survival time (MST; 80 days). (D) T-ALL cell expansion in the periphery assessed by NGFR+ cells in total PBLs. FACS analysis on PBLs is shown at the indicated time points p.tr., the percentage of NGFR+ cells was measured gating on live cells, and mouse numbers are indicated in brackets. Symbols represent mean ± standard deviation (SD) for mice analyzed per time point. (E) Representative flow cytometric analysis of NGFR+ PBLs of MigR1-hN1ICD-NGFR transduced Dicer1lox/lox (red) or Dicer1Δ/ΔCD4-Cre (green) BM chimeras at day 31 p.tr. (left panel, histogram and contour plots) and day 39 p.tr. (right panel, histogram and contour plots). The CD4+ vs CD8+ profile is shown gated on NGFR+ cells based on the histograms. (F) Left bar graph indicates the percentage of mice within the Dicer1lox/lox control cohort with >3% NGFR+ cells (T-ALL detectable, ▪) or <3% (T-ALL free, ☐) in total PBLs at 31 days p.tr. (62%; n = 9/13) and 39 days p.tr. (100%; n = 12/12). Dicer1Δ/Δ CD4-Cre chimeras are shown in the right bar graph. At day 31 p.tr., 42% (n = 5/12) of these mice have >3% of NGFR+ cells in the PBLs, but 0% (n = 0/10) at day 39 p.tr. (G) Dicer1 deletion PCR on total PBLs of MigR1-hN1ICD-NGFR transduced Dicer1lox/lox and Dicer1Δ/Δ CD4-Cre chimeras at day 31 (left panel) and day 39 p.tr. (right panel). (H) Expression (qRT-PCR) of oncomiRs miR-19b, miR-20a, miR-26a, miR-92a, and miR-451 is shown at day 31 p.tr. in RNA samples isolated from sorted CD45.2+ NGFR+ cells from splenocytes of MigR1-hN1ICD-NGFR transduced Dicer1Δ/Δ CD4-Cre BM chimeras (n = 4) and normalized to the expression of sorted NGFR+ cells from MigR1-hN1ICD-NGFR transduced Dicer1lox/lox BM chimeras (n = 5). (I) Dicer1 deletion was assessed in samples from panel H by PCR. Data (D-H) represent 2 independent experiments. *P < .05; **P < .01.
Dicer1 is essential for Notch-mediated T-ALL induction. (A) Lin− BM cells from CD45.2+ donors (Dicer1lox/lox or Dicer1Δ/Δ CD4-Cre) were transduced with MigR1-hN1ICD-NGFR RV or MigR1-NGFR RV and transplanted into CD45.1+ congenic recipients, and hosts were monitored for T-ALL development by PBL analysis. (B) Dicer1 deletion PCR performed on thymocyte DNA from MIGR1-hN1ICD-NGFR transduced chimeric Dicer1lox/lox or Dicer1Δ/Δ CD4-Cre mice 8 weeks posttransplantation (p.tr.). wt, wild-type Dicer1, 351 bp; lox, loxP Dicer1 allele, 420 bp; Δ, recombined Dicer1 allele, 550 bp. (C) Kaplan-Meier survival plot of BM chimeras as outlined in panel A. Depicted are 3 experimental cohorts pooled from 2 independent experiments: (1) Dicer1lox/lox transduced with NGFR control RV particles, Dicer1lox/loxNGFR; (2) Dicer1lox/lox transduced with hN1ICD-NGFR RV particles, Dicer1lox/lox hN1ICD-NGFR; and (iii) Dicer1Δ/ΔCD4-Cre transduced with hN1ICD-NGFR RV particles, Dicer1Δ/ΔCD4-Cre hN1ICD-NGFR. The dashed line indicates median survival time (MST; 80 days). (D) T-ALL cell expansion in the periphery assessed by NGFR+ cells in total PBLs. FACS analysis on PBLs is shown at the indicated time points p.tr., the percentage of NGFR+ cells was measured gating on live cells, and mouse numbers are indicated in brackets. Symbols represent mean ± standard deviation (SD) for mice analyzed per time point. (E) Representative flow cytometric analysis of NGFR+ PBLs of MigR1-hN1ICD-NGFR transduced Dicer1lox/lox (red) or Dicer1Δ/ΔCD4-Cre (green) BM chimeras at day 31 p.tr. (left panel, histogram and contour plots) and day 39 p.tr. (right panel, histogram and contour plots). The CD4+ vs CD8+ profile is shown gated on NGFR+ cells based on the histograms. (F) Left bar graph indicates the percentage of mice within the Dicer1lox/lox control cohort with >3% NGFR+ cells (T-ALL detectable, ▪) or <3% (T-ALL free, ☐) in total PBLs at 31 days p.tr. (62%; n = 9/13) and 39 days p.tr. (100%; n = 12/12). Dicer1Δ/Δ CD4-Cre chimeras are shown in the right bar graph. At day 31 p.tr., 42% (n = 5/12) of these mice have >3% of NGFR+ cells in the PBLs, but 0% (n = 0/10) at day 39 p.tr. (G) Dicer1 deletion PCR on total PBLs of MigR1-hN1ICD-NGFR transduced Dicer1lox/lox and Dicer1Δ/Δ CD4-Cre chimeras at day 31 (left panel) and day 39 p.tr. (right panel). (H) Expression (qRT-PCR) of oncomiRs miR-19b, miR-20a, miR-26a, miR-92a, and miR-451 is shown at day 31 p.tr. in RNA samples isolated from sorted CD45.2+ NGFR+ cells from splenocytes of MigR1-hN1ICD-NGFR transduced Dicer1Δ/Δ CD4-Cre BM chimeras (n = 4) and normalized to the expression of sorted NGFR+ cells from MigR1-hN1ICD-NGFR transduced Dicer1lox/lox BM chimeras (n = 5). (I) Dicer1 deletion was assessed in samples from panel H by PCR. Data (D-H) represent 2 independent experiments. *P < .05; **P < .01.
Previous studies using solid tumor models (eg, lung cancer23 and retinoblastoma24 ) or a mouse AML model7 have demonstrated that Dicer1 can function as a haploinsufficient tumor suppressor.7,23,24 Therefore, BM chimeras reconstituted with Dicer1wt/lox or Dicer1wt/ΔCD4-Cre expressing hNICD-NGFR were established to assess whether Dicer1 may play a similar role in Notch-driven T-ALL (supplemental Figure 2A). Dicer1wt/ΔCD4-Cre and Dicer1wt/lox developed T-ALL at a similar rate (supplemental Figure 2B-E), with an MST of 56 days posttransplantation. This suggests that Dicer1 haploinsufficiency does not influence Notch-driven T-ALL development.
Dicer1 is essential for early and late-stage T-ALL
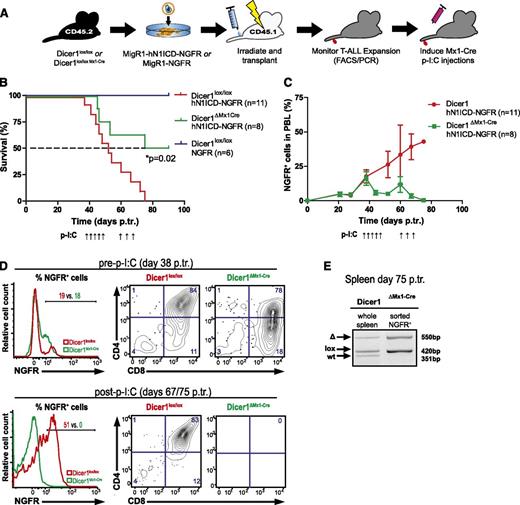
Our results so far clearly indicate that Dicer1 is essential for the induction of Notch-driven T-ALL. However, whether established T-ALL cells also depend on Dicer1 function is currently unclear. To assess whether Dicer1 is required for the maintenance of early and late-stage T-ALL cells, we generated Dicer1lox/loxMx1-Cre mice. Chimeras reconstituted with Dicer1lox/loxMx1-Cre BM cells expressing either hN1ICD-NGFR or NGFR alone (control) were subsequently established (Figure 2A). Once ∼20% NGFR+ cells were detected in PBLs of both cohorts, Cre-mediated inactivation of Dicer1 was induced. Strikingly, this resulted in an overall survival rate of 50% in Dicer1ΔMx1-Cre chimeras (Figure 2B). Remaining mice in this cohort had an MST of 82.5 days, whereas Dicer1-proficient chimeras all died of T-ALL by day 75 posttransplantation with an MST of 52 days (Figure 2B and supplemental Figure 3A). Although NGFR+ T-ALL cells continuously accumulated in PBLs of Dicer1lox/lox chimeras, the frequency of these cells dropped to 4% in surviving Dicer1ΔMx1-Cre chimeras before rising again to 10% at day 60 posttransplantation (Figure 2C), presumably because of cells that escaped Cre-mediated Dicer1 inactivation. To counteract T-ALL relapse, all BM chimeras were subjected to a second p-I:C cycle, which led to the eradication of NGFR+ leukemic cells and survival of the remaining mice (Figure 2C-D). Analysis of NGFR+ leukemic cells sorted from spleens of leukemic Dicer1ΔMx1-Cre chimeras (day 75 posttransplantation) revealed that the floxed Dicer1 allele was retained by the majority of tumor cells (Figure 2E). This indicated that T-ALL relapse in Dicer1ΔMx1-Cre chimeras was because of inefficient deletion of Dicer1, resulting in the persistence and expansion of leukemic Dicer1-proficient cells. As expected, analysis of the BM compartment of surviving Dicer1ΔMx1-Cre chimeras at 90 days posttransplantation revealed the complete loss of the CD45.2+ transplant confirming previous reports (supplemental Figure 3B-E).5
Efficient inactivation of Dicer1 rescues mice from early stage T-ALL. (A) Lin− BM cells from CD45.2+ donors (Dicer1lox/lox or Dicer1lox/lox Mx1-Cre) were retrovirally transduced with hN1ICD-NGFR or NGFR alone and transplanted into CD45.1+ congenic hosts. Once NGFR+ cells were detected in PBLs (at day 38 p.tr.), Mx1-Cre was activated using p-I:C. A second round of 3 p-I:C injections was initiated at day 60 p.tr. in order to activate Mx1-Cre in residual Dicer1lox/lox T-ALL cells in Dicer1∆Mx1-Cre BM chimeras. (B) Kaplan-Meier survival plot of chimeric animals pooled from 2 independent experiments as outlined in panel A. Depicted are 3 experimental groups: (1) Dicer1lox/lox transduced with NGFR control RV particles, Dicer1lox/loxNGFR; (2) Dicer1lox/lox transduced with hNICD-NGFR RV particles, Dicer1lox/loxhNICD-NGFR; and (3) Dicer1lox/lox Mx1-Cre transduced with hNICD-NGFR RV particles, Dicer1ΔMx1-Cre hNICD-NGFR . The dashed line indicates MST: red= 52 days, green = 82.5 days. (C) Expansion of peripheral T-ALL cells measured by the percentage of NGFR+ cells in total PBLs from panel B. (D) Representative FACS plots of NGFR+ PBLs of Dicer1lox/lox or Dicer1lox/lox Mx1-Cre at day 38 p.tr. (upper panel) and at day 67 p.tr. Dicer1lox/lox and day 75 p.tr. Dicer1ΔMx1-Cre (lower panel). The CD4+ vs CD8+ profile is shown gated on NGFR+ cells based on the positive fraction in the histograms. (E) Dicer1 deletion PCR on DNA extracted from total splenocytes of a representative Dicer1ΔMx1-Cre BM chimeric animal with incomplete Dicer1 deletion at day 75 p.tr. (left) and on sorted (CD45.2+ NGFR+) T-ALL cells from the same spleen (right).
Efficient inactivation of Dicer1 rescues mice from early stage T-ALL. (A) Lin− BM cells from CD45.2+ donors (Dicer1lox/lox or Dicer1lox/lox Mx1-Cre) were retrovirally transduced with hN1ICD-NGFR or NGFR alone and transplanted into CD45.1+ congenic hosts. Once NGFR+ cells were detected in PBLs (at day 38 p.tr.), Mx1-Cre was activated using p-I:C. A second round of 3 p-I:C injections was initiated at day 60 p.tr. in order to activate Mx1-Cre in residual Dicer1lox/lox T-ALL cells in Dicer1∆Mx1-Cre BM chimeras. (B) Kaplan-Meier survival plot of chimeric animals pooled from 2 independent experiments as outlined in panel A. Depicted are 3 experimental groups: (1) Dicer1lox/lox transduced with NGFR control RV particles, Dicer1lox/loxNGFR; (2) Dicer1lox/lox transduced with hNICD-NGFR RV particles, Dicer1lox/loxhNICD-NGFR; and (3) Dicer1lox/lox Mx1-Cre transduced with hNICD-NGFR RV particles, Dicer1ΔMx1-Cre hNICD-NGFR . The dashed line indicates MST: red= 52 days, green = 82.5 days. (C) Expansion of peripheral T-ALL cells measured by the percentage of NGFR+ cells in total PBLs from panel B. (D) Representative FACS plots of NGFR+ PBLs of Dicer1lox/lox or Dicer1lox/lox Mx1-Cre at day 38 p.tr. (upper panel) and at day 67 p.tr. Dicer1lox/lox and day 75 p.tr. Dicer1ΔMx1-Cre (lower panel). The CD4+ vs CD8+ profile is shown gated on NGFR+ cells based on the positive fraction in the histograms. (E) Dicer1 deletion PCR on DNA extracted from total splenocytes of a representative Dicer1ΔMx1-Cre BM chimeric animal with incomplete Dicer1 deletion at day 75 p.tr. (left) and on sorted (CD45.2+ NGFR+) T-ALL cells from the same spleen (right).
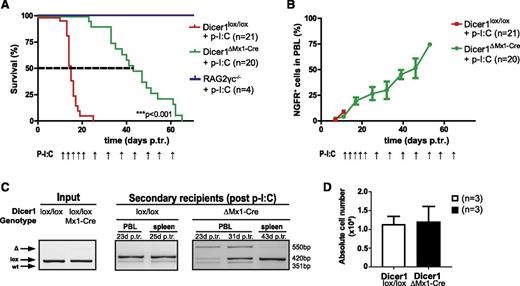
To investigate whether Dicer1 is essential for late-stage T-ALL, we established BM chimeras using donor hN1ICD-NGFR transformed Dicer1lox/lox and Dicer1lox/loxMx1-Cre BM cells. Once T-ALL was established, clonal NGFR+ leukemic cells were transplanted into secondary Rag2−/−γc−/− hosts, and continuous p-I:C administration was initiated 12 days posttransplantation (supplemental Figure 4A-C). Ablation of Dicer1 significantly prolonged the survival (2.8-fold increase; P < .001) of corresponding animals (Figure 3A). Despite the attempts to continuously inactivate Dicer1, NGFR+ T-ALL cells rapidly accumulated and recipients died of disease (Figure 3B and supplemental Figure 4D). To determine whether late-stage Dicer1-deficient T-ALL cells contributed to the tumor bulk but had impaired tumorigenic potential or continuous counterselection and elimination of late-stage Dicer1-deficient T-ALL cells occurred in recipients, Dicer1 recombination status in PBLs of secondary recipients was analyzed at different time points. Incomplete deletion of Dicer1 was detected in Dicer1ΔMx1-Cre recipient PBLs at day 23 posttransplantation with undeleted cells accumulating over time (day 31 posttransplantation; Figure 3C), eventually becoming the only detectable tumor fraction by PCR in splenocytes at end point analysis (day 43 posttransplantation; Figure 3C). At this stage, tumor burden in the spleen was comparable between control and Dicer1 mutant cohorts (Figure 3D), indicating complete counterselection against Dicer1-deficient T-ALL cells. To confirm the hypothesis of counterselection of Dicer1-deficient T-ALL cells, 2 independent monoclonal T-ALL cell lines carrying floxed alleles of Dicer1 (M295 and R112) were established (Figure 4 and supplemental Figure 5A-C). Following infection with a lentivirus encoding tamoxifen-inducible CreERT to induce Dicer1 deletion, the cells were serially passaged in the absence of tamoxifen and the maintenance of Dicer1 pro- and deficient cells was monitored over time (Figure 4A). Approximately 50% Dicer1-deficient cells were present 3 days post–tamoxifen treatment. However, Dicer1-deficient cells were progressively lost with serial passages in vitro, indicated by the gradual disappearance of the Dicer1 deletion PCR band 3 and 7 days post–tamoxifen treatment (Figure 4B and supplemental Figure 5D). These results demonstrate that Dicer1-deficient T-ALL cells have a growth disadvantage compared with Dicer1-proficient T-ALL cells.
Late-stage T-ALL cells counterselect against loss of Dicer1 function. (A) Kaplan-Meier survival plot of secondary RAG2γc−/− recipients from 3 pooled independent experiments. Depicted are 3 experimental groups: (1) RAG2γc−/− injected with p-I:C only, RAG2γc−/− + p-I:C; (2) RAG2γc−/− transplanted with late-stage T-ALL cells (Dicer1lox/lox) and injected with p-I:C, Dicer1lox/lox + p-I:C; and (3) RAG2γc−/− transplanted with late-stage T-ALL cells (Dicer1lox/lox Mx1-Cre) and injected with p-I:C, Dicer1lox/lox Mx1-Cre + p-I:C . The dashed line indicates MST: red = 15 days, green = 43 days. P-I:C and arrows indicate p-I:C injections. (B) T-ALL progression measured by the percentage of NGFR+ cells in total PBLs. P-I:C and arrows indicate p-I:C injections. (C) Dicer1 deletion PCR. Shown are the results on DNA samples of representative tumor cells prior to transplantation into RAG2γc−/− recipients (“input”, Dicer1lox/lox and Dicer1lox/lox Mx1-Cre; left panel), PBLs and splenocytes of corresponding representative secondary recipient animals after transplantation (“secondary recipients”; middle and right panels) 23 to 43 days p.tr. (D) Bar graph depicts the absolute cell numbers of splenocytes isolated from late-stage secondary T-ALL recipient animals.
Late-stage T-ALL cells counterselect against loss of Dicer1 function. (A) Kaplan-Meier survival plot of secondary RAG2γc−/− recipients from 3 pooled independent experiments. Depicted are 3 experimental groups: (1) RAG2γc−/− injected with p-I:C only, RAG2γc−/− + p-I:C; (2) RAG2γc−/− transplanted with late-stage T-ALL cells (Dicer1lox/lox) and injected with p-I:C, Dicer1lox/lox + p-I:C; and (3) RAG2γc−/− transplanted with late-stage T-ALL cells (Dicer1lox/lox Mx1-Cre) and injected with p-I:C, Dicer1lox/lox Mx1-Cre + p-I:C . The dashed line indicates MST: red = 15 days, green = 43 days. P-I:C and arrows indicate p-I:C injections. (B) T-ALL progression measured by the percentage of NGFR+ cells in total PBLs. P-I:C and arrows indicate p-I:C injections. (C) Dicer1 deletion PCR. Shown are the results on DNA samples of representative tumor cells prior to transplantation into RAG2γc−/− recipients (“input”, Dicer1lox/lox and Dicer1lox/lox Mx1-Cre; left panel), PBLs and splenocytes of corresponding representative secondary recipient animals after transplantation (“secondary recipients”; middle and right panels) 23 to 43 days p.tr. (D) Bar graph depicts the absolute cell numbers of splenocytes isolated from late-stage secondary T-ALL recipient animals.
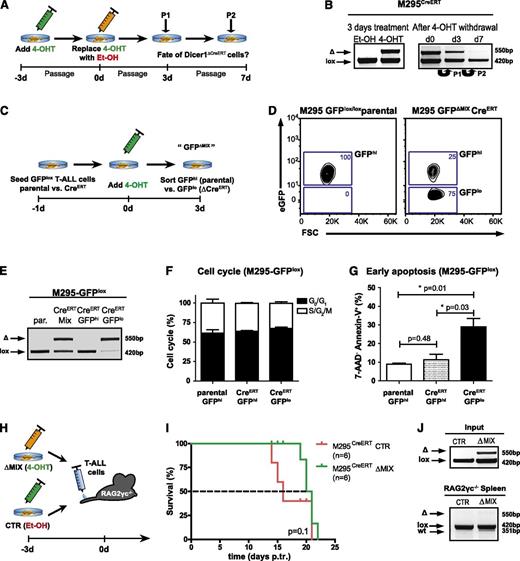
Loss of miRNAs leads to induction of apoptosis in T-ALL cells. (A) Experimental strategy to study Dicer1 deletion in vitro. CreERT was activated in the CreERT transgenic murine T-ALL cell line M295CreERT (Dicer1lox/lox) for 3 days by addition of tamoxifen (4-OHT) to inactivate Dicer1. Control T-ALL cells (M295CreERT) were treated with ethanol (Et-OH) only. T-ALL cells were passaged twice (P1, P2), maintained under 4-OHT-free conditions, and Dicer1 deletion was assessed by deletion PCR. (B) Dicer1 deletion PCRs on T-ALL cell line M295CreERT after initial deletion of Dicer1 in vitro as outlined in panel A. Shown is 1 representative out of 2 performed experiments. Initial deletion after 3 days of 4-OHT treatment (left). Dicer1 deletion at day 0, 3 (P1, passage 1) and 7 (P2, passage 2) after initial CreERT activation (right). (C) Experimental strategy to lineage trace Dicer1-deficient T-ALL cell populations. Both T-ALL cell lines M295 (parental) and M295CreERT were lentivirally transduced with a loxP flanked eGFP reporter construct (M295CreERT-GFPlox). After administration of 4-OHT for 3 days, parental GFP expressing (GFPhi) and CreERT transgenic GFPlox transduced GFP positive and negative (ΔMIX, GFPhi and GFPlo) cells were assessed by FACS for cell cycle analysis and induction of apoptosis. (D) Representative flow cytometric analysis of M295CreERT-GFPlox 3 days after 4-OHT treatment compared with parental M295-GFPlox T-ALL cells. (E) Representative Dicer1 deletion PCR on DNA derived from parental M295-GFPlox (par.), unsorted M295CreERT-GFPlox (CreERT MIX) and FACS-sorted GFPhi vs GFPlo (CreERT-GFPhi vs CreERT-GFPlo) cells 3 days after 4-OHT treatment. (F) Assessment of cell cycle status of GFPlox transduced T-ALL cells 3 days after 4-OHT treatment (M295-GFPhi parental vs M295CreERT-GFPhi vs M295CreERT-GFPlo). (G) Quantification of early apoptosis (7-AAD− Annexin-V+) in M295-GFPlox (parental GFPhi) and M295CreERT-GFPhi vs M295CreERT-GFPlo cells. Data (F-G) are pooled from 2 independent experiments. (H) Experimental approach to assess the fate of Dicer1-deleted T-ALL cells in vivo. M295CreERT cells were treated for 3 days with 4-OHT yielding deleted and undeleted cells (ΔMIX) or Et-OH only as control (CTR), these cells were subsequently transplanted into RAG2γc−/− recipient animals. (I) Kaplan-Meier survival plot of RAG2γc−/− recipients receiving M295CreERT CTR T-ALL cells or ΔMIX cells. The dashed line indicates MST: red = 16 days, green = 20.5 days. (J) Dicer1 deletion PCR on DNA from cells of the M295CreERT T-ALL line prior to transplantation (CTR or ΔMIX, upper panel, “input”, same experiment as in panel I) and on splenocytes of RAG2γc−/− recipient animals with late-stage T-ALL (lower panel).
Loss of miRNAs leads to induction of apoptosis in T-ALL cells. (A) Experimental strategy to study Dicer1 deletion in vitro. CreERT was activated in the CreERT transgenic murine T-ALL cell line M295CreERT (Dicer1lox/lox) for 3 days by addition of tamoxifen (4-OHT) to inactivate Dicer1. Control T-ALL cells (M295CreERT) were treated with ethanol (Et-OH) only. T-ALL cells were passaged twice (P1, P2), maintained under 4-OHT-free conditions, and Dicer1 deletion was assessed by deletion PCR. (B) Dicer1 deletion PCRs on T-ALL cell line M295CreERT after initial deletion of Dicer1 in vitro as outlined in panel A. Shown is 1 representative out of 2 performed experiments. Initial deletion after 3 days of 4-OHT treatment (left). Dicer1 deletion at day 0, 3 (P1, passage 1) and 7 (P2, passage 2) after initial CreERT activation (right). (C) Experimental strategy to lineage trace Dicer1-deficient T-ALL cell populations. Both T-ALL cell lines M295 (parental) and M295CreERT were lentivirally transduced with a loxP flanked eGFP reporter construct (M295CreERT-GFPlox). After administration of 4-OHT for 3 days, parental GFP expressing (GFPhi) and CreERT transgenic GFPlox transduced GFP positive and negative (ΔMIX, GFPhi and GFPlo) cells were assessed by FACS for cell cycle analysis and induction of apoptosis. (D) Representative flow cytometric analysis of M295CreERT-GFPlox 3 days after 4-OHT treatment compared with parental M295-GFPlox T-ALL cells. (E) Representative Dicer1 deletion PCR on DNA derived from parental M295-GFPlox (par.), unsorted M295CreERT-GFPlox (CreERT MIX) and FACS-sorted GFPhi vs GFPlo (CreERT-GFPhi vs CreERT-GFPlo) cells 3 days after 4-OHT treatment. (F) Assessment of cell cycle status of GFPlox transduced T-ALL cells 3 days after 4-OHT treatment (M295-GFPhi parental vs M295CreERT-GFPhi vs M295CreERT-GFPlo). (G) Quantification of early apoptosis (7-AAD− Annexin-V+) in M295-GFPlox (parental GFPhi) and M295CreERT-GFPhi vs M295CreERT-GFPlo cells. Data (F-G) are pooled from 2 independent experiments. (H) Experimental approach to assess the fate of Dicer1-deleted T-ALL cells in vivo. M295CreERT cells were treated for 3 days with 4-OHT yielding deleted and undeleted cells (ΔMIX) or Et-OH only as control (CTR), these cells were subsequently transplanted into RAG2γc−/− recipient animals. (I) Kaplan-Meier survival plot of RAG2γc−/− recipients receiving M295CreERT CTR T-ALL cells or ΔMIX cells. The dashed line indicates MST: red = 16 days, green = 20.5 days. (J) Dicer1 deletion PCR on DNA from cells of the M295CreERT T-ALL line prior to transplantation (CTR or ΔMIX, upper panel, “input”, same experiment as in panel I) and on splenocytes of RAG2γc−/− recipient animals with late-stage T-ALL (lower panel).
To investigate the fate of Dicer1-deficient T-ALL cells, mouse T-ALL cell lines were infected with a lentivirus encoding a floxed eGFP reporter gene allowing lineage tracing of tamoxifen-activated CreERT infected cells (Figure 4C-D and supplemental Figure 5E-F). Following tamoxifen treatment, the parental cell lines remained GFPhi, whereas eGFPhi and eGFPlo cells were present in CreERT-expressing cultures. Efficient Dicer1 deletion in sorted eGFPlo was confirmed by PCR analysis (Figure 4E). The cell cycle status of Dicer1-deficient T-ALL cells (GFPlo) was comparable to that of undeleted Dicer1-proficient (GFPhi) T-ALL cells (Figure 4F). However, a significant increase in apoptotic cells was detected in the GFPlo fraction compared with the GFPhi fraction and parental T-ALL cells (Figure 4F-G and supplemental Figure 5F-H), strongly suggesting that Dicer1 is necessary for T-ALL cell survival in vitro.
Prosurvival factors provided by the cellular microenvironment in vivo could potentially influence T-ALL cell survival. Therefore, equal numbers of M295CreERTDicer1-proficient and Dicer-deficient cells (ΔMix 4-OHT) or fully Dicer1-proficient M295CreERT cells (CTR Et-OH) were transplanted into Rag2−/−γc−/− recipients to assess Dicer1-deficient T-ALL cell maintenance in vivo (Figure 4H). Recipients of a mixture of Dicer1-proficient and mutant cells survived slightly, but not significantly, longer than those transplanted with fully Dicer1-proficient cells (MST 20.5 days vs 16 days) (Figure 4I). Importantly, only floxed Dicer1 alleles were detected in leukemic cells derived from moribund recipients, demonstrating that Dicer1 ablated T-ALL cells were also outcompeted by Dicer1-proficient cells in vivo (Figure 4J), strongly suggesting that Dicer1-mediated miRNA biogenesis is required for survival of T-ALL cells.
Abundantly expressed miR-21 prevents apoptosis in mouse and human T-ALL
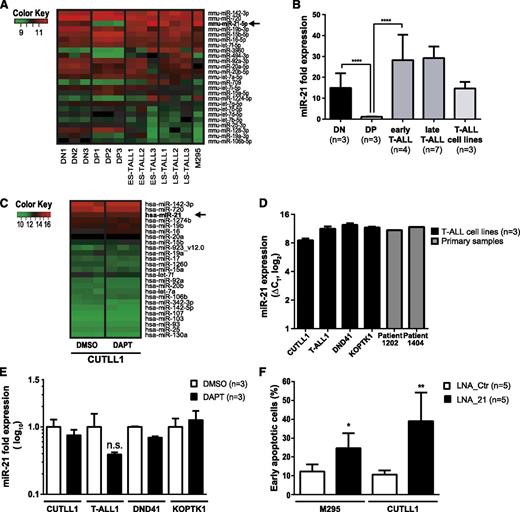
Induction of apoptosis following conditional loss of Dicer1 in the context of Notch1-driven T-ALL suggested that miRNAs exert an essential prosurvival function. Thus, we reasoned that T-ALL induction and/or maintenance may rely on abundantly expressed miRNAs. Furthermore, miRNAs are differentially regulated during T-cell development25,26 as well as in cancer.27 We hypothesized that certain prosurvival miRNA candidates may be abundantly expressed at early compared with later stages of T-cell development, and that these may be hijacked in T-ALL cells. The miRNA expression profiles of mouse DN and DP thymocytes were compared with those of primary mouse early stage and late-stage T-ALL samples as well as the M295 mouse T-ALL cell line in order to identify potential deregulated miRNAs in Notch1-driven T-ALL (Figure 5A). The miRNA profile of the human T-ALL cell line CUTLL1 was also assessed. Interestingly, out of 409 detected candidates, miR-21 was found to be the third most abundantly expressed miRNA in mouse T-ALL samples (Figure 5A-B). miR-21 expression was downregulated 15-fold in DP thymocytes compared with DN thymocytes (Figure 5B). Moreover, miR-21 levels were increased 30-fold in both early and late-stage primary murine T-ALL cells compared with DP thymocytes. miR-21 was also abundantly expressed in 2 primary human T-ALL samples as well as in CUTLL1, T-ALL1, DND41, and KOPTK1 cell lines suggesting that miR-21 expression is conserved in both mouse and human T-ALL (Figure 5C-D). Treatment of NOTCH1-dependent CUTLL1, T-ALL1, DND41, and KOPTK1 cells28,29 for 3 days with the γ-secretase inhibitor DAPT revealed no significant change in the most abundantly expressed miRNAs including miR-21 indicating that these are unlikely to be under the control of Notch signaling (Figure 5C,E). This is in agreement with publically available chromatin immunoprecipitation sequencing data for NOTCH1 and CSL,30 which do not show any binding of NOTCH1 or CSL 10 kb up- or downstream of the transcription start side of the MIR-21 gene (data not shown).
KD of miR-21 in T-ALL induces apoptosis. (A) Heatmap of the 25 most abundantly expressed miRNAs as assessed by microarray in FACS-sorted mouse CD4−CD8− DN (n = 3) and CD4+CD8+ DP (n = 3) thymocytes compared with early stage (ES-TALL, n = 3) and late-stage (LS-TALL, n = 3) primary mouse T-ALL samples and mouse T-ALL cell line M295. (B) qRT-PCR validation of miR-21 expression in independent mouse samples compared with CD4+CD8+ DP thymocytes and normalized to sn-U6. Samples used: DN, sorted DN thymocytes; DP, sorted DP thymocytes; early T-ALL, polyclonal early stage mouse T-ALL; late T-ALL, oligoclonal late-stage mouse T-ALL samples; T-ALL cell lines, 3 independent mouse T-ALL cell lines. (C) Heatmap of the 25 most abundantly expressed miRNAs assessed by microarray in a human T-ALL cell line CUTLL1. Cells (n = 3 independent samples per condition) were treated either with dimethylsulfoxide (DMSO) or difluorophenylacetyl-L-alanyl-S-phenylglycine t-butyl ester (DAPT) (10 µM) for 3 days prior to analysis. (D) miR-21 expression analysis by qRT-PCR on human T-ALL cell lines CUTLL1, T-ALL1, DND41, and KOPTK1 (black bars, n = 3 independent samples per cell line) normalized to sn-U6 as illustrated by ΔCT values. Furthermore, 2 primary T-ALL patient samples were analyzed for miR-21 expression (gray bars). (E) qRT-PCR was performed on RNA samples extracted from the gamma secretase inhibitor (GSI)-sensitive T-ALL cell lines CUTLL1, T-ALL1, DND41, and KOPTK1. Cells were treated with DMSO or DAPT (10 µM) for 3 days. qRT-PCR for miR-21 was performed, and expression levels were normalized to sn-U6 expression. (F) Quantification of early apoptosis (7-AAD− Annexin-V+) in fluorescein (FAM)+ T-ALL cell lines M295 and CUTLL1 3 days posttransfection with FAM-LNA anti-miR-21 (▪, LNA_21, n = 5) or control (□, LNA_Ctr, n = 5). Data (E-F) are pooled from 2 independent experiments. *P < .05; **P < .01; ***P < .001.
KD of miR-21 in T-ALL induces apoptosis. (A) Heatmap of the 25 most abundantly expressed miRNAs as assessed by microarray in FACS-sorted mouse CD4−CD8− DN (n = 3) and CD4+CD8+ DP (n = 3) thymocytes compared with early stage (ES-TALL, n = 3) and late-stage (LS-TALL, n = 3) primary mouse T-ALL samples and mouse T-ALL cell line M295. (B) qRT-PCR validation of miR-21 expression in independent mouse samples compared with CD4+CD8+ DP thymocytes and normalized to sn-U6. Samples used: DN, sorted DN thymocytes; DP, sorted DP thymocytes; early T-ALL, polyclonal early stage mouse T-ALL; late T-ALL, oligoclonal late-stage mouse T-ALL samples; T-ALL cell lines, 3 independent mouse T-ALL cell lines. (C) Heatmap of the 25 most abundantly expressed miRNAs assessed by microarray in a human T-ALL cell line CUTLL1. Cells (n = 3 independent samples per condition) were treated either with dimethylsulfoxide (DMSO) or difluorophenylacetyl-L-alanyl-S-phenylglycine t-butyl ester (DAPT) (10 µM) for 3 days prior to analysis. (D) miR-21 expression analysis by qRT-PCR on human T-ALL cell lines CUTLL1, T-ALL1, DND41, and KOPTK1 (black bars, n = 3 independent samples per cell line) normalized to sn-U6 as illustrated by ΔCT values. Furthermore, 2 primary T-ALL patient samples were analyzed for miR-21 expression (gray bars). (E) qRT-PCR was performed on RNA samples extracted from the gamma secretase inhibitor (GSI)-sensitive T-ALL cell lines CUTLL1, T-ALL1, DND41, and KOPTK1. Cells were treated with DMSO or DAPT (10 µM) for 3 days. qRT-PCR for miR-21 was performed, and expression levels were normalized to sn-U6 expression. (F) Quantification of early apoptosis (7-AAD− Annexin-V+) in fluorescein (FAM)+ T-ALL cell lines M295 and CUTLL1 3 days posttransfection with FAM-LNA anti-miR-21 (▪, LNA_21, n = 5) or control (□, LNA_Ctr, n = 5). Data (E-F) are pooled from 2 independent experiments. *P < .05; **P < .01; ***P < .001.
We next decided to assess the biological function of miR-21 in mouse and human T-ALL by miR-21 KD using fluorescein (FAM)–coupled LNAs.31 KD of miR-21 in both murine and human T-ALL cell lines did not affect cell cycle regulation (supplemental Figure 6A,C-D) but instead induced apoptosis (Figure 5F and supplemental Figure 6B). These results indicate that miR-21-controlled target genes are involved in regulating the survival of T-ALL cells.
Pdcd4 is an important miR-21 target in T-ALL
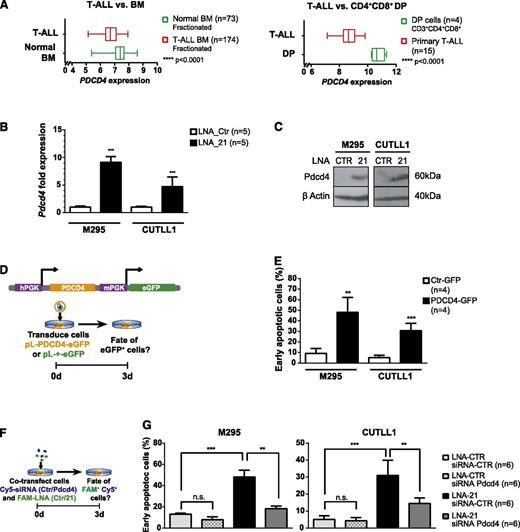
In various solid tumors, miR-21 has been suggested to function as an oncomiR.32 A comparison of predicted target genes between mouse and human identified 149 conserved miR-21 target genes (supplemental Figure 6E), among which apoptotic regulators have been identified.33 Using Leukemia Gene Atlas,34 the expression pattern of the 149 conserved miR-21 target genes in BM aspirates of 174 T-ALL patients compared with 73 normal controls or compared with patient derived CD4CD8+ DP thymocytes was reanalyzed34,35 (supplemental Table 1 and supplemental Figure 7). This analysis allowed clustering the samples into 2 groups, which correlated with disease status (T-ALL vs normal BM, and T-ALL vs CD4+CD8+ DP thymocytes). miR-21 target-gene repression correlated mostly with patient samples from T-ALL compared with control including DP thymocytes. Among these miR-21 target genes, several candidates implicated in apoptosis were identified, including PDCD436-40 (Figure 6A). To determine whether miR-21 regulates the expression of some of these conserved genes in T-ALL cells, expression of several miR-21 target genes was assessed by qRT-PCR in mouse and human T-ALL cell lines following miR-21 KD. In murine and human T-ALL cell lines, Pdcd4 was found the most consistently derepressed gene after miR-21 KD (Figure 6B-C and supplemental Figure 8A-C). To assess whether derepression of Pdcd4 is functionally involved in the induction of apoptosis, Pdcd441 was ectopically expressed in T-ALL cell lines using an eGFP-traceable Pdcd4 expression vector (Figure 6D). Interestingly, Pdcd4 expression was sufficient in both mouse and human T-ALL cells to induce apoptosis (Figure 6E and supplemental Figure 8D).
Overexpression of the miR-21 target gene PDCD4 induces apoptosis in T-ALL. (A) Box plot expression analysis of PDCD4 on RNA extracted from fractionated BM aspirates of 174 T-ALL patients and 73 normal controls (left panel), expression data publicly accessible through Leukemia Gene Atlas40 and on RNA extracted of FACS-sorted CD3+CD4+CD8+ primary human thymocytes (n = 4) compared with primary T-ALL specimens (n = 15), expression data publicly accessible through gene expression omnibus (GEO) data set browser (GSE6200635 ) (right panel). (B) Pdcd4 qRT-PCR performed on FAM+ mouse M295 and human CUTLL1 T-ALL cells after FAM-LNA-mediated miR-21 KD (or control transfection). Data were pooled from 2 independent experiments. (C) Western blot analysis for PDCD4 in M295 and CUTLL1 T-ALL cells was performed 3 days p.tr. T-ALL cells were transfected with FAM-LNA-Ctr or FAM-LNA-21 for miR-21 KD, and protein lysates of FACS-sorted cells were assessed for PDCD4 expression. (D) Overexpression of PDCD4 in T-ALL. A lentivirus-based system was employed to overexpress human PDCD4 in T-ALL cells. PDCD4 and eGFP contain 2 independent PGK promoters (upper panel). Experimental protocol: T-ALL cells were transduced with control virus (pL-+-eGFP) or PDCD4 overexpression virus (pL-PDCD4-eGFP) and analyzed for apoptosis induction 3 days following transduction (lower panel). (E) T-ALL cell lines M295 and CUTLL1 strongly induce early apoptosis (eGFP+, 7-AAD− Annexin-V+ cells) after 3 days in PDCD4 transduced cells compared with control. (F) Double KD experiment to simultaneously downregulate miR-21 and Pdcd4 to quantify apoptosis induction in T-ALL cells. T-ALL cells were cotransfected with Cy5-coupled siRNAs (Ctr or a mix of 2 independent siRNAs targeting human or mouse Pdcd4) and FAM-coupled LNAs (Ctr or miR-21). Three days p.tr., Cy5+FAM+ cells were assessed for apoptosis induction (G). Cy5+FAM+ T-ALL cells generated through the experimental protocol in panel F were assessed for early apoptosis (7-AAD− Annexin-V+ cells). miR-21 and Pdcd4 double KD cells were compared with cells that were transfected only with miR-21 KD (LNA-21 and siRNA-Ctr) or transfected with control oligonucleotides (LNA-Ctr and siRNA-Ctr) and cells where only Pdcd4 was downregulated (LNA-Ctr and siRNA-Pdcd4). Data (B, E, G) are pooled from 2 independent experiments. *P < .05; **P < .01; ***P < .001.
Overexpression of the miR-21 target gene PDCD4 induces apoptosis in T-ALL. (A) Box plot expression analysis of PDCD4 on RNA extracted from fractionated BM aspirates of 174 T-ALL patients and 73 normal controls (left panel), expression data publicly accessible through Leukemia Gene Atlas40 and on RNA extracted of FACS-sorted CD3+CD4+CD8+ primary human thymocytes (n = 4) compared with primary T-ALL specimens (n = 15), expression data publicly accessible through gene expression omnibus (GEO) data set browser (GSE6200635 ) (right panel). (B) Pdcd4 qRT-PCR performed on FAM+ mouse M295 and human CUTLL1 T-ALL cells after FAM-LNA-mediated miR-21 KD (or control transfection). Data were pooled from 2 independent experiments. (C) Western blot analysis for PDCD4 in M295 and CUTLL1 T-ALL cells was performed 3 days p.tr. T-ALL cells were transfected with FAM-LNA-Ctr or FAM-LNA-21 for miR-21 KD, and protein lysates of FACS-sorted cells were assessed for PDCD4 expression. (D) Overexpression of PDCD4 in T-ALL. A lentivirus-based system was employed to overexpress human PDCD4 in T-ALL cells. PDCD4 and eGFP contain 2 independent PGK promoters (upper panel). Experimental protocol: T-ALL cells were transduced with control virus (pL-+-eGFP) or PDCD4 overexpression virus (pL-PDCD4-eGFP) and analyzed for apoptosis induction 3 days following transduction (lower panel). (E) T-ALL cell lines M295 and CUTLL1 strongly induce early apoptosis (eGFP+, 7-AAD− Annexin-V+ cells) after 3 days in PDCD4 transduced cells compared with control. (F) Double KD experiment to simultaneously downregulate miR-21 and Pdcd4 to quantify apoptosis induction in T-ALL cells. T-ALL cells were cotransfected with Cy5-coupled siRNAs (Ctr or a mix of 2 independent siRNAs targeting human or mouse Pdcd4) and FAM-coupled LNAs (Ctr or miR-21). Three days p.tr., Cy5+FAM+ cells were assessed for apoptosis induction (G). Cy5+FAM+ T-ALL cells generated through the experimental protocol in panel F were assessed for early apoptosis (7-AAD− Annexin-V+ cells). miR-21 and Pdcd4 double KD cells were compared with cells that were transfected only with miR-21 KD (LNA-21 and siRNA-Ctr) or transfected with control oligonucleotides (LNA-Ctr and siRNA-Ctr) and cells where only Pdcd4 was downregulated (LNA-Ctr and siRNA-Pdcd4). Data (B, E, G) are pooled from 2 independent experiments. *P < .05; **P < .01; ***P < .001.
To ensure that miR-21 KD-driven induction of apoptosis is mediated by derepression of Pdcd4, we performed double KD experiments for miR-21 and Pdcd4 in T-ALL cell lines. Simultaneous KD of miR-21 and Pdcd4 rescued to a large extent the miR-21 KD-induced apoptotic phenotype in T-ALL cell lines (Figure 6F-G and supplemental Figure 8E). Pdcd4 has been proposed to act at least in part through translational inhibition of BCL-xL,39,42 which has been implicated in the survival of human T-ALL lines and primary tumors.43 Interestingly, BCL-xL protein levels were downregulated in miR-21 KD T-ALL cells and increased in cells in which miR-21 and Pdcd4 were knocked down simultaneously (supplemental Figure 8F). This strongly suggests that miR-21, through suppression of its downstream target Pdcd4, acts in a prosurvival manner in T-ALL at least in part through stabilizing the protein levels of BCL-xL.
Discussion
Although several miRNA candidates have been shown to modulate the onset of Notch1-driven T-ALL in mouse models,8,10,11,13 it remains unclear whether miRNAs are essential for onset and/or maintenance of this disease. Here, we show that CD4-Cre-mediated LOF of Dicer1 prevents the development of Notch-induced T-ALL in BM chimeras in vivo. Although Dicer1 mutant mice did not develop disease, short-lived preleukemic DP T cells were transiently detected. These findings demonstrate that aberrant Notch1 signaling can induce the development of preleukemic cells in the absence of Dicer1, but complete cellular transformation and/or survival is strictly dependent on Dicer1-mediated miRNA biogenesis. Whereas a similar essential role for Dicer1 has been reported for the development of myc-induced B-cell lymphomas,9 its requirement in myeloid leukemia seems to differ. miRNAs in myeloid progenitors appear to be involved in regulating the switch of self-renewal vs differentiation,7 whereas in ALL they regulate proliferation and/or survival.9 Although Dicer1 mutant myeloid progenitors exhibited increased self-renewal, Dicer1-deficient dysplastic cells did not give rise to AML. Interestingly, mono- but not biallelic Dicer1 deletion in myeloid committed progenitors in a p53 mutant background led to the development of AML-like leukemia.7 These data suggest that Dicer1 in AML acts as a haploinsufficient tumor suppressor, and loss of both alleles of Dicer1 is sufficient to induce a preleukemic disease stage. Similar observations have also been reported in a Kras-driven model of lung cancer23 and in a mouse model of retinoblastoma.24 In both models, heterozygous deletion of Dicer1 led to accelerated tumor formation. However, in this study, mice with heterozygous deletion of Dicer1 were found to develop T-ALL like Dicer1-proficient animals. This suggests that Dicer1 does not function as a haploinsufficient tumor suppressor in Notch-driven T-ALL. These results are consistent with the notion that miRNAs in N1ICD-driven T-ALL predominantly act in an oncogenic fashion during progenitor cell transformation, whereas development of AML might rely more on tumor-suppressive miRNAs.
Analysis of global miRNA levels in multiple human tumors revealed a general trend of decreased miRNA content.44-46 Thus, it is conceivable that a certain number of Dicer1-processed miRNAs are needed for transformation of progenitor cells but may no longer be required in established tumors. Indeed, the requirement for Dicer1 in tumor maintenance appears to vary depending on the type of cancer.23 A recent study showed that Dicer1−/− sarcoma cells, completely devoid of miRNAs, continue proliferating in vitro and maintain tumorigenicity in vivo, suggesting that in some types of cancer, Dicer1 activity is not required for tumor maintenance.47,48 We assessed the role of Dicer1 in the maintenance of early and late-stage T-ALL. Successful ablation of Dicer1 in early established leukemic cells resulted in complete regression of T-ALL cells, and relapse did not occur, in contrast to incomplete Dicer1 inactivation. These Dicer1-proficient T-ALL cell clones continued to proliferate and animals died of disease. Corresponding splenic tumors were primarily composed of Dicer1-proficient cells, indicating that they quickly outcompeted Dicer1-deficient cells. These results imply that Dicer1 LOF may be deleterious for the progression of established T-ALL. This hypothesis was confirmed by studying Mx1-Cre-mediated ablation of Dicer1 in oligoclonal T-ALL cells following serial transplantation into RAG2γc−/− mice. Animals died of T-ALL because of a strong selective pressure for cells escaping Mx1-Cre-mediated Dicer1 deletion.
Collectively, these data demonstrate that Dicer1 is essential for the progression and maintenance of N1ICD-induced T-ALL
The fact that Dicer1-deficient T-ALL cells are rapidly outcompeted indicated an important role for miRNAs in processes relevant for T-ALL maintenance.49,50 Therefore, we established N1ICD-induced T-ALL cell lines and performed linage-tracing experiments to follow the fate of Dicer1-deficient T-ALL cells. These experiments revealed that Dicer1 has no detectable influence on cell cycle control but is absolutely necessary for survival. This observation is again similar to findings in B-ALL in which Dicer1 ablation in vitro resulted in induction of apoptosis in the majority of B-ALL cells, whereas residual Dicer1-proficient escaper cells continued to proliferate.9
The findings suggested that certain abundantly expressed miRNAs in T-ALL may execute important prosurvival functions. Thus, we aimed to identify novel prosurvival miRNAs potentially deregulated in T-ALL. We focused on miRNAs that are expressed in DN thymocytes, downregulated in DP thymocytes but abundant in T-ALL cells. Interestingly, only miR-21 fulfilled these criterions. To date there are no reports on the role of miR-21 in thymocyte development or T-ALL; miR-21 has been described as an oncogene in several solid tumors.51-53 Importantly, miR-21 function has been linked to cancer-related processes in hepatocellular, glioblastoma, and breast cancer.33,54-56 Furthermore, microarray analysis of miR-21 target gene expression in BM aspirates of T-ALL patients and normal controls revealed that many miR-21 targets were repressed in T-ALL patient samples, indicative of a clinical relevance of miR-21 target gene repression in T-ALL. KD of miR-21 in T-ALL cells led to the induction of apoptosis. Although this largely mimics the Dicer1 LOF effects in T-ALL, miR-21 is certainly not the only miRNA exerting important survival functions. Other miRNAs such as the miR-17-92 gene cluster (data not shown) may be of similar importance. Nevertheless, our data imply that miR-21 functions as an important oncomiR in T-ALL promoting survival of T-ALL cells.
Microarray analysis of miR-21 target gene expression led to the identification of several candidate target genes implicated in survival and apoptosis pathways. Among these, we focused on Pdcd4 because it was consistently downregulated in all murine and human T-ALL samples examined. Pdcd4 was originally characterized as an inhibitor of cellular transformation in mouse epidermal cells.57 Subsequent studies showed that Pdcd4−/− mice spontaneously develop B-cell lymphomas58 and are more sensitive to carcinogen-induced skin cancer.59,60 Moreover, Pdcd4 is a direct miR-21 target and is downregulated in tumors with high miR-21 expression, thus contributing to tumorigenesis including suppression of apoptosis.39,41,61-64 To test whether derepression of Pdcd4 is sufficient to induce apoptosis, Pdcd4 was ectopically expressed in T-ALL cell lines. Pdcd4 was sufficient to induce apoptosis in T-ALL cells, suggesting that apoptosis of T-ALL cells in which miR-21 expression is downregulated is at least partly caused by derepression of Pdcd4. Recent structural studies on PDCD4 revealed that it inhibits initiation of cap-dependent protein translation by binding to the RNA helicase elongation initiation factor 4A (eIF4A) and the scaffold protein elF4G65 or by blocking internal ribosomal entry site–mediated translation of the prosurvival genes Bcl-XL and Xiap.42 Interestingly, our correlative studies on BCL-xL protein levels in murine and human T-ALL cell lines confirms that miR-21 is regulating BCL-xL through the repression of Pdcd4.
Collectively, we demonstrate that miRNAs are essential for development and maintenance of Notch1-driven T-ALL. Furthermore, we identified miR-21 as an important regulator of T-ALL cell survival, which mediates its function in part via repression of the tumor suppressor Pdcd4.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors acknowledge Christelle Dubey, Laure Bardouillet, and Marianne Nkosi for technical assistance; Michele de Palma and Mario Squadrito for fruitful discussions; the Flow Cytometry Core Facility for cell sorting; and the DNA array facility of the University of Lausanne for microarray preparation and technical analysis. The authors would also like to thank Marion Leleu from the Bioinformatics Department (Ecole Polytechnique Fédérale de Lausanne) for assistance in bioinformatic data analysis and Dr Jean-Pierre Bourquin, Kinderspital Zürich, and Viktoras Frismantas for providing primary human T-ALL samples.
This work was supported in part by the Swiss National Science Foundation and the Swiss Cancer League (F.R.).
Authorship
Contribution: F.J. performed experiments, analyzed data, and wrote the manuscript; A.C. performed experiments and analyzed data; U.K. performed experiments, analyzed data, and wrote the manuscript; and F.R. conceived the study, analyzed data, and wrote the manuscript.
Conflict of interest disclosure: The authors declare no competing financial interests.
Correspondence: Freddy Radtke, Ecole Polytechnique Fédérale de Lausanne (EPFL), Swiss Institute for Experimental Cancer Research (ISREC), Station 19, 1015 Lausanne, Switzerland; e-mail: freddy.radtke@epfl.ch.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal