Key Points
Platelets stimulate proliferation of HepG2 cells, which requires uptake of platelets by the HepG2 cell.
Platelets stimulate HepG2 cell proliferation in part by transfer of RNA from the anucleate platelet to the nucleated HepG2 cell.
Abstract
Liver regeneration is stimulated by blood platelets, but the molecular mechanisms involved are largely unexplored. Although platelets are anucleate, they do contain coding or regulatory RNAs that can be functional within the platelet or, after transfer, in other cell types. Here, we show that platelets and platelet-like particles (PLPs) derived from the megakaryoblastic cell line MEG-01 stimulate proliferation of HepG2 cells. Platelets or PLPs were internalized within 1 hour by HepG2 cells and accumulated in the perinuclear region of the hepatocyte. Platelet internalization also occurred following a partial hepatectomy in mice. Annexin A5 blocked platelet internalization and HepG2 proliferation. We labeled total RNA of MEG-01 cells by incorporation of 5-ethynyluridine (EU) and added EU-labeled PLPs to HepG2 cells. PLP-derived RNA was detected in the cytoplasm of the HepG2 cell. We next generated PLPs containing green fluorescent protein (GFP)-tagged actin messenger RNA. PLPs did not synthesize GFP, but in coculture with HepG2 cells, significant GFP protein synthesis was demonstrated. RNA-degrading enzymes partly blocked the stimulating effect of platelets on hepatocyte proliferation. Thus, platelets stimulate hepatocyte proliferation via a mechanism that is dependent on platelet internalization by hepatocytes followed by functional transfer of RNA stored in the anucleate platelet. This mechanism may contribute to platelet-mediated liver regeneration.
Introduction
Blood platelets have essential roles in hemostasis and thrombosis, inflammation, host defense, and wound healing.1-4 Emerging evidence from recent in vitro and in vivo studies suggests that platelets have a pivotal role in liver regeneration.5-8 In experimental animal models in which platelets were depleted or functionally impaired, liver regeneration after a partial liver resection was substantially delayed.6 Conversely, following a partial liver resection in animals with a drug-induced thrombocytosis, liver regeneration was accelerated.9,10 In a clinical study, we showed that a low platelet count is an independent predictor of delayed postoperative liver function recovery following a partial liver resection, suggesting that platelets stimulate liver regeneration also in humans.11
The molecular mechanisms of platelet-mediated stimulation of liver regeneration are largely unexplored. Platelets contain 2 distinct types of storage organelles: α granules and dense granules. The α granules contain, among many proteins, a number of growth factors that have an established role in liver regeneration (platelet-derived growth factor [PDGF], hepatocyte growth factor [HGF], insulin-like growth factor [IGF], and vascular endothelial growth factor [VEGF]).8,12 In addition, the dense granules contain serotonin, which is also an established mediator of liver regeneration.5,6 It seems plausible that local release of these factors following stimulation of the platelets contributes to platelet-mediated stimulation of regeneration. Indeed, it has been demonstrated that platelets stimulate hepatocyte proliferation in vitro, and it has been suggested that direct contact of platelets and hepatocytes is required for this effect.8 Also, in vivo studies have demonstrated that platelets accumulate in the liver parenchyma following partial liver resection.13 Recently, a novel mechanism by which platelets may communicate with their environment has been described, which involves de novo protein synthesis. Although platelets lack a nucleus, they do contain a wide array of (pre–)messenger RNAs, which may be translated to protein.14-17 Protein synthesis by platelets may occur in particular following stimulation of the platelet.14-16 In addition, it has been convincingly demonstrated that platelets are capable of transferring their messenger RNA (mRNA) to other cell types, including monocytic and endothelial cells.17 It was shown that the recipient cell is capable of translating platelet-derived mRNA, which may have biologically relevant effects on the recipient cell. Platelet also contain microRNAs (miRNAs),18,19 and functional transfer of platelet miRNA to endothelial cells has recently been described.20
Here, we studied the fate of platelets during platelet-mediated stimulation of hepatocyte proliferation in vitro. We observed that platelets were internalized by hepatocytes. Based on our observation that platelets are, after internalization, directed toward the hepatocyte nucleus, we hypothesized that platelets deliver their RNA content to the hepatocyte and that this RNA transfer contributes to platelet-mediated hepatocyte proliferation.
Material and methods
Study approval
Institutional review board and animal care and use committee approval was granted from local committees.
Cell culture
HepG2 cells (ATCC, Georgetown, WA) were cultured in Dulbecco’s modified Eagle medium (Lonza, Basel, Switzerland) supplemented with 10% (vol/vol) fetal bovine serum (Invitrogen, Carlsbad, CA). MEG-01 cells (Health Protection Agency Culture Collections, London, United Kingdom) were grown in suspension using Dulbecco’s modified RPMI 1640 medium (Lonza) supplemented with 4.5% (vol/vol) l-glutamine and 10% (vol/vol) fetal bovine serum (Invitrogen). Cells were cultured without the addition of antibiotics to the culture medium.
MEG-01 cells were differentiated to mature megakaryocytes and stimulated to form platelet-like particles (PLPs) according to previously described methods, with some modifications.17,21 In short, for differentiation, cells were treated for at least 10 days with 5 mM valproic acid (Sigma, St. Louis, MO). Subsequently, PLP production was stimulated by treatment with recombinant human thrombopoietin (rTPO; Life Technologies, Carlsbad, CA; 100 ng/mL, diluted in culture medium) for 72 hours. Cell culture medium from differentiated, rTPO-stimulated MEG-01 cells was centrifuged at 100g for 10 minutes to remove nucleated cells from the medium. The supernatant was centrifuged at 1000g for 10 minutes, and the pellet containing PLPs was resuspended in culture medium. The PLP preparation contained no detectable nucleated cells as evidenced by flow cytometry using the nuclear dye Draq5 (Thermo Fisher Scientific, Etten Leur, The Netherlands). PLPs stained positive for glycoprotein Ibα and integrin αIIbβ3 by flow cytometry using antibodies from BD Biosciences (Franklin Lakes, NJ) (data not shown).
In selected experiments, we used MEG-01 cells that, following differentiation with valproic acid, were transfected with CellLight Actin-GFP (Molecular Probes, Carlsbad, CA), a modified baculovirus containing an actin and green fluorescent protein (GFP) mRNA construct. After 48 hours of incubation, virus-containing medium was replaced by standard culture medium containing rTPO. PLPs were isolated 72 hours following rTPO addition. Actin-GFP–expressing PLPs were cocultured up to 48 hours with HepG2 cells in 96-well plates to assess transfer of actin-GFP mRNA to HepG2 cells. GFP synthesis was quantified by measuring fluorescence intensity (emission and excitation wavelengths were 485 nm and 535 nm, respectively) using a Victor3 plate reader (Perkin Elmer, Waltham, MA). A standard curve of purified GFP (Cell Biolabs, San Diego, CA) was used to quantify the amount of GFP in the cells, and values were corrected for background fluorescence of cells without actin-GFP.
In another set of experiments, PLPs or isolated human platelets (200 000/µL; see the section on platelet isolation below) were treated with RNaseA (10 or 100 U/mL; Sigma) for 1 hour at 37°C. Subsequently, RNaseA was inhibited by incubation with SUPERase In RNAse (0.1-10 U/µL) for 30 minutes (Invitrogen). Platelet RNA was fully degraded by this procedure (supplemental Figure 1 available at the Blood Web site), but platelet functionality, as assessed by flow cytometry and flow-based platelet adhesion assays, was fully preserved (supplemental Figure 2). PLPs were washed twice with culture medium after RNaseA and RNase-inhibitor treatment.
Partial hepatectomy in mice
Male C57Bl6 mice (Harlan Laboratories, Venray, The Netherlands) of 8 to 10 weeks of age underwent a 70% partial hepatectomy according to published protocols.22 One hour after hepatectomy, mice were terminated by exsanguination. Livers were flushed with saline and processed for transmission electron microscopy as indicated below.
Platelet isolation, activation, and labeling
Blood from healthy volunteers, who claimed not to have used aspirin or other nonsteroidal anti-inflammatory drugs for the preceding 10 days, was drawn into one-tenth volume of 3.4% sodium citrate. The local institutional review board approved the study, and written informed consent was obtained from each blood donor. Washed platelets were isolated as described previously23 and resuspended in cell culture medium, and platelet count was adjusted to 200 000/μL. Platelet preparations contained <1% of CD45-positive cells as assessed by flow cytometry. Platelets were activated by addition of 15 μg/mL adenosine diphosphate (Stago BNL, Leiden, The Netherlands) and 15 μg/mL thrombin receptor activating peptide (Bachem, Bubendorf, Switzerland) and incubated for 30 minutes at 37°C. In selected experiments, isolated platelets or PLPs (200 000/μL) were fluorescently labeled with 2.5 µM CellTracker green CMFDA (Life Technologies, Carlsbad, CA). This dye was added to the platelet suspension for 30 minutes, during which time it is taken up by the platelet spontaneously. Platelets were washed with cell culture medium and diluted to a platelet count of 200 000/μL.
In a separate set of experiments, platelet microparticles were isolated from resting or activated platelet preparations as described previously.24 In short, platelet preparations were centrifuged thrice at 1250g for 15 minutes. The final supernatant contained only platelet microparticles as evidenced by size (<1 µm) and presence of glycoprotein Ibα and integrin αIIbβ3 by flow cytometry.
Quantification of cell proliferation
The proliferation rate of HepG2 cells was estimated by quantification of 5-bromo-2′-deoxyuridine (BrdU) incorporation using a commercially available enzyme-linked immunosorbent assay (ELISA) (Cell proliferation ELISA, BrdU; Roche, Basel, Switzerland). HepG2 cells were plated in a 96-well plate at 15 000 cells/well and cultured overnight. Subsequently, cells were washed twice with serum-free culture medium and vehicle, platelets, or PLPs (both at 200 000/µL) or platelet microparticles were added and incubated under serum-free conditions for 48 hours. Alternatively, platelets were removed at earlier time points by gentle washing with culture medium, and hepatocytes were incubated with serum-free medium alone. Platelet integrity after 48 hours was preserved as evidenced by only a marginal increase in phosphatidylserine exposure (5.7% ± 1.7% of platelets were Annexin A5 positive by flow cytometry at baseline, compared with 9.4% ± 1.9% after 48 hours). In selected experiments, platelets were treated for 30 minutes at 37°C with 30 mM Annexin A5 (eBioscience, Santa Clara, CA) or O-sialyglycoprotein endopeptidase (OSE) (30 µg/mL) (Cedarlane, Burlington, ON, Canada) as described previously25 before adding them to the HepG2 cells. Alternatively, HepG2 cells were treated with 100 µg/mL asialofetuin (Asf) (Sigma) for 30 minutes at 37°C prior to the addition of platelets as described previously.26 All compounds were diluted in cell culture medium.
After 48 hours of incubation of HepG2 cells with platelets or PLPs, cell culture medium was replaced by fresh culture medium containing BrdU. After 2 hours, HepG2 cells were washed 3 times with phosphate-buffered saline (PBS). Cells were subsequently fixed with 4% paraformaldehyde (PFA) diluted in distilled H2O for 10 minutes, after which BrdU incorporation was measured by ELISA. In these experiments, HepG2 cells cultured in medium containing 10% serum were used as positive control and HepG2 cells cultured in serum-free medium were used as negative control. In selected experiments, we ascertained that the BrdU incorporation assay gives an accurate estimate of cell proliferation by comparing BrdU incorporation with manual cell counting. Whereas the number of cells per well clearly increased by platelets, PLPs, or fetal calf serum (FCS)-containing medium, serum-free conditions only led to a marginal increase in the number of cells in each well, confirming BrdU test results.
Labeling of total RNA of PLPs
Differentiated MEG-01 cells were incubated for 48 hours with 0.5 mM 5-ethynyluridine (EU) (Life Technologies) in combination with rTPO. This procedure results in production of PLPs that have incorporated EU (a modified form of uridine) in their RNA. Isolated PLPs were washed twice in RPMI 1640 medium (Lonza, Walkersville, MI). EU was visualized using the Click-iT RNA Alexa Fluor 488 Imaging Kit (Life Technologies). EU incorporation was subsequently analyzed by flow cytometry on a BD FACSCalibur flow cytometer (BD Biosciences) or immunofluorescence microscopy. Flow cytometry data acquisition was performed using FlowJo X (Tree Star, Ashland, OR).
Immunofluorescence staining
For immunofluorescence experiments, HepG2 cells were cultured overnight and plated to obtain ∼50% confluence on 0.1 mg/mL poly-l-lysine–coated (Santa Cruz Biotechnology, Dallas, TX) glass coverslips (Thermo Scientific, Waltham, MA) in a 24-well plate (Corning, Corning, NY). Coverslips were treated with poly-l-lysine for 30 minutes at 37°C and washed 3 times with PBS before HepG2 cells were seeded. After addition of CellTracker green CMFDA–labeled platelets, the culture medium was removed after various time points and cells were washed twice with PBS to remove nonadherent platelets or PLPs. Cells were subsequently fixed with 4% PFA for 10 minutes.
HepG2 cells were stained for actin with Alexa 594–labeled phalloidin (Life Technologies) or with a rabbit anti-actin antibody (Sigma) followed by an Alexa 594–labeled goat anti-rabbit antibody (Life Technologies). Nuclei were identified using DNA staining with 4,6 diamidino-2-phenylindole (DAPI) by using VECTASHIELD Hard-set mounting medium with DAPI (Vector Laboratories, Burlingame, CA). Imaging was performed on a Leica SP2 AOBS confocal microscope (Leica Microsystems, Solms, Germany) and on a Leica DMI4000B LED. Images were captured using Leica Confocal Version 2.5 software or Leica Application Suite Advanced Fluorescence software (LAS AF). Image processing was performed using Imaris 7.1.1 (Bitplane Scientific Software, Zurich, Switzerland).
Electron microscopy
For ultrastructural analysis, HepG2 cells were cocultured with platelets and fixed with 2.5% glutaraldehyde and 1% PFA in 0.1 M sodium cacodylate buffer (pH 7.4) for 24 hours. Pieces (∼1 mm3) of remnant livers from mice that underwent a 70% partial hepatectomy were also fixed in this buffer. The fixed cells or tissue were then rinsed with 0.1 M sodium cacodylate buffer and postfixed with 2% osmium tetroxide for 1 hour at 4°C. Cells were washed 3 times in double-distilled H2O and dehydrated with a graded series of ethanol (50%, 70%, and 100%; 3 × 10 minutes each) followed by embedding in Epon and ultrathin sectioning. After uranyl acetate and lead citrate staining, ultrathin sections were examined by the transmission electron microscope FEI CM100 (Philips, Amsterdam, The Netherlands). Images were captured with an Advantage CCD camera using iTEM software (Olympus, Tokyo, Japan).
Statistical analysis
Statistical analysis was performed with the GraphPad Prism 5 (San Diego, CA) software package. Continuous variables were expressed as mean ± standard deviation (SD) or median and range. Values are representative of at least 3 independent experiments performed in triplicate. Continuous data were tested for normality and analyzed by Student t test, Mann-Whitney U test, or 1-way analysis of variance, as appropriate. A P value < .05 was considered statistically significant. Images are representative for at least 3 independent experiments.
Results
Platelets stimulate hepatocyte proliferation
To investigate the proliferative effect of platelets on hepatocytes, we cocultured freshly isolated human platelets for 48 hours with HepG2 cells under serum-free conditions. Serum-free conditions were well tolerated by HepG2 cells for 48 hours, as evidenced by a lack of trypan blue uptake (not shown). Addition of resting platelets resulted in a 1.5-fold increase of BrdU incorporation into HepG2 cell DNA (Figure 1A). When activated platelets were added to the HepG2 cells, a 2.2-fold increase in BrdU incorporation was seen. Subsequently, we added resting platelets to hepatocytes, removed platelets at various time points, and assessed hepatocyte proliferation at 48 hours. A maximal effect on hepatocyte proliferation was observed when platelets were present for at least 24 hours (Figure 1B). We next investigated whether platelet microparticles also stimulated HepG2 cell proliferation. Whereas resting or activated platelet preparations potently stimulated hepatocyte proliferation, microparticles isolated from these preparations had no stimulatory effects (Figure 1A).
Isolated platelets increase proliferation of hepatocytes. (A) Resting or activated platelets (plt) or microparticles (MP) isolated from these platelet preparations were added to HepG2 cells and incubated under serum-free conditions. After 48 hours, the cell proliferation rate was estimated by quantifying BrdU incorporation. Control groups represent HepG2 cells cultured in presence or absence of FCS. *P < .05 compared with −FCS. Data represent the mean of 3 independent triplicate experiments. Error bars indicate SD. (B) Resting platelets were added to HepG2 cells and incubated under serum free conditions. After various time points, platelets were removed by gentle washing and replaced by serum-free medium. After 48 hours, the cell proliferation rate was estimated by quantifying BrdU incorporation. Control groups represent HepG2 cells cultured in the presence or absence of FCS. *P < .05 compared with −FCS. Data represent the mean of 3 independent triplicate experiments. Error bars indicate SD. OD, optical density.
Isolated platelets increase proliferation of hepatocytes. (A) Resting or activated platelets (plt) or microparticles (MP) isolated from these platelet preparations were added to HepG2 cells and incubated under serum-free conditions. After 48 hours, the cell proliferation rate was estimated by quantifying BrdU incorporation. Control groups represent HepG2 cells cultured in presence or absence of FCS. *P < .05 compared with −FCS. Data represent the mean of 3 independent triplicate experiments. Error bars indicate SD. (B) Resting platelets were added to HepG2 cells and incubated under serum free conditions. After various time points, platelets were removed by gentle washing and replaced by serum-free medium. After 48 hours, the cell proliferation rate was estimated by quantifying BrdU incorporation. Control groups represent HepG2 cells cultured in the presence or absence of FCS. *P < .05 compared with −FCS. Data represent the mean of 3 independent triplicate experiments. Error bars indicate SD. OD, optical density.
Platelets are internalized during hepatocyte proliferation
To study whether platelets stimulate proliferation of HepG2 cells by direct contact, we added CellTracker green CMFDA–labeled platelets to HepG2 cells. After 30 minutes of coincubation, platelets were present at the HepG2 cell membrane as shown by fluorescent imaging (Figure 2A-B) and electron microscopy (Figure 2B). After 1 hour, platelets were observed inside the HepG2 cells (Figure 2A). Confocal laser scanning microscopy confirmed that platelets were located inside the cell and not attached to the outer cell membrane (Figure 2C). After 1 hour, platelets appeared predominantly located in the perinuclear region of the HepG2 cells, which was confirmed by electron microscopy as shown in Figure 2D. Platelets were found within a few nanometers of the nucleus and were surrounded by endoplasmic reticulum. Internalized platelets were observed in >50% of HepG2 cells. Platelet internalization in hepatocytes was also demonstrated in mice that underwent a 70% partial hepatectomy. One hour after hepatectomy, platelets were found within hepatocytes (Figure 2E).
Isolated platelets are internalized by hepatocytes. (A) Activated platelets (green) were labeled with CellTracker green CMFDA and incubated under serum-free conditions with HepG2 cells. Fluorescent images were taken after 5 minutes, 30 minutes, and 1 hour. HepG2 cells were stained for actin (red), and nuclei were stained with DAPI (blue). Original magnification ×200. Scale bar, 20µm. (B) (i) High magnification fluorescence image of single platelets (arrows) attached to the HepG2 cell membrane after 30 minutes of coculturing. Original magnification ×400. Scale bar, 10 µm. (ii) Electron micrograph of a single platelet attached to a HepG2 membrane. Original magnification ×9700; scale bar, 1 µm. (C) Confocal microscopy image of HepG2 cells that have been exposed to platelets. Images were captured after 1-hour incubation with CMFDA-labeled platelets (green). The HepG2 cells were stained for actin (red). The image shows a slice from the middle of the confocal stack. Original magnification ∼400. Scale bar, 20 μm. (D) Transmission electron microscopy imaging of hepatocytes that have been exposed to platelets. (i) Lower-magnification (×9700) image of a group of HepG2 cells showing platelets within hepatocytes indicated by arrows. Scale bar, 5 µm. (ii) High-magnification image of the same region (×24 500) shows the internalized platelet (*) surrounded by endoplasmic reticulum and located close to the nucleus (#). The arrow indicates the plasma membrane of the HepG2 cell. Scale bar, 1 µm. Images are representative of at least 3 independent experiments. (E) Transmission electron microscopy imaging of mouse liver 1 hour after hepatectomy. (i) A section of liver tissue taken from a mouse that underwent a 70% hepatectomy. The arrow indicates a platelet within a hepatocyte. Original magnification ×5800. Scale bar, 10 µm. (ii) High-magnification image of the same region (×33 000) shows the internalized platelet by the hepatocyte. Scale bar, 1 µm.
Isolated platelets are internalized by hepatocytes. (A) Activated platelets (green) were labeled with CellTracker green CMFDA and incubated under serum-free conditions with HepG2 cells. Fluorescent images were taken after 5 minutes, 30 minutes, and 1 hour. HepG2 cells were stained for actin (red), and nuclei were stained with DAPI (blue). Original magnification ×200. Scale bar, 20µm. (B) (i) High magnification fluorescence image of single platelets (arrows) attached to the HepG2 cell membrane after 30 minutes of coculturing. Original magnification ×400. Scale bar, 10 µm. (ii) Electron micrograph of a single platelet attached to a HepG2 membrane. Original magnification ×9700; scale bar, 1 µm. (C) Confocal microscopy image of HepG2 cells that have been exposed to platelets. Images were captured after 1-hour incubation with CMFDA-labeled platelets (green). The HepG2 cells were stained for actin (red). The image shows a slice from the middle of the confocal stack. Original magnification ∼400. Scale bar, 20 μm. (D) Transmission electron microscopy imaging of hepatocytes that have been exposed to platelets. (i) Lower-magnification (×9700) image of a group of HepG2 cells showing platelets within hepatocytes indicated by arrows. Scale bar, 5 µm. (ii) High-magnification image of the same region (×24 500) shows the internalized platelet (*) surrounded by endoplasmic reticulum and located close to the nucleus (#). The arrow indicates the plasma membrane of the HepG2 cell. Scale bar, 1 µm. Images are representative of at least 3 independent experiments. (E) Transmission electron microscopy imaging of mouse liver 1 hour after hepatectomy. (i) A section of liver tissue taken from a mouse that underwent a 70% hepatectomy. The arrow indicates a platelet within a hepatocyte. Original magnification ×5800. Scale bar, 10 µm. (ii) High-magnification image of the same region (×33 000) shows the internalized platelet by the hepatocyte. Scale bar, 1 µm.
Inhibition of platelet uptake reduces hepatocyte proliferation
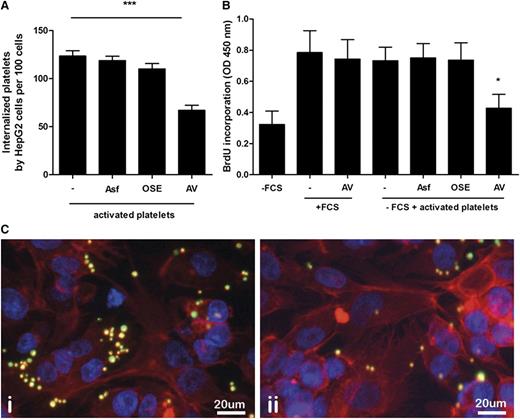
It has been well established that clustering of platelet glycoprotein Ibα is responsible for clearance of chilled platelets by hepatocytes in vivo and that this process is mediated by the hepatocyte Ashwell-Morell receptor.27 Additionally, the Ashwell-Morell receptor is responsible for clearance of human platelets by porcine liver endothelial cells in a xenotransplantation context.28 Based on these data, we tested the involvement of platelet glycoprotein Ibα and the hepatocyte Ashwell-Morell receptor in platelet internalization by HepG2 cells. Treatment with OSE, which removes glycoprotein Ibα from the platelet surface, or Asf, a competitive inhibitor of the Ashwell-Morell receptor, did not reduce platelet uptake by HepG2 cells (Figure 3A). In contrast, incubation of activated platelets with Annexin A5 prior to 2 hours of coculturing with HepG2 cells resulted in a significant reduction of platelet internalization by ∼65% (Figure 3A).
Annexin A5 inhibits platelet uptake and platelet-mediated hepatocyte proliferation. (A) Activated platelets were added to HepG2 cells in presence of Annexin A5 (AV), OSE, or asialofetuin (Asf). Internalized platelets were quantified based on fluorescence microscopy. Platelets were manually counted in at least 5 high-power fields and expressed as number of platelets/100 cells. ***P < .001. Data represent the mean of 3 independent triplicate experiments. Error bars indicate SD. (B) Activated platelets were added to HepG2 cells in the presence or absence of various inhibitors and incubated under serum-free conditions. After 48 hours, the cell proliferation rate was estimated by quantifying BrdU incorporation. Control groups represent HepG2 cells cultured in the presence or absence of FCS. In addition, the effect of Annexin A5 on BrdU incorporation in the absence of platelets, but in the presence of FCS, is shown. *P < .05 compared with −FCS. Data represent the mean of 3 independent triplicate experiments. Error bars indicate SD. (C) Representative fluorescence microscopy image from activated platelets incubated with HepG2 cells in the absence (i) or presence (ii) of Annexin A5 for 2 hours. HepG2 cells were stained for actin (red). Platelets were labeled with CMFDA (green). Original magnification ×200. Scale bar, 20 µm.
Annexin A5 inhibits platelet uptake and platelet-mediated hepatocyte proliferation. (A) Activated platelets were added to HepG2 cells in presence of Annexin A5 (AV), OSE, or asialofetuin (Asf). Internalized platelets were quantified based on fluorescence microscopy. Platelets were manually counted in at least 5 high-power fields and expressed as number of platelets/100 cells. ***P < .001. Data represent the mean of 3 independent triplicate experiments. Error bars indicate SD. (B) Activated platelets were added to HepG2 cells in the presence or absence of various inhibitors and incubated under serum-free conditions. After 48 hours, the cell proliferation rate was estimated by quantifying BrdU incorporation. Control groups represent HepG2 cells cultured in the presence or absence of FCS. In addition, the effect of Annexin A5 on BrdU incorporation in the absence of platelets, but in the presence of FCS, is shown. *P < .05 compared with −FCS. Data represent the mean of 3 independent triplicate experiments. Error bars indicate SD. (C) Representative fluorescence microscopy image from activated platelets incubated with HepG2 cells in the absence (i) or presence (ii) of Annexin A5 for 2 hours. HepG2 cells were stained for actin (red). Platelets were labeled with CMFDA (green). Original magnification ×200. Scale bar, 20 µm.
We next investigated whether platelet uptake is required for platelet-mediated HepG2 cell proliferation. Annexin A5–treated platelets were cocultured for 48 hours with HepG2 cells, and cell proliferation was estimated by a BrdU incorporation assay. Annexin A5–treated activated platelets showed a significantly reduced proliferation of HepG2 cells compared with untreated platelets (Figure 3B-C). Treatment with OSE or Asf did not reduce HepG2 cell proliferation (Figure 3B).
Transfer of RNA from PLPs following internalization by hepatocytes
We next used PLPs generated from the megakaryoblastic cell line MEG-01 to further assess the mechanism behind platelet-induced hepatocyte proliferation. PLPs had an identical potency to stimulate HepG2 proliferation compared with isolated platelets (Figure 4A). CFMDA-labeled PLPs were also internalized by HepG2 cells and once internalized were located in the perinuclear region (Figure 4B). To assess potential transfer of platelet RNA to the hepatocyte following uptake of platelets, we labeled RNA in MEG-01 cells by incorporation of EU and generated PLPs from these labeled megakaryocytes. EU-modified PLPs were isolated 48 hours after rTPO stimulation from differentiated MEG-01 cells and analyzed by flow cytometry using a fluorescently labeled probe that specifically binds EU. Approximately 37% of the PLPs were positive for EU (Figure 4C). Next, EU-containing PLPs were cocultured with HepG2 cells and assessed by confocal microscopy. EU-positive PLPs were observed in the perinuclear region of HepG2 cells. EU staining was predominantly present in structures resembling PLPs. Importantly, EU accumulated over time in the cytoplasm of the recipient HepG2 cell (Figure 4D), indicative of transfer of PLP-derived RNA to HepG2 cells.
PLPs stimulate proliferation of and are internalized by HepG2 cells. (A) Activated platelets or PLPs were added to HepG2 cells and incubated under serum-free conditions. After 48 hours, cell proliferation rate was estimated by quantifying BrdU incorporation. *P < .05. Data represent the mean of 3 independent triplicate experiments. Error bars indicate SD. (B) PLPs (green) were labeled with CMFDA and were incubated under serum-free conditions for 4 hours with HepG2 cells. Original magnification ×400. Scale bar, 10 µm. (C) Quantification of EU incorporation into PLP RNA by flow cytometry. PLPs were generated from MEG01 cells that had been exposed to EU. EU in PLPs was visualized using Click-iT RNA Alexa Fluor 488. Histogram plot overlay shows control PLPs (red line) and PLPs generated from EU-treated MEG01 cells (blue line). (D) EU-labeled PLPs were incubated under serum-free conditions for 2, 4, or 8 hours with HepG2 cells. Dashed lines represent the HepG2 cell membrane. Structures resembling PLPs (arrows) as well as parts of the cytoplasm of the HepG2 cell are EU positive (green, arrowheads). Original magnification ×400. Scale bar, 10 µm.
PLPs stimulate proliferation of and are internalized by HepG2 cells. (A) Activated platelets or PLPs were added to HepG2 cells and incubated under serum-free conditions. After 48 hours, cell proliferation rate was estimated by quantifying BrdU incorporation. *P < .05. Data represent the mean of 3 independent triplicate experiments. Error bars indicate SD. (B) PLPs (green) were labeled with CMFDA and were incubated under serum-free conditions for 4 hours with HepG2 cells. Original magnification ×400. Scale bar, 10 µm. (C) Quantification of EU incorporation into PLP RNA by flow cytometry. PLPs were generated from MEG01 cells that had been exposed to EU. EU in PLPs was visualized using Click-iT RNA Alexa Fluor 488. Histogram plot overlay shows control PLPs (red line) and PLPs generated from EU-treated MEG01 cells (blue line). (D) EU-labeled PLPs were incubated under serum-free conditions for 2, 4, or 8 hours with HepG2 cells. Dashed lines represent the HepG2 cell membrane. Structures resembling PLPs (arrows) as well as parts of the cytoplasm of the HepG2 cell are EU positive (green, arrowheads). Original magnification ×400. Scale bar, 10 µm.
RNA from PLPs is translated by hepatocytes following PLP-to-hepatocyte RNA transfer
To test whether RNA transferred by the platelet to the HepG2 cell may be translated to protein by the recipient cell, we transfected MEG-01 cells with a GFP-tagged actin mRNA construct. By flow cytometry and fluorescence microscopy, we demonstrated expression of actin-GFP protein in MEG-01 cells (Figure 5A-B). Approximately 26% of all PLPs generated from these transfected MEG01 cells were GFP positive (Figure 5B). Incubation of HepG2 cells with these GFP-tagged actin-mRNA–containing PLPs resulted in GFP expression throughout the hepatocyte cytoskeleton, suggesting translation of PLP-derived mRNA by the hepatocyte (Figure 5C). To confirm that the GFP-tagged actin is synthesized by the hepatocyte from platelet mRNA, we quantified GPF protein content of hepatocytes over time. As shown in Figure 5D, actin-GFP protein increased over time in HepG2 cells coincubated with PLPs. Importantly, the GFP content of PLPs cultured in the absence of HepG2 cells did not increase over time, indicating that the GFP present in the PLPs was already synthesized by the megakaryocyte. Subsequently, we treated actin-GFP–containing PLPs with RNA-degrading enzymes. GFP protein production by these RNase-treated PLPs was almost fully blunted, confirming that HepG2 cells translate the actin-GFP mRNA transferred by PLPs to protein.
Actin-GFP mRNA from PLPs is transferred to and translated by HepG2 cells. (A) Representative fluorescence microscopy image from actin-GFP–transfected MEG-01 cells 48 hours after transfection. Shown is a bright-field image combined with actin-GFP signal. Original magnification ×100. Scale bar, 25 µm. (B) Representative flow cytometry analysis of isolated PLPs from actin-GFP–transfected MEG-01 cells. Histogram plot of actin-GFP-expressing MEG-01 cells (green line) compared with control MEG-01 cells (black line) (left). Histogram plot of PLPs derived from actin-GFP–expressing MEG-01 cells (green line) compared with control PLPs (black line) (right). (C) PLPs from actin-GFP–transfected MEG-01 cells (green, indicated by arrowheads) were cocultured with HepG2 cells for 4 hours. Internalization was assessed by confocal microscopy. (i) Total actin inside the HepG2 cell is stained in red. (ii) Actin-GFP from PLPs (green) was localized throughout the cytoplasm of the HepG2 cell. (iii) Total actin in HepG2 cells that were not treated with PLPs is shown for comparison. Original magnification ×400. Scale bar, 5 µm. (D) Actin-GFP–expressing PLPs were cultured alone or in combination with HepG2 cells for up to 48 hours. PLPs that were cocultured with HepG2 cells were pretreated with 100 U/ml RNaseA or vehicle prior to addition to the HepG2 cells. Actin-GFP expression was quantified at the indicated time points. *P < .05. Data represent the mean of 3 independent triplicate experiments. Error bars indicate SD. (E) Activated platelets treated with vehicle or different concentrations of RNaseA were added to HepG2 cells and incubated under serum-free conditions. After 48 hours, the cell proliferation rate was estimated by quantifying BrdU incorporation. Control groups represent HepG2 cells cultured in the presence or absence of FCS. *P < .05. Data represent the mean of 3 independent triplicate experiments. Error bars indicate SD.
Actin-GFP mRNA from PLPs is transferred to and translated by HepG2 cells. (A) Representative fluorescence microscopy image from actin-GFP–transfected MEG-01 cells 48 hours after transfection. Shown is a bright-field image combined with actin-GFP signal. Original magnification ×100. Scale bar, 25 µm. (B) Representative flow cytometry analysis of isolated PLPs from actin-GFP–transfected MEG-01 cells. Histogram plot of actin-GFP-expressing MEG-01 cells (green line) compared with control MEG-01 cells (black line) (left). Histogram plot of PLPs derived from actin-GFP–expressing MEG-01 cells (green line) compared with control PLPs (black line) (right). (C) PLPs from actin-GFP–transfected MEG-01 cells (green, indicated by arrowheads) were cocultured with HepG2 cells for 4 hours. Internalization was assessed by confocal microscopy. (i) Total actin inside the HepG2 cell is stained in red. (ii) Actin-GFP from PLPs (green) was localized throughout the cytoplasm of the HepG2 cell. (iii) Total actin in HepG2 cells that were not treated with PLPs is shown for comparison. Original magnification ×400. Scale bar, 5 µm. (D) Actin-GFP–expressing PLPs were cultured alone or in combination with HepG2 cells for up to 48 hours. PLPs that were cocultured with HepG2 cells were pretreated with 100 U/ml RNaseA or vehicle prior to addition to the HepG2 cells. Actin-GFP expression was quantified at the indicated time points. *P < .05. Data represent the mean of 3 independent triplicate experiments. Error bars indicate SD. (E) Activated platelets treated with vehicle or different concentrations of RNaseA were added to HepG2 cells and incubated under serum-free conditions. After 48 hours, the cell proliferation rate was estimated by quantifying BrdU incorporation. Control groups represent HepG2 cells cultured in the presence or absence of FCS. *P < .05. Data represent the mean of 3 independent triplicate experiments. Error bars indicate SD.
Platelet RNA plays a critical role in platelet-mediated stimulation of hepatocyte proliferation
To test whether transfer of platelet RNA to the HepG2 cell is required for platelet-mediated stimulation of HepG2 cell proliferation, we performed proliferation experiments in which we compared proliferative activity of intact and RNA-depleted platelets. Treatment of platelets with an RNA-degrading enzyme substantially and significantly decreased platelet-mediated stimulation of HepG2 proliferation (Figure 5E).
Discussion
This study shows that platelet-mediated stimulation of HepG2 cell proliferation requires platelet internalization by hepatocytes. Following this internalization, platelets transfer RNA to the hepatocyte, and we demonstrated protein synthesis from platelet-derived mRNA. Importantly, platelet RNA contributes substantially to platelet-mediated hepatocyte proliferation, suggesting that transfer of platelet RNA to the hepatocyte with subsequent protein synthesis by the recipient cell is key in this process. Because we also demonstrated platelet internalization by hepatocytes following a partial hepatectomy in mice, it appears plausible that functional platelet RNA transfer is also relevant for platelet-mediated liver regeneration.
The molecular mechanisms of platelet-mediated stimulation of liver regeneration are only poorly understood. Local release of growth factors such as PDGF, HGF, IGF, VEGF, and serotonin from platelet granules may explain the stimulatory effect of platelets on liver regeneration.6,8,12 Nevertheless, it has yet to be demonstrated that (1) local release of growth factors occurs within the liver parenchyma and (2) growth factor release from platelets is the major mechanism responsible for platelet-mediated liver regeneration. Our results indicate that transfer of platelet RNA to the hepatocyte cytoplasm may be a crucial mechanism for platelet-mediated liver regeneration in addition to release of growth factor proteins stored in platelet granules. As degradation of all platelet RNA did not completely reduce the proliferative effect on hepatocytes, it appears that both release of growth factors by the platelet and transfer of platelet RNA are important for platelet-mediated hepatocyte proliferation. Whether both mechanisms are also relevant for platelet-mediated liver regeneration in vivo remains to be established.
Platelets contain ∼8500 individual mRNA species, and it has been convincingly demonstrated that several of those mRNAs can be translated by the platelet itself into protein, as the platelet contains the full machinery required for protein synthesis.14,16,29 In line with previously published data, we demonstrate that this platelet mRNA is transferred to and translated by nucleated cells.17 This “parasitic protein synthesis” may be much more efficient compared with protein synthesis by the platelet itself. Platelets also contain ∼500 miRNA species, and transfer of regulatory RNAs in addition to or instead of transfer of coding RNA could also be required for platelet-mediated hepatocyte proliferation.19 We demonstrate that transfer of platelet RNA to the nucleated hepatocyte has direct effects on the functional properties of the recipient cell, and it is tempting to speculate that transfer of platelet RNA to the recipient cell is directly responsible for the increased cellular proliferation. Which of the 8500 platelet mRNA or ∼500 miRNA species are required for platelet-mediated hepatocyte proliferation remains to be determined but may include mRNAs encoding growth factors, transcription factors, or cell-cycle genes and/or regulatory RNAs.
It has been previously shown that transfer of mRNA and miRNA between 2 types of nucleated cells occurs via vesicular transport.30-32 Importantly, mRNA transfer between 2 nucleated cells has functional consequences for the recipient cell. Moreover, it has been demonstrated that microvesicles derived from human liver stem cells were internalized by hepatocytes, resulting in transfer of mRNA. This vesicular RNA supported hepatocyte proliferation and induced apoptosis resistance.33
Given the small size of the anucleate platelet, platelet RNA transfer to nucleated cells resembles the RNA transport via microvesicles between 2 nucleated cells.32,34 Because vesicular RNA transport between 2 nucleated cells appears to be a common and widespread phenomenon, it is not unlikely that transfer of platelet RNA to nucleated cells with resulting biological changes in the recipient cell is also a common physiological or pathophysiological phenomenon. Interestingly, platelet-derived microparticles had no effect on hepatocyte proliferation, suggesting that platelet-specific functions are required for this platelet to hepatocyte communication.
It has now been well established that platelets have biological functions that by far exceed their well-recognized role in thrombosis and hemostasis.2-4 Transfer of platelet RNA to nucleated cells resulting in parasitic protein synthesis with consequent changes in the recipient cell may be relevant in the role of platelets in processes such as inflammation, angiogenesis, and repair of tissue other than the liver.
To our knowledge, we are the first to show that platelets that are taken up by cells preferentially accumulate at the nucleus. This peculiar phenomenon may be required for hepatocyte proliferation in a mechanism involving transfer of platelet pre-mRNA to the hepatocyte nucleus with subsequent nuclear splicing, export of mature mRNA to the cytosol, and eventually protein synthesis. Earlier studies on platelet uptake by hepatocytes were performed in the context of removal of aged or dysfunctional platelets from circulation.26,35 Platelet uptake by hepatocytes in this context is assumed to result in platelet breakdown, presumably in lysosomes, although the fate of platelets after uptake by hepatocytes has not been studied in this context. Platelet clearance by hepatocytes has been shown to occur in a process dependent on platelet glycoprotein Ib and the hepatocyte Ashwell-Morell receptor.26,36 The platelet uptake described in our studies was independent of these receptors. Rather, platelets were taken up in a process dependent on negatively charged phospholipids, which may explain why activated platelets are better stimulators of HepG2 proliferation compared with resting platelets. The trafficking of the platelet toward the hepatocyte nucleus following platelet internalization appears to be a process that is distinct from platelet uptake in the context of platelet clearance. The mechanisms responsible for platelet uptake and translocation to the nucleus remain unknown but may resemble translocation of plasma membrane proteins such as epidermal growth factor receptors, the insulin receptor, and the HGF receptor cMet to the nucleus of the hepatocyte.37-39
In conclusion, our combined results demonstrate that platelets stimulate hepatocyte proliferation in a mechanism that is, at least in part, dependent on platelet internalization by hepatocytes followed by transfer of RNA stored in the anucleate platelet. Although in vivo confirmation of this mechanism is required, these studies provide fundamentally new insights in the stimulatory effects of platelets on liver regeneration. In addition, platelet communication with nucleated cells by the transfer of RNA may be relevant in other processes in which platelets play key modulatory roles.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Susanne Veldhuis for expert technical assistance with the animal experiment.
This work was supported in part by a grant from The Netherlands Organisation for Scientific Research (VIDI, 917.11.304) (T.L.).
Authorship
Contribution: M.K., G.K., and J.A. performed experiments and interpreted data; B.N.G.G. supervised microscopy experiments and interpreted data; R.J.P. interpreted data; T.L. designed and supervised the study, interpreted data, and obtained funding; M.K. and T.L. wrote the manuscript; and all authors revised and approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ton Lisman, Surgical Research Laboratory, Department of Surgery BA44, University Medical Center Groningen, Hanzeplein 1, 9713 GZ Groningen, The Netherlands; e-mail: j.a.lisman@umcg.nl.