Key Points
CD19+CD27+ memory B cells are detectable at supranormal frequencies in patients with high-level EBV DNAemia following allogeneic HSCT.
These memory B cells are frequently positive for EBV genomes and bear many of the hallmarks of lymphoblastoid transformation.
Abstract
Allogeneic stem cell transplantation (allo-HSCT) provides a unique opportunity to track Epstein-Barr virus (EBV) infection in the context of the reconstituting B-cell system. Although many allo-HSCT recipients maintain low or undetectable levels of EBV DNA posttransplant, a significant proportion exhibit elevated and rapidly increasing EBV loads which, if left untreated, may lead to potentially fatal EBV-associated posttransplant lymphoproliferative disease. Intriguingly, this high-level EBV reactivation typically arises in the first 3 months posttransplant, at a time when the peripheral blood contains low numbers of CD27+ memory cells which are the site of EBV persistence in healthy immunocompetent donors. To investigate this apparent paradox, we prospectively monitored EBV levels and B-cell reconstitution in a cohort of allo-HSCT patients for up to 12 months posttransplant. In patients with low or undetectable levels of EBV, the circulating B-cell pool consisted predominantly of transitional and naive cells, with a marked deficiency of CD27+ memory cells which lasted >12 months. However, among patients with high EBV loads, there was a significant increase in both the proportion and number of CD27+ memory B cells. Analysis of sorted CD27+ memory B cells from these patients revealed that this population was preferentially infected with EBV, expressed EBV latent transcripts associated with B-cell growth transformation, had a plasmablastic phenotype, and frequently expressed the proliferation marker Ki-67. These findings suggest that high-level EBV reactivation following allo-HSCT may drive the expansion of latently infected CD27+ B lymphoblasts in the peripheral blood.
Introduction
Epstein-Barr virus (EBV) is a widespread B-lymphotropic gammaherpesvirus with potent B-cell growth transforming activity. Following primary infection, the virus replicates in the oropharynx while establishing latency in a small number of infected memory B lymphocytes.1 In healthy individuals, this lifelong viral persistence is usually asymptomatic, as the proliferation of EBV-infected B cells is strictly controlled by host T-cell immunity.2 However, in immunocompromised individuals, EBV can drive the opportunistic outgrowth of virus-transformed B cells which may subsequently develop into lymphoproliferative lesions.3,4 For instance, patients undergoing allogeneic hematopoietic stem cell transplantation (allo-HSCT), an intervention used to treat a wide range of hematologic conditions, remain profoundly T-cell compromised for many months posttransplant. Consequently a significant proportion of allo-HSCT patients develop high levels of circulating EBV DNA, referred to as EBV reactivation or DNAemia.5-11 These viral reactivations usually occur within the first few months posttransplant11-17 and, if left untreated, can progress to life-threatening posttransplant lymphoproliferative disease (PTLD). Accordingly, most transplant centers routinely monitor the levels of EBV DNA in the blood of allo-HSCT recipients for several months after transplant and preemptively administer rituximab, an anti-CD20 monoclonal antibody, to those individuals who exhibit rapidly increasing viral loads.
Although posttransplant monitoring has led to an improvement in the early detection of patients at risk of developing PTLD, the pathophysiology of EBV reactivation in the context of allo-HSCT remains poorly understood. Given that memory B cells are the normal reservoir of EBV persistence in immunocompetent individuals,18-20 we were particularly intrigued by existing reports in the literature that EBV reactivation following allo-HSCT usually occurs at a time when the newly reconstituting B-cell system consists predominantly of transitional and naive B cells.21-25 To investigate this apparent paradox, here we have explored the relationship between immune reconstitution and EBV reactivation in a cohort of allo-HSCT recipients and ask whether the well-documented pattern of immune reconstitution26-28 following allo-HSCT is perturbed in patients with high-level EBV reactivation. We have also characterized the phenotype of EBV-infected cells in patients with high-level EBV reactivation and ask whether, in this situation, EBV can colonize the numerically dominant transitional and naive B cells rather than memory B cells.
Patients, materials, and methods
See supplemental Methods (available on the Blood Web site) for additional materials and methods.
Patients and control donors
Blood samples and clinical data were collected from patients undergoing T-cell–deplete allo-HSCT at University Hospital Birmingham (Birmingham, United Kingdom) between May 2009 and September 2012. Control blood samples were obtained from healthy laboratory donors. The study was approved by the National Research Ethics Service (REC references 05/Q2707/148 and 14/WM/0001) and participants gave written informed consent in accordance with the Declaration of Helsinki.
All but 1 patient received reduced-intensity conditioning comprising fludarabine (30 mg/m2 daily for 5 days) and melphalan (140 mg/m2 for 1 day). A single patient received myeloablative conditioning comprising cyclophosphamide (60 mg/kg daily for 2 days) and total body irradiation (14.4 Gy in 8 fractions). All patients received a peripheral blood stem cell graft from either a human leukocyte antigen matched sibling or unrelated donor. For graft-versus-host disease (GVHD) prophylaxis, all patients received alemtuzumab (Campath) 10 mg daily IV for 5 days prior to transplant and ciclosporin; the patient who underwent myeloablative conditioning also received methotrexate (8 mg/m2 on alternate days for 4 days). All patients received aciclovir for a minimum of 3 months posttransplant.
Recipient whole-blood samples were subjected to routine clinical monitoring of EBV DNA with testing performed every 1 to 2 weeks for the first 6 months posttransplant and intermittently thereafter (Health Protection Agency Laboratory, Heartlands Hospital, United Kingdom). Subsequently, 13 consecutive allo-HSCT patients were identified with high-level EBV reactivation (defined as any EBV load over 2 × 104 genomes per mL; Table 1). These “high EBV” patients provided additional blood samples for research purposes at, or close to, the time of peak reactivation prior to starting preemptive rituximab treatment. Blood samples were also collected at around 3, 6, and 12 months posttransplant from a control “no/low EBV” group of 16 age- and sex-matched allo-HSCT patients with either undetectable or low-level EBV reactivation (defined as <5000 EBV genomes per mL). Patient peripheral blood mononuclear cells (PBMCs) were isolated using density gradient centrifugation and cryopreserved for later analysis.
Patient characteristics
| Patient . | Age, y . | Sex . | Diagnosis . | Donor . | Regimen . | Acute GVHD grade . | Peak EBV, copies per mL* . | PTLD . |
|---|---|---|---|---|---|---|---|---|
| High EBV | ||||||||
| 1 | 34 | M | AML | MUD | Cy TBI C | II+ | 4.35 × 106 | + |
| 2 | 57 | M | AML | MUD | FMC | II+ | 2.16 × 105 | |
| 3 | 57 | M | AML | MUD | FMC | I | 1.60 × 105 | |
| 4 | 55 | M | AML | MUD | FMC | II+ | 3.31 × 104 | |
| 5 | 64 | M | MDS | MUD | FMC | II+ | 1.81 × 106 | + |
| 6 | 51 | M | AML | MUD | FMC | I | 1.41 × 105 | |
| 7 | 61 | M | AML | MUD | FMC | 9.73 × 104 | ||
| 8 | 48 | M | HL | MUD | FMC | II+ | 2.69 × 104 | |
| 9 | 57 | F | CLL | MUD | FMC | II+ | 1.54 × 106 | |
| 10 | 57 | M | AML | Sibling | FMC | I | 9.49 × 104 | + |
| 11 | 67 | F | MDS | MUD | FMC | 1.67 × 106 | ||
| 12 | 61 | M | AML | MUD | FMC | II+ | 5.36 × 105 | |
| 13 | 50 | F | AML | Sibling | FMC | I | 2.50 × 105 | |
| No/low EBV | ||||||||
| 14 | 55 | M | AML | MUD | FMC | 9.67 × 102 | ||
| 15 | 61 | M | AML | MUD | FMC | 1.80 × 103 | ||
| 16 | 65 | M | AML | Sibling | FMC | II+ | 3.20 × 103 | |
| 17 | 51 | F | ALL | Sibling | FMC | I | 3.65 × 103 | |
| 18 | 57 | M | MDS | MUD | FMC | I | <5 × 102 | |
| 19 | 54 | M | ALL | MUD | FMC | I | 3.29 × 103 | |
| 20 | 54 | F | HL | Sibling | FMC | II+ | 4.24 × 103 | |
| 21 | 58 | M | T-PLL | Sibling | FMC | 3.80 × 103 | ||
| 22 | 62 | M | AML | MUD | FMC | 1.34 × 103 | ||
| 23 | 64 | M | AML | MUD | FMC | I | 1.23 × 103 | |
| 24 | 70 | F | AML | MUD | FMC | <5 × 102 | ||
| 25 | 66 | M | AML | MUD | FMC | II+ | 1.37 × 103 | |
| 26 | 53 | M | AML | Sibling | FMC | <5 × 102 | ||
| 27 | 61 | M | MDS | Sibling | FMC | 5.50 × 103 | ||
| 28 | 44 | M | MF | MUD | FMC | <5 × 102 | ||
| 29 | 47 | F | AML | MUD | FMC | I | <5 × 102 |
| Patient . | Age, y . | Sex . | Diagnosis . | Donor . | Regimen . | Acute GVHD grade . | Peak EBV, copies per mL* . | PTLD . |
|---|---|---|---|---|---|---|---|---|
| High EBV | ||||||||
| 1 | 34 | M | AML | MUD | Cy TBI C | II+ | 4.35 × 106 | + |
| 2 | 57 | M | AML | MUD | FMC | II+ | 2.16 × 105 | |
| 3 | 57 | M | AML | MUD | FMC | I | 1.60 × 105 | |
| 4 | 55 | M | AML | MUD | FMC | II+ | 3.31 × 104 | |
| 5 | 64 | M | MDS | MUD | FMC | II+ | 1.81 × 106 | + |
| 6 | 51 | M | AML | MUD | FMC | I | 1.41 × 105 | |
| 7 | 61 | M | AML | MUD | FMC | 9.73 × 104 | ||
| 8 | 48 | M | HL | MUD | FMC | II+ | 2.69 × 104 | |
| 9 | 57 | F | CLL | MUD | FMC | II+ | 1.54 × 106 | |
| 10 | 57 | M | AML | Sibling | FMC | I | 9.49 × 104 | + |
| 11 | 67 | F | MDS | MUD | FMC | 1.67 × 106 | ||
| 12 | 61 | M | AML | MUD | FMC | II+ | 5.36 × 105 | |
| 13 | 50 | F | AML | Sibling | FMC | I | 2.50 × 105 | |
| No/low EBV | ||||||||
| 14 | 55 | M | AML | MUD | FMC | 9.67 × 102 | ||
| 15 | 61 | M | AML | MUD | FMC | 1.80 × 103 | ||
| 16 | 65 | M | AML | Sibling | FMC | II+ | 3.20 × 103 | |
| 17 | 51 | F | ALL | Sibling | FMC | I | 3.65 × 103 | |
| 18 | 57 | M | MDS | MUD | FMC | I | <5 × 102 | |
| 19 | 54 | M | ALL | MUD | FMC | I | 3.29 × 103 | |
| 20 | 54 | F | HL | Sibling | FMC | II+ | 4.24 × 103 | |
| 21 | 58 | M | T-PLL | Sibling | FMC | 3.80 × 103 | ||
| 22 | 62 | M | AML | MUD | FMC | 1.34 × 103 | ||
| 23 | 64 | M | AML | MUD | FMC | I | 1.23 × 103 | |
| 24 | 70 | F | AML | MUD | FMC | <5 × 102 | ||
| 25 | 66 | M | AML | MUD | FMC | II+ | 1.37 × 103 | |
| 26 | 53 | M | AML | Sibling | FMC | <5 × 102 | ||
| 27 | 61 | M | MDS | Sibling | FMC | 5.50 × 103 | ||
| 28 | 44 | M | MF | MUD | FMC | <5 × 102 | ||
| 29 | 47 | F | AML | MUD | FMC | I | <5 × 102 |
ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; C, Campath; CLL, chronic lymphocytic leukemia; Cy, cyclophosphamide; F, female; FMC, fludarabine, melphalan, and Campath; HL, Hodgkin lymphoma; M, male; MDS, myelodysplastic syndrome; MF, myelofibrosis; MUD, matched unrelated donor; T-PLL, T-cell prolymphocytic leukemia; TBI, total body irradiation.
Clinical EBV load measured as EBV DNA copies per mL whole blood.
Statistical analysis
Statistical comparisons between the high EBV group at the time of reactivation (median, 109 days posttransplant; interquartile range [IQR], 92-141 days) and the no/low EBV group using combined data from the 3 and 6 months posttransplant samples (median, 112 days; IQR, 84-169 days) were made using the Mann-Whitney U test. GraphPad was used to perform statistical analyses and generate graphical plots.
Results
Patient characteristics and kinetics of EBV reactivation
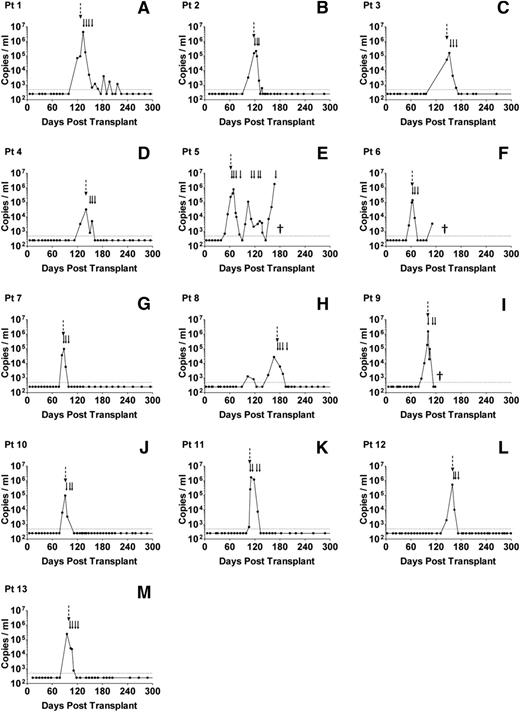
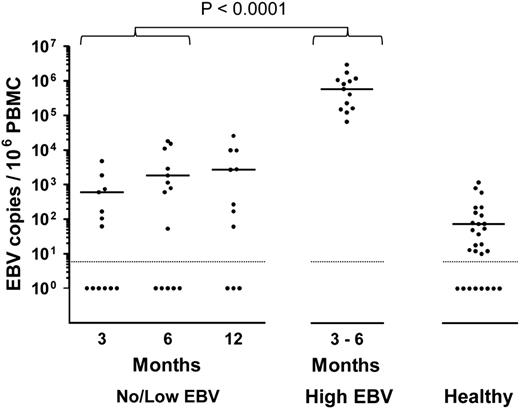
From routine clinical monitoring, we identified 13 allo-HSCT patients who developed high-level EBV reactivation during the first 6 months posttransplant (high EBV group) and 16 age- and sex-matched patients who either had undetectable or low viral loads at any time posttransplant (no/low EBV group). The clinical characteristics and viral loads of these 2 patient groups are summarized in Table 1. All 13 high EBV patients showed similar kinetics of reactivation (Figure 1), with a median interval from transplant to first EBV quantitative polymerase chain reaction (Q-PCR) positivity (>500 genomes per mL) of 99 days (IQR, 84-119 days). This was followed by a rapid expansion in circulating EBV DNA, with a median time from first Q-PCR positivity to high-level DNAemia of 7 days (IQR, 0-14 days) and a median peak viral load of 2.2 × 105 genomes per mL (Table 1). By contrast, 5 of 16 patients in the no/low EBV group remained Q-PCR–negative although the others showed much lower viral burdens with a median peak load of 1.8 × 103 genomes per mL (Table 1). Because measurement of EBV DNA in whole blood does not distinguish between cell-associated EBV genomes or viral DNA in plasma, we also quantified the number of EBV genomes in PBMC DNA isolated from the high EBV group at the time of peak EBV reactivation and from the no/low EBV patients bled at around 3, 6, and 12 months posttransplant. Matching for time posttransplant, there was a highly significant increase in cell-associated EBV DNA from the high EBV cohort at peak reactivation (Figure 2).
Serial analysis of EBV loads in patients with high-level EBV reactivation. (A-M) Routine Q-PCR monitoring of whole-blood samples, collected every 1 to 2 weeks for at least 6 months following allo-HSCT, was used to identify 13 patients with high-level EBV reactivation, defined as an EBV load >20 000 copies per mL blood. The dotted horizontal lines represent the limit of detection of the assay (500 EBV copies per mL); values below this were assigned a value of 250 copies per mL. The dashed vertical arrows indicate the time points at which samples were collected; the solid arrows indicate the time points of rituximab infusion. (E) Patient 5 died of PTLD, (F) patient 6 died of relapsed AML, and (I) patient 9 died of pneumonia (indicated by †).
Serial analysis of EBV loads in patients with high-level EBV reactivation. (A-M) Routine Q-PCR monitoring of whole-blood samples, collected every 1 to 2 weeks for at least 6 months following allo-HSCT, was used to identify 13 patients with high-level EBV reactivation, defined as an EBV load >20 000 copies per mL blood. The dotted horizontal lines represent the limit of detection of the assay (500 EBV copies per mL); values below this were assigned a value of 250 copies per mL. The dashed vertical arrows indicate the time points at which samples were collected; the solid arrows indicate the time points of rituximab infusion. (E) Patient 5 died of PTLD, (F) patient 6 died of relapsed AML, and (I) patient 9 died of pneumonia (indicated by †).
Cellular EBV loads in allo-HSCT patients. Cell-associated EBV loads were determined by Q-PCR screening of PBMC DNA from no/low EBV patients at around 3, 6, and 12 months posttransplant, and from high EBV patients at the time of peak EBV DNAemia. Viral loads from healthy control donors are included as a reference. The solid horizontal lines represent the medians of the positive values for each group. For the no/low EBV group, the median EBV loads from samples available at 3, 6, and 12 months posttransplant were 600, 1800, and 2700 EBV copies per 106 PBMCs, respectively. The median value for the high EBV group, collected at a median of 109 days posttransplant (IQR, 92-141 days), was 5.9 × 105 EBV copies per 106 PBMCs. Of the healthy controls with detectable EBV, the median viral load was 73 EBV copies per 106 PBMCs. For purposes of statistical analysis, comparison with the no/low EBV group was made with samples combined from the 3- and 6-month time points collected at a median of 112 days posttransplant (IQR, 84-169 days). EBV loads in the high EBV group were significantly higher than those of the no/low group (P < .0001). The dotted horizontal lines represent the limit of detection of the assay (10 EBV copies per 106 PBMCs) and samples with undetectable virus DNA were assigned a value of 1.
Cellular EBV loads in allo-HSCT patients. Cell-associated EBV loads were determined by Q-PCR screening of PBMC DNA from no/low EBV patients at around 3, 6, and 12 months posttransplant, and from high EBV patients at the time of peak EBV DNAemia. Viral loads from healthy control donors are included as a reference. The solid horizontal lines represent the medians of the positive values for each group. For the no/low EBV group, the median EBV loads from samples available at 3, 6, and 12 months posttransplant were 600, 1800, and 2700 EBV copies per 106 PBMCs, respectively. The median value for the high EBV group, collected at a median of 109 days posttransplant (IQR, 92-141 days), was 5.9 × 105 EBV copies per 106 PBMCs. Of the healthy controls with detectable EBV, the median viral load was 73 EBV copies per 106 PBMCs. For purposes of statistical analysis, comparison with the no/low EBV group was made with samples combined from the 3- and 6-month time points collected at a median of 112 days posttransplant (IQR, 84-169 days). EBV loads in the high EBV group were significantly higher than those of the no/low group (P < .0001). The dotted horizontal lines represent the limit of detection of the assay (10 EBV copies per 106 PBMCs) and samples with undetectable virus DNA were assigned a value of 1.
According to local clinical protocol, all patients with high-level EBV reactivation were preemptively treated with rituximab and assessed for possible PTLD. Patients 1, 5, and 10 were subsequently diagnosed with PTLD based on radiographic evidence of disease at 16, 9, and 1 day(s), respectively, after the onset of reactivation. Following rituximab infusion, 12 patients showed a complete response with lasting resolution of EBV DNAemia and disease where evident (Figure 1). Patient 5 died of progressive PTLD despite showing an initial reduction in EBV load following rituximab therapy (Figure 1).
Reconstitution of main lymphocyte subsets
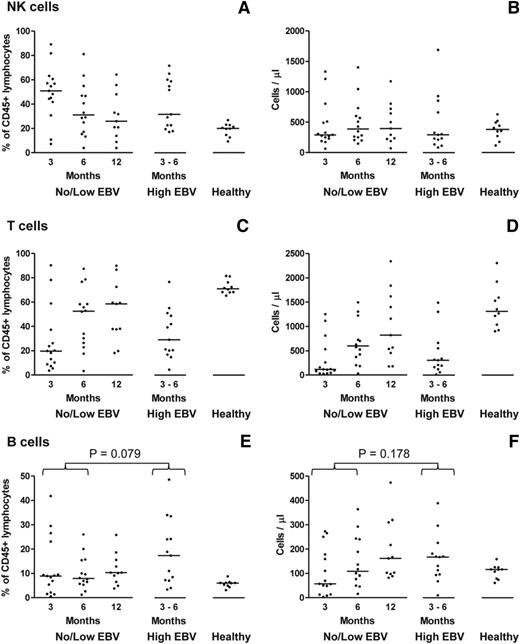
To characterize reconstitution of the main lymphocyte subsets following allo-HSCT, CD56+CD3− natural killer (NK) cells, CD3+ T cells, and CD19+ B cells were identified by flow cytometry and quantified both as a proportion of total CD45+ lymphocytes and as absolute numbers derived from the corresponding lymphocyte counts (Figure 3). The pattern of immune reconstitution among the no/low EBV patients at 3, 6, and 12 months posttransplant was in accord with earlier studies.26-28 Thus, there was marked numerical deficiency of all 3 lymphocyte subsets relative to healthy donors, with an initial predominance of NK cells that was gradually replaced by recovering T cells. Although the proportion of B cells in most no/low patients was normal (or supranormal) by 3 months posttransplant, the absolute number of B cells did not fully recover until 6 to 12 months (Figure 3). Matching for time posttransplant, a similar pattern was seen in the high EBV patients (Figure 3) indicating that high-level EBV reactivation did not significantly affect the reconstitution of the main lymphocyte subsets.
Characterization of immune reconstitution of the main lymphocyte subsets following allo-HSCT. (A-F) Multicolor flow cytometry was used to enumerate CD56+CD3− NK cells, CD3+ T cells, and CD19+ B cells in PBMC samples from allo-HSCT patients, either without (no/low EBV) or with (high EBV) high-level EBV reactivation, and from healthy controls. In each case, the left plot reports values as a percentage of CD45+ lymphocytes; the right plot shows the values as absolute cell numbers per microliter of blood, based on the corresponding lymphocyte counts. The solid horizontal lines represent the median values for each group. No significant differences were seen for both the NK-cell and T-cell subsets between the no/low EBV group at 3 and 6 months posttransplant and the high EBV group at the time of peak reactivation. The proportion of B cells in the high EBV group was greater than in the no/low EBV group with borderline significance (P = .079), but there was a nonsignificant increase (P = .178) in absolute B-cell numbers.
Characterization of immune reconstitution of the main lymphocyte subsets following allo-HSCT. (A-F) Multicolor flow cytometry was used to enumerate CD56+CD3− NK cells, CD3+ T cells, and CD19+ B cells in PBMC samples from allo-HSCT patients, either without (no/low EBV) or with (high EBV) high-level EBV reactivation, and from healthy controls. In each case, the left plot reports values as a percentage of CD45+ lymphocytes; the right plot shows the values as absolute cell numbers per microliter of blood, based on the corresponding lymphocyte counts. The solid horizontal lines represent the median values for each group. No significant differences were seen for both the NK-cell and T-cell subsets between the no/low EBV group at 3 and 6 months posttransplant and the high EBV group at the time of peak reactivation. The proportion of B cells in the high EBV group was greater than in the no/low EBV group with borderline significance (P = .079), but there was a nonsignificant increase (P = .178) in absolute B-cell numbers.
High-level EBV reactivation is associated with increased numbers of CD27+ B cells
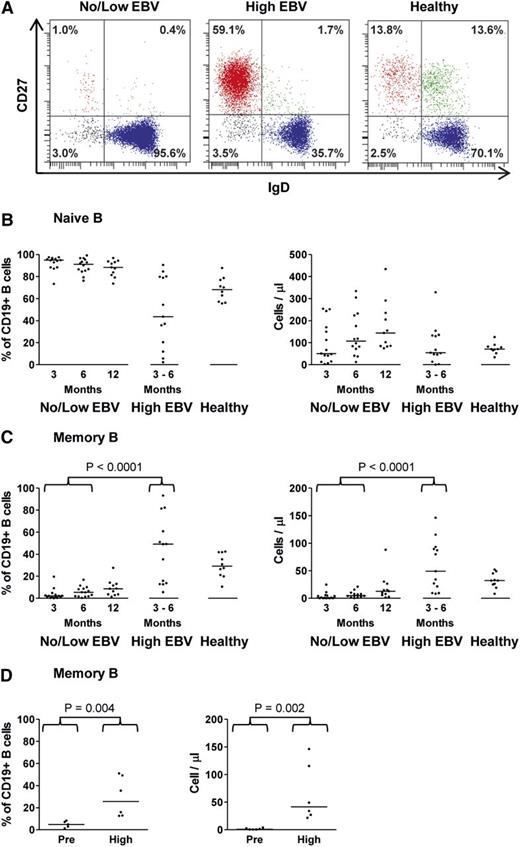
The detailed immunophenotype of the emerging B cells posttransplant was then compared between the no/low EBV and high EBV cohorts. B-cell reconstitution in the no/low EBV group was characterized by an initial predominance of CD27−IgD+ naive B cells, which lasted for >12 months posttransplant, and a marked delay in the recovery of CD27+ memory B cells (Figure 4A-C; supplemental Figure 1). As expected, naive B cells in such patients showed a mostly transitional cell phenotype (CD38hiCD24hiCD10+) that gradually declined over time (supplemental Figure 2).29 However, among the high EBV patients, we observed a significant increase in both the proportion and number of CD27+ B cells (Figure 4A-C; supplemental Figure 1). Notably, these expanded CD27+ B-cell populations were almost entirely IgD−, consistent with a class-switched memory B-cell phenotype (Figure 4A; supplemental Figure 1). Additional staining for surface κ and λ immunoglobulin light (IgL) chains revealed that the CD27+ B cells from the majority of high EBV patients had a polytypic IgL phenotype, although 4 patients showed some evidence of light-chain restriction (supplemental Figure 3).
Alterations in B-cell subset frequencies between no/low EBV and high EBV patients following allo-HSCT. (A) Representative staining of PBMCs to identify CD27−IgD+ naive, CD27+IgD− isotype-switched memory, CD27+IgD+ nonswitched memory, and CD27−IgD− B-cell populations in no/low EBV and high EBVallo-HSCT recipients, and in a healthy control. The no/low EBV patient was bled 98 days posttransplant when the EBV load was undetectable in whole blood. The high EBV patient was bled at 101 days posttransplant during high-level DNAemia (1.54 × 106 EBV copies per mL blood). (B) Enumeration of CD27−IgD+ naive B cells expressed as a percentage of CD45+ lymphocytes (left panel) or absolute numbers (right panel) in allo-HSCT patients and healthy controls. (C) Enumeration of CD27+IgD− memory B cells expressed as a percentage of CD45+ lymphocytes (left panel) or absolute numbers (right panel) in allo-HSCT patients and healthy controls. In panels B and C, the solid horizontal lines represent the median values for each group. Both the percentage (P < .0001) and number (P < .0001) of memory B cells were significantly higher in the high EBV group compared with the no/low EBV group. (D) Enumeration of CD19+CD27+IgD− memory B cells in PBMC samples prereactivation and at the point of high-level EBV DNAemia. There was a significant increase in both the proportion (P < .004) and absolute number (P < .002) of memory B cells during high-level EBV reactivation.
Alterations in B-cell subset frequencies between no/low EBV and high EBV patients following allo-HSCT. (A) Representative staining of PBMCs to identify CD27−IgD+ naive, CD27+IgD− isotype-switched memory, CD27+IgD+ nonswitched memory, and CD27−IgD− B-cell populations in no/low EBV and high EBVallo-HSCT recipients, and in a healthy control. The no/low EBV patient was bled 98 days posttransplant when the EBV load was undetectable in whole blood. The high EBV patient was bled at 101 days posttransplant during high-level DNAemia (1.54 × 106 EBV copies per mL blood). (B) Enumeration of CD27−IgD+ naive B cells expressed as a percentage of CD45+ lymphocytes (left panel) or absolute numbers (right panel) in allo-HSCT patients and healthy controls. (C) Enumeration of CD27+IgD− memory B cells expressed as a percentage of CD45+ lymphocytes (left panel) or absolute numbers (right panel) in allo-HSCT patients and healthy controls. In panels B and C, the solid horizontal lines represent the median values for each group. Both the percentage (P < .0001) and number (P < .0001) of memory B cells were significantly higher in the high EBV group compared with the no/low EBV group. (D) Enumeration of CD19+CD27+IgD− memory B cells in PBMC samples prereactivation and at the point of high-level EBV DNAemia. There was a significant increase in both the proportion (P < .004) and absolute number (P < .002) of memory B cells during high-level EBV reactivation.
De novo expansion of CD27+ B cells is coincident with high-level EBV DNAemia
We next asked whether the increased numbers of CD27+ B cells observed in patients with high-level reactivation existed prior to the onset of EBV DNAemia. Immunophenotyping of PBMCs available from 6 high EBV patients collected a median of 32 days (16-73 days) prior to peak EBV reactivation revealed that these earlier samples contained both a significantly lower proportion and frequency of CD27+ memory B cells (Figure 4D). Further analysis combining all available data from the high EBV patients at the time of reactivation and the no/low EBV patients at 3, 6, and 12 months posttransplant revealed no correlation between PBMC viral loads and the number of NK cells, T cells, or total B cells (supplemental Figure 4). However, there was a highly significant correlation between EBV load and the number of CD27+ B cells, supporting the view that memory B cells are the site of EBV persistence following allo-HSCT.
EBV selectively resides within the CD27+ memory B-cell population
To obtain direct evidence that EBV is present in CD27+ memory B cells following reactivation, PBMCs from 4 high EBV patients were sorted with a fluorescence-activated cell sorter (FACS) into CD19+CD27−IgD+ naive B, CD19+CD27+ memory B, and CD19− non-B-cell subsets (supplemental Figure 5) and then assayed for EBV DNA using Q-PCR (Figure 5A). In all 4 cases, EBV was highly enriched in CD27+ memory B cells but was almost absent from other lymphocyte populations. The extremely high viral loads seen in these memory B cells (median, 1.2 × 107 copies per 106 CD27+ B cells) also suggested either there was a high frequency of EBV infection or EBV replication was occurring in a proportion of cells. To distinguish between these 2 possibilities, we estimated the fraction of infected cells by assaying for viral DNA in single cells isolated by FACS. We first validated our approach by sorting individual cells from mixtures containing known ratios of EBV-positive Kem Burkitt lymphoma cells and an isogenic EBV-negative derivative and then screening for the presence of EBV DNA by PCR. Using mixtures containing 25%, 50%, and 100% EBV-positive cells, we observed EBV DNA signals in 15 of 48 (31%), 23 of 48 (52%), and 39 of 48 (81%) wells, respectively, values which closely approximated the expected results. Applying the same technique to naive and memory B cells sorted by FACS from 4 high load patients, naive B cells were consistently EBV-negative but viral DNA was detected in a high proportion (81%-92%) of memory B cells in all 4 cases (Figure 5B). Notably, the median copy number of 19 EBV genomes per infected memory B-cell (IQR, 10-29) is consistent with viral latency and is in marked contrast to the much higher viral loads seen in productively infected B cells (median load, 2.4 × 104 EBV copies per cell) included here as a comparator (Lytic Akata, Figure 5C). In summary, the relatively low viral load and narrow distribution of viral genome copy number strongly suggest that high-level EBV reactivation following allo-HSCT is due to an accumulation of latently infected CD27+ memory B cells.
EBV loads in different B-cell subsets in patients with high-level reactivation. (A) Cellular EBV loads determined by Q-PCR in bulk populations of unsorted total CD19+ B cells and CD19− non-B cells sorted by FACS, CD27−IgD+ naive B cells and CD27+ memory B cells from 4 patients with high-level EBV reactivation. (B) Proportion of EBV-positive CD27−IgD+ naive and CD27+ memory B cells in the same 4 patients as above, determined by single-cell Q-PCR for EBV DNA. Gray shading represents the fraction of EBV-positive cells in the sorted naive and memory B-cell populations for each patient. (C) Distribution of EBV genome copy number per infected CD27+ memory B cell. Values are only shown for EBV-positive cells, with the solid horizontal lines indicating the median value for each patient. As a comparator, we also determined the EBV genome copy number in EBV-positive AKBM Akata-GFP cells induced into lytic cycle (Lytic Akata). Following induction, cells that had entered virus lytic cycle were single-cell sorted on the basis of GFP expression before determining the genome load per cell by Q-PCR, as above. Although the majority of sorted GFP+ cells have in excess of 1000 EBV genomes per cell, note that a small proportion of GFP+ cells have much lower (latent) viral loads because viral replication is slightly delayed with respect to GFP expression. GFP, green fluorescent protein.
EBV loads in different B-cell subsets in patients with high-level reactivation. (A) Cellular EBV loads determined by Q-PCR in bulk populations of unsorted total CD19+ B cells and CD19− non-B cells sorted by FACS, CD27−IgD+ naive B cells and CD27+ memory B cells from 4 patients with high-level EBV reactivation. (B) Proportion of EBV-positive CD27−IgD+ naive and CD27+ memory B cells in the same 4 patients as above, determined by single-cell Q-PCR for EBV DNA. Gray shading represents the fraction of EBV-positive cells in the sorted naive and memory B-cell populations for each patient. (C) Distribution of EBV genome copy number per infected CD27+ memory B cell. Values are only shown for EBV-positive cells, with the solid horizontal lines indicating the median value for each patient. As a comparator, we also determined the EBV genome copy number in EBV-positive AKBM Akata-GFP cells induced into lytic cycle (Lytic Akata). Following induction, cells that had entered virus lytic cycle were single-cell sorted on the basis of GFP expression before determining the genome load per cell by Q-PCR, as above. Although the majority of sorted GFP+ cells have in excess of 1000 EBV genomes per cell, note that a small proportion of GFP+ cells have much lower (latent) viral loads because viral replication is slightly delayed with respect to GFP expression. GFP, green fluorescent protein.
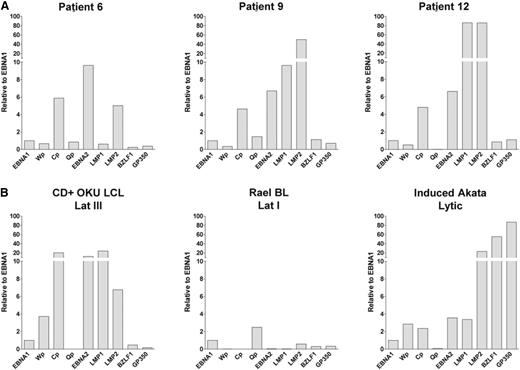
CD27+ memory B cells express EBV transcripts associated with growth transformation
EBV can adopt 1 of several forms of latent infection which can be distinguished by alternative patterns of virus gene expression and promoter usage.30 To characterize the viral gene expression profile in CD27+ memory B cells from high EBV patients, we used Q-PCR to quantify EBV transcripts encoding the latent genes EBNA1, EBNA2, LMP1, and LMP2, the immediate early gene BZLF1, and the late lytic gene GP350/BLLF1; in addition, we also screened for transcripts initiated from 3 latent promoters, Wp, Cp, and Qp (Figure 6A). After normalizing the data to EBNA1 expression, all 3 patients tested showed readily detectable levels of Cp-initiated transcripts (with accompanying lower levels of Wp activity) and high levels of EBNA2 and LMP2 messenger RNA, whereas 2 cases also showed lymphoblastoid cell line (LCL)-like levels of LMP1 expression. This pattern of viral gene expression closely resembles the latency III pattern seen in an EBV-transformed B LCL (CD+Oku LCL; Figure 6B) but is clearly different to the more restricted latency I pattern seen in a representative Burkitt lymphoma cell line Rael-BL (Figure 6B). Expression of the lytic cycle transcripts BZLF1 and GP350 was much lower, ranging from 0.5% to 2% of the values seen in productively infected BL cells (Lytic Akata, Figure 6B). Overall, these results confirm that CD27+ memory B cells in allo-HSCT patients with high-level DNAemia predominantly harbor EBV as a latent infection and suggest that a proportion of infected cells express viral transcripts associated with B-cell growth transformation.
EBV gene expression in CD27+ memory B cells in high load allo-HSCT patients. Absolute numbers of EBV transcripts were quantified using a Q-PCR array. All data were first normalized to cellular PGK1 expression and then expressed relative to the levels of EBNA1 transcript. (A) EBV gene expression in CD27+ memory B cells sorted by FACS from patients 6, 9, and 12. (B) EBV gene expression in EBV-positive reference cell lines. CD+Oku LCL is a latently infected EBV-transformed B lymphoblastoid cell line characterized by activity of the Wp- and Cp promoters, and expression of EBNA1, EBNA2, LMP1, and LMP2 latent genes (a form of infection termed latency III). Rael-BL is a latently infected Burkitt lymphoma cell line which expresses a single latent gene EBNA1 transcribed from an alternative promoter Qp. Because both CD+Oku LCL and Rael-BL are tightly latent with only low-level expression of lytic cycle transcripts (exemplified by the immediate early gene BZLF1 and the late gene gp350), we also included a third reference line, AKBM Akata-GFP cells induced into lytic cycle by IgG crosslinking, to illustrate the much higher levels (typically >100-fold) of viral lytic gene transcription in productively infected cells.
EBV gene expression in CD27+ memory B cells in high load allo-HSCT patients. Absolute numbers of EBV transcripts were quantified using a Q-PCR array. All data were first normalized to cellular PGK1 expression and then expressed relative to the levels of EBNA1 transcript. (A) EBV gene expression in CD27+ memory B cells sorted by FACS from patients 6, 9, and 12. (B) EBV gene expression in EBV-positive reference cell lines. CD+Oku LCL is a latently infected EBV-transformed B lymphoblastoid cell line characterized by activity of the Wp- and Cp promoters, and expression of EBNA1, EBNA2, LMP1, and LMP2 latent genes (a form of infection termed latency III). Rael-BL is a latently infected Burkitt lymphoma cell line which expresses a single latent gene EBNA1 transcribed from an alternative promoter Qp. Because both CD+Oku LCL and Rael-BL are tightly latent with only low-level expression of lytic cycle transcripts (exemplified by the immediate early gene BZLF1 and the late gene gp350), we also included a third reference line, AKBM Akata-GFP cells induced into lytic cycle by IgG crosslinking, to illustrate the much higher levels (typically >100-fold) of viral lytic gene transcription in productively infected cells.
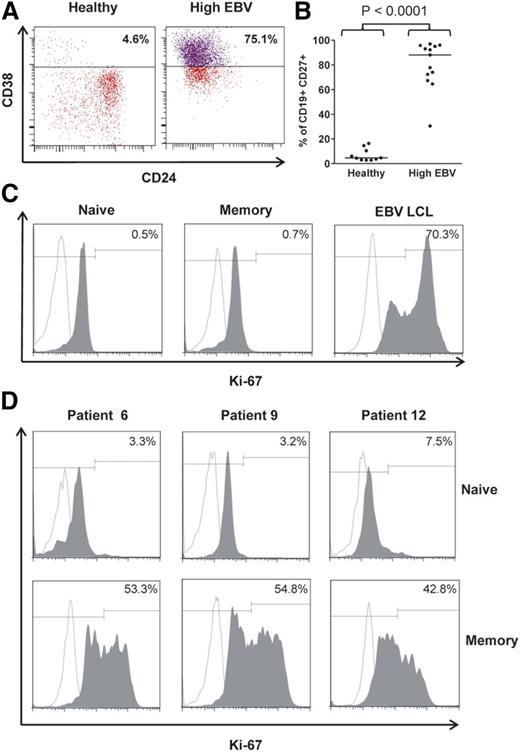
CD27+ memory B cells exhibit a proliferative phenotype
Given that CD27+ memory B cells in patients with high-level EBV reactivation express viral genes that can drive B-cell proliferation, the final experiments sought to characterize the activation and proliferation status of these CD27+ B cells in more detail. Flow cytometry revealed that a significant proportion of CD19+CD27+ B cells exhibited a CD24−CD38hi plasmablastic immunophenotype (Figure 7A). Importantly, this CD24−CD38hi population was significantly expanded in high EBV patients when compared with healthy controls (Figure 7B). We then compared the proliferation status of CD27+ B cells from allo-HSCT patients with healthy donor B cells by means of intracellular staining for Ki-67 expression. As expected, both naive and memory B cells from a healthy individual had a predominantly resting Ki-67− phenotype (Figure 7C), as did CD27−IgD+ naive B cells from 3 high EBV patients (Figure 7D). By contrast, CD27+ memory B cells from the same high EBV patients contained a substantial proportion (∼50% in each case) of Ki-67+ cells, indicative of cell proliferation (Figure 7C).
Proliferation status of CD27+ B cells in high load allo-HSCT patients. (A) Representative staining of PBMCs from a high EBV patient and a healthy control donor to identify CD38hi plasmablasts within the CD19+CD27+ memory B-cell population. (B) Proportion of CD19+CD27+ memory B cells exhibiting a CD38hi plasmablastic phenotype in high EBV patients and healthy controls. The proportion of plasmablasts in the high EBV group was highly significantly increased (P < .0001) compared with the healthy controls. (C) Intracellular staining for Ki-67 expression. Naive and memory B cells, sorted by FACS, from a healthy individual and a representative EBV-transformed LCL (included as a positive control) were stained with an anti-Ki-67 antibody (shaded histogram) or isotype control antibody (open histogram) followed by flow cytometric analysis. (D) Ki-67 expression in naive and memory B cells sorted by FACS from 3 allo-HSCT patients with high-level EBV reactivation.
Proliferation status of CD27+ B cells in high load allo-HSCT patients. (A) Representative staining of PBMCs from a high EBV patient and a healthy control donor to identify CD38hi plasmablasts within the CD19+CD27+ memory B-cell population. (B) Proportion of CD19+CD27+ memory B cells exhibiting a CD38hi plasmablastic phenotype in high EBV patients and healthy controls. The proportion of plasmablasts in the high EBV group was highly significantly increased (P < .0001) compared with the healthy controls. (C) Intracellular staining for Ki-67 expression. Naive and memory B cells, sorted by FACS, from a healthy individual and a representative EBV-transformed LCL (included as a positive control) were stained with an anti-Ki-67 antibody (shaded histogram) or isotype control antibody (open histogram) followed by flow cytometric analysis. (D) Ki-67 expression in naive and memory B cells sorted by FACS from 3 allo-HSCT patients with high-level EBV reactivation.
Discussion
Although EBV reactivation following allo-HSCT is an important clinical problem, it also provides a unique opportunity to study the biology of EBV infection in the context of newly emerging B- and T-cell systems. The present work was prompted by the apparent contradiction that EBV DNAemia usually occurs in the first few months posttransplant when there is a marked deficiency in circulating CD27+ memory B cells, the natural reservoir for EBV in immunocompetent individuals. Here, we sought to explore the pattern of immune reconstitution following allo-HSCT and, for the first time, characterize the detailed phenotype of EBV-infected cells in patients with high-level EBV reactivation. Notably, our approach differed from the previous studies in 2 important aspects. First, by prospectively monitoring EBV loads posttransplant, we were able to bleed patients at, or close to, their peak of EBV DNAemia. Second, we have compared the reconstitution of the main lymphocyte populations in patients with high-level reactivation (sampled at the time of peak viral load) and control patients with undetectable or low level EBV DNAemia. Importantly, among patients with high-level reactivation, we show that EBV maintains its selectivity for isotype-switched CD27+ memory B cells, but that high viral loads are associated with the early emergence of such cells. Several lines of evidence support the idea that this memory B-cell expansion is an EBV-driven event. Thus, further analysis of the CD27+ B-cell population from high load patients revealed that the vast majority of cells are virus-positive, the population expresses viral genes associated with B-cell growth transformation, and a significant proportion express Ki-67 consistent with cell proliferation.
The overall pattern of immune reconstitution following allo-HSCT has been well documented.26-28 In general, cells of the innate immune system including monocytes, macrophages, and NK cells are produced first and recover to normal frequencies within a few weeks posttransplant. By contrast, adaptive immunity returns much more slowly, with functional recovery of T and B lymphocytes taking up to 2 years.29,31 Immune reconstitution data from our no/low EBV patients sampled at 3, 6, and 12 months posttransplant are in accord with these previous studies. Intriguingly, our data suggest that the reconstitution of the major lymphocyte populations was not markedly different between this no/low EBV group and patients with high-level EBV reactivation, although we acknowledge that subtle differences in immune reconstitution may have been missed due to the relatively small cohort sizes. For example, future studies could explore the possibility that the numbers of EBV-specific CD8+ T cells may be an informative marker to identify patients at risk of EBV reactivation.13,32-34
Notably, our data highlighted an important difference in B-cell reconstitution between the 2 transplant groups. The nascent B-cell system among the no/low EBV patients was dominated by transitional and naive cells for the first 6 to 12 months, in agreement with earlier studies,21-25,35 and consistent with the view that mature B cells reconstitute from newly engrafted donor stem cells. By contrast, high EBV patients showed a rapid increase in both the proportion and number of B cells expressing the memory marker CD27.36 This expansion consisted almost exclusively of IgG+ or IgA+ class-switched CD27+ memory B cells, with few detectable IgD+CD27+ nonswitched memory cells. Interestingly, not all patients sampled at the point of high-level EBV DNAemia had particularly raised levels of CD27+ memory B cells. This variation may reflect the magnitude of reactivation because we observed a good correlation between cellular EBV loads and the absolute number of circulating CD27+ memory B cells.
Given these findings, we infer that the accumulation of isotype-switched CD27+ memory B cells in allo-HSCT patients is likely to result from de novo expansion associated with high-level EBV reactivation. This hypothesis is supported by our cell sorting data which indicated that EBV was almost exclusively present in the CD27+ memory B cells in high load patients. Although a small fraction of EBV DNA (<1%) was detected within the naive B-cell and non-B-cell subsets, these results could be explained by low-level contamination with CD27+ memory cells. Notably, a lack of sufficient numbers of CD27+ B cells precluded a similar analysis of this population in the no/low EBV patients. In summary, although we had anticipated that EBV might be present in the numerically dominant transitional and naive B-cell populations in allo-HSCT recipients, the present data argue that EBV is restricted to CD27+ memory B cells, just as in immunocompetent individuals.18,20 This is not to deny that EBV can also access other lymphocytes in certain situations, such as in T- and NK-cell lymphoproliferations.37 Indeed, a recent study by Calattini et al38 of patients with EBV-associated lymphoproliferation reported that >20% of the EBV-positive cells were phenotypically non-B, non-T, and nonmonocyte.
The single-cell analysis of sorted naive and memory B cells from high load patients also provided valuable new insights into EBV reactivation following allo-HSCT. Remarkably, we estimated that 80% to 90% of the CD27+ memory B cells were virus-positive, a frequency which is several orders of magnitude higher than the 1 in 5000 to 1 in 200 000 infected cells typically seen in healthy seropositive individuals.20,39,40 Moreover, we estimated that infected CD27+ B cells typically carried 10 to 30 EBV genome copies. These values are consistent with viral loads found in latently infected EBV-positive cell lines41-43 and are also in accord with previous fluorescence in situ hybridization studies of allo-HSCT38 and solid organ transplant (SOT) patients.44 Crucially, we never found any cells with high EBV genome loads indicative of virus replication. Taken together, these observations indicate that high-level EBV DNAemia following allo-HSCT is fundamentally associated with an accumulation of latently infected B cells rather than due to EBV replication. Interestingly, the genome loads of individual infected B cells may also shed light on the proliferative history of this population. Thus, a recent study, in which resting B cells were infected in vitro with EBV at <1 virus particle per cell, found that the virus genome was amplified to around 20 to 40 copies per cell after 3 to 4 weeks in culture.42 We infer that the EBV-positive B cells in our high load transplant patients must have undergone multiple rounds of proliferation postinfection and therefore the initial infection event is likely to have occurred several weeks prior to the peak of reactivation. This hypothesis is consistent with our finding that numbers of CD27+ memory B cells were much lower 2 to 3 weeks prior to the onset of reactivation.
Further evidence that the accumulation of CD27+ memory cells is an EBV-driven transformation event came from EBV gene expression profiling which detected several EBV latent transcripts associated with B-cell growth transformation45-47 Thus, our Q-PCR analysis revealed LCL-like levels of EBNA1, EBNA2, and LMP2 transcripts in all 3 patients tested, whereas 2 patients also had high levels of LMP1 expression. However, we cannot exclude the possibility of heterogeneity within the infected B-cell pool, and that 2 distinct populations of virus-positive memory B cells may simultaneously exist: 1 characterized by expression of the full spectrum of EBV latent genes (so called latency III) and the second characterized by a more restricted pattern of viral gene expression (latency I/latency 0).30 Indeed, this notion agrees with our proliferation data showing that around half of the memory B cells were Ki-67+.
Although there is little information on the expression of EBV genes in the blood of allo-HSCT recipients, previous studies have examined EBV RNA profiling in the context of SOT.17,48-50 Notably, these studies have not identified a common pattern of EBV gene expression associated with reactivation. For example, 1 large-scale study of healthy immunosuppressed cardiothoracic transplant patients revealed multiple patterns of EBV transcription which included LMP2 only, coexpression of LMP1 and LMP2, and coexpression of LMP1, LMP2, and the lytic gene GP350.48 However, around 30% of all posttransplant samples in this study did show evidence of latency III-associated transcripts and this finding correlated with increased viral load. Meanwhile, a smaller study of pediatric liver transplant patients with high chronic EBV loads revealed expression of only LMP2.49 It is unclear to what extent such heterogeneity reflects technical differences in the methods used to detect EBV transcripts in different studies. Comparison between SOT and allo-HSCT may also be further complicated by the fact that SOT patients often exhibit a more chronic and lower-level viral DNAemia and are less T-cell compromised than allo-HSCT patients. The timing of sample collection in relation to the emergence of EBV reactivation may also be important. These factors may explain the finding of a study by Babcock et al51 which reported that EBV-infected memory cell B cells that accumulate in the blood of healthy immunosuppressed SOT recipients are resting rather than proliferating.
Finally, it is interesting to speculate how opportunistic expansion of EBV-transformed B cells leads to the exaggerated numbers of CD27+ memory B cells observed in allo-HSCT patients. In immunocompetent individuals, it is proposed that EBV gains access to the long-lived memory B-cell pool by initially infecting naive B cells and then driving them to differentiate through the natural physiologic route of the germinal center.1 However, such a scenario seems unlikely in the context of allo-HSCT patients, given that normal germinal center activity is likely to be disrupted for many months after transplant due to deficiency of CD4+ T cells and other supporting cells.52 Notably, microsatellite analysis of CD27+ B cells sorted by FACS from patients 6, 9, and 12 confirmed that these cells were all of 100% donor origin (data not shown), and in light of this we propose 2 alternative explanations. In the first, EBV infects mature donor B cells which are transferred in the stem cell graft.53 This mechanism is favored by studies showing that graft B-cell depletion with alemtuzumab (an anti-CD52 monoclonal antibody that depletes both T cells and B cells) is associated with a reduced incidence of EBV reactivation compared with antithymocyte globulin–conditioned transplants in which B cells are retained.11,16 The second model proposes that EBV infects newly emerging mature B cells which have differentiated from donor stem cells. This idea is supported by the observation that EBV reactivation is often delayed until 2 to 3 months after transplant, concomitant with the onset of lymphopoiesis. Whether EBV drives the differentiation of emerging naive B cells, either by germinal center transit1 or by inducing a memory phenotype via a GC-independent pathway,54 or through the direct infection of memory B cells,55 remains to be tested.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Roger Bird for help with cell sorting. The authors also thank the patients who contributed to this study.
D.M.B. was supported by a Wellcome Trust Clinical Training Fellowship (097439/Z/11/Z). A.B.R., M.R., and A.I.B. were supported by a Cancer Research UK Programme Grant award (C5575/A15032).
Authorship
Contribution: D.M.B., R.T., C.S.-L., and A.I.B. designed and performed the experiments; D.M.B., R.T., C.S.-L., M.R., and A.I.B. analyzed and interpreted data; J.C., C.I., B.A., S.N., S.C., and P.M. provided patient samples and data; D.M.B. and A.I.B. wrote the manuscript; and D.M.B., C.P.F., S.C., C.F.C., A.B.R., A.I.B., and M.R. designed and directed the study
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Andrew I. Bell, School for Cancer Sciences, University of Birmingham, Edgbaston, Birmingham B15 2TT, United Kingdom; e-mail: a.i.bell@bham.ac.uk.