Abstract
Infant B-cell acute lymphoblastic leukemia (B-ALL) accounts for 10% of childhood ALL. The genetic hallmark of most infant B-ALL is chromosomal rearrangements of the mixed-lineage leukemia (MLL) gene. Despite improvement in the clinical management and survival (∼85-90%) of childhood B-ALL, the outcome of infants with MLL-rearranged (MLL-r) B-ALL remains dismal, with overall survival <35%. Among MLL-r infant B-ALL, t(4;11)+ patients harboring the fusion MLL-AF4 (MA4) display a particularly poor prognosis and a pro-B/mixed phenotype. Studies in monozygotic twins and archived blood spots have provided compelling evidence of a single cell of prenatal origin as the target for MA4 fusion, explaining the brief leukemia latency. Despite its aggressiveness and short latency, current progress on its etiology, pathogenesis, and cellular origin is limited as evidenced by the lack of mouse/human models recapitulating the disease phenotype/latency. We propose this is because infant cancer is from an etiologic and pathogenesis standpoint distinct from adult cancer and should be seen as a developmental disease. This is supported by whole-genome sequencing studies suggesting that opposite to the view of cancer as a “multiple-and-sequential-hit” model, t(4;11) alone might be sufficient to spawn leukemia. The stable genome of these patients suggests that, in infant developmental cancer, one “big-hit” might be sufficient for overt disease and supports a key contribution of epigenetics and a prenatal cell of origin during a critical developmental window of stem cell vulnerability in the leukemia pathogenesis. Here, we revisit the biology of t(4;11)+ infant B-ALL with an emphasis on its origin, genetics, and disease models.
Introduction
B-cell acute lymphoblastic leukemia (B-ALL) is the most frequent cancer in the childhood.1 B-ALL is characterized by an uncontrolled expansion of immature B-cell precursors within the bone marrow (BM).2,3 There are different subtypes of B-ALL attending to both the specific differentiation stage where B-cell precursors are stalled and to the cytogenetic/molecular diagnosis.4,5 Over the last decades, improved understanding of the disease biology, precise risk stratification, improved supportive care, and personalized chemotherapy has increased the cure rate of childhood B-ALL close to 90%.3,6 However, leukemia in infants (<1 year of age) is rare but captures a lot of interest due to its aggressive clinical presentation in a uniquely vulnerable host. Of special interest is the infant B-ALL mediated by mixed-lineage leukemia rearrangement (MLL-r), which possesses unique clinical and biological features, representing an outlier B-ALL with unfavorable prognosis.
Germline MLL (also known as KMT2A) on chromosome 11q23 is required for normal hematopoiesis and the expression of the HOXA cluster gene.7-11 The MLL gene is rearranged most often in de novo infant leukemia and in adult secondary therapy-related leukemia.12-14 MLL-r functions as the initiating, and perhaps as the sole driving, oncogenic event by dysregulating epigenetic/transcriptional programs.15,16 The most common rearrangement is a reciprocal translocation between MLL and a partner gene resulting in a chimeric protein composed of the N terminus domain of MLL and C terminus domain of the partner gene.17,18 Many partners have been identified in MLL-r leukemia, the most common being AF4 (also known as AFF1), AF9, and ENL.19
A biologically and clinically intriguing MLL-r is MLL-AF4 (MA4, also referred as KMT2A-AFF1), which results from t(4;11)(q21;q23) and is the hallmark genetic abnormality of infant t(4;11) pro-B/mixed B-ALL. Infant MA4+ B-ALL is associated with a dismal prognosis,20,21 therapy refractoriness, central nervous system (CNS) infiltration, and short latency.22 Our understanding of the disease pathogenesis, oncogenic insults, and cellular origin is very limited. We speculate this is largely because infant cancer is, from an etiologic and pathogenesis standpoint, very distinct to adult cancer, and it should be studied as a developmental disease.20,23 In fact, compelling evidence of a single cell of prenatal origin as the target cell for MA4 fusion has been extensively reported, with important implications in the way we understand and study developmental cancer.20,23 Despite its aggressiveness, current disease models have failed to faithfully recapitulate the disease, which may be explained from a developmental angle because some developmental cues and the prenatal nature of the target cell remain elusive. In addition, another controversial question is whether MA4 fusion alone functions as a single “big-hit” sufficient to induce an overt B-ALL.24 Whole-genome sequencing (WGS) studies reported a silent mutational landscape in MLL-r infant B-ALL,25 suggesting that a single driver mutation (MLL-r) suffices to spawn this aggressive B-ALL, thus contradicting the conventional dogma of multiple and sequential genetic changes needed for leukemia initiation. Here, we revisit the biology of normal MLL and MA4+ infant B-ALL, with an emphasis on its cellular origin, genetics, and disease models.
Clinical features of t(4;11)+ infant B-ALL
Infant MA4+ B-ALL is among the most vexing clinical problems in pediatric hematology/oncology. The incidence of MA4+ B-ALL in infant is between 1 and 2 logs lower than in children aged 1 to 14 years old.1 Phenotypically, MA4+ B-ALL is a CD34+CD19+ pro-B/mixed leukemia lacking CD10 and coexpressing aberrant myeloid markers such as CD15 and CD65 (Table 1).26 The majority of MA4+ B-ALLs express the neural glial marker 2 (NG2).27 Although it might be associated with the high propensity of this leukemia to infiltrate the CNS, its exact contribution to disease evolution remains a mystery.28-30
Main clinico-biological features of t(4;11) MA4+ B-ALL
| . | . |
|---|---|
| Characteristic . | Comment . |
| Incidence* | Estimated 1 case/1 million newborns |
| Phenotype | Pro-B/mixed: CD34+CD19+CD10− with expression of the myeloid markers CD15 and CD65 and NG2. |
| EFS | 5-year EFS is 34% for Children’s Cancer Group protocol CCG-1953 |
| 4-year EFS is 37% for Interfant-99 protocol | |
| State of the art treatment† | Initial glucocorticoid-based treatment (prednisone, l-asparaginase), followed by induction (dexamethasone, cytarabine, vincristine, daunorubicin, and mitoxantrone) and by myeloid-like (cytarabine, daunorubicin/mitoxantrone, and etoposide) or lymphoid-like consolidation (cyclophosphamide, cytarabine, and 6-mercaptopurine) |
| CNS involvement† | Common: up to 50% accumulative at diagnosis and during evolution |
| Age at diagnosis | <1 year |
| Negative prognostic factors | High WBC counts (>300 000 WBC/µL) |
| Age < 6 months | |
| CNS infiltration | |
| Early poor response to prednisone | |
| RAS mutations | |
| High expression level of FLT3 | |
| Low expression level of HoxA cluster |
| . | . |
|---|---|
| Characteristic . | Comment . |
| Incidence* | Estimated 1 case/1 million newborns |
| Phenotype | Pro-B/mixed: CD34+CD19+CD10− with expression of the myeloid markers CD15 and CD65 and NG2. |
| EFS | 5-year EFS is 34% for Children’s Cancer Group protocol CCG-1953 |
| 4-year EFS is 37% for Interfant-99 protocol | |
| State of the art treatment† | Initial glucocorticoid-based treatment (prednisone, l-asparaginase), followed by induction (dexamethasone, cytarabine, vincristine, daunorubicin, and mitoxantrone) and by myeloid-like (cytarabine, daunorubicin/mitoxantrone, and etoposide) or lymphoid-like consolidation (cyclophosphamide, cytarabine, and 6-mercaptopurine) |
| CNS involvement† | Common: up to 50% accumulative at diagnosis and during evolution |
| Age at diagnosis | <1 year |
| Negative prognostic factors | High WBC counts (>300 000 WBC/µL) |
| Age < 6 months | |
| CNS infiltration | |
| Early poor response to prednisone | |
| RAS mutations | |
| High expression level of FLT3 | |
| Low expression level of HoxA cluster |
EFS, event-free survival; FLT3, fms-like tyrosine kinase 3; RAS, rat sarcoma; WBC, white blood cell.
Estimated in Europe/United States.
Based on Interfant protocol.
In infant B-ALL, MLL-r is associated with poorer outcome. In the Children's Cancer Group protocol CCG-1953, the 5-year event-free survival (EFS) for MLL-r infants was 34% compared with 60% with germline MLL.31 In the Interfant protocol, the 4-year EFS in MLL-r and germline MLL infants was 37% and 74%, respectively (Table 1).31 Conversely, in infant acute myeloid leukemia (AML), MLL-r is not a significant risk factor. The standard care for infant MA4+ B-ALL includes intensive multiagent chemotherapy to induce remission, followed by consolidation chemotherapy (for favorable prognosis) or allogeneic hematopoietic stem cell transplantation (for unfavorable prognosis).32-35 (Table 1). The major cooperative groups conducting clinical trials for infant ALL are Interfant (Interfant-06), The Children’s Oncology Group, and the Japanese Pediatric Leukemia Study Group. All have adopted an identical induction strategy based on Interfant-99, and they use prospective risk-stratified approaches that incorporate MLL-r status. The Interfant protocol gives a 7-day single-agent prednisone before intensive induction chemotherapy and is testing whether consolidation with myeloid-like chemotherapy will prove superior to lymphoid-like consolidation in MLL-r infants. The rationale is that infant MA4+ B-ALL may derive from an early hematopoietic precursor with myeloid differentiation potential and may respond better to myeloid chemotherapy regimens. The Children’s Oncology Group is testing FLT3 tyrosine kinase inhibitors for postinduction chemotherapy (Table 1).
Even though infant MLL-r B-ALL displays, by default, an unfavorable prognosis, several additional independent factors have been identified (Table 1). The most important are age and WBC count at diagnosis, with younger infants and higher WBC count associated with poorer outcomes.21,33-37 A poor response to prednisone (≥1000 blasts/μL in peripheral blood on day 8) is also an independent negative prognostic factor.37,38 Furthermore, the presence of CNS infiltration, RAS mutations, high levels of FLT3, and low levels of HOX-A genes have also been reported negative prognostic factors in independent studies.21,39-41 A retrospective multivariate analysis encompassing both clinico-biological parameters with molecular data still needs to be undertaken in MLL-r infant B-ALL to ascertain the independent robustness of these molecular markers.
Molecular pathogenesis of MLL-r: the importance of the MLL protein
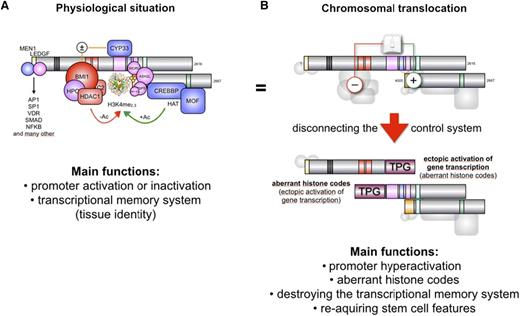
Given the large variety of MLL fusions diagnosed thus far in infant, childhood, and adult acute leukemia, one may consider the possibility that a first oncogenic hit derives from the particular truncation of the MLL protein, whereas a potential secondary hit may derive from the fused partner protein. The MLL protein is processed by Taspase1,42 resulting in 2 protein fragments (p320/p180) that bind each other, forming a molecular hub for the assembly of a large nuclear complex (Figure 1). Binding proteins are MEN1, LEDGF, GADD34, PP2A, the polymerase associated factor (PAF) elongation complex, a polycomb group complex (BMI1, HPC2, HDAC1/2, and CtBP), CYP33, CREBP, MOF, and the SET domain core proteins (WDR5, RbBP5, ASH2L, and SRY-30).11,43-52 The N-terminal portion of the MLL protein is functionally linked to bind-and-read chromatin signatures, whereas the C-terminal portion of the MLL protein associates to enzymatic functions, namely acetylation and methylation of histone core particles. Thus, the MLL complex binds to promoter regions of active genes, marking these regions by covalent histone modifications.53
Proposed model for the oncogenic conversion of MLL fusions. (A) The physiologic situation of MLL functions. Taspase1-cleaved MLL is assembled into the holo-complex and binds to target promoter regions. This occurs via the N-terminally bound MEN1/LEDGF protein complex that allows binding to many transcription factors. The PHD domain is able to read histone core particles, whereas the SET domain allows it to write epigenetic signatures (H3K4me2/3). Associated CREBP and MOF are able to acetylate nucleosomes. CYP33 allows switching into the repressor mode by enabling the docking of a Polycomb group complex composed of BMI1, HPC2, CtBP, and several HDACs. This enables the removal of acetyl groups from nucleosomes or transcription factors to shut down gene transcription. (B) In the case of a chromosomal translocation, the intrinsic regulatory mechanism of MLL becomes destroyed. The disrupted MLL portions are fused to protein sequences deriving from a large amount of different partner genes (n > 80). The N-terminal portion of MLL retains the ability to bind MEN1 and LEDGF, and thus, to bind to target promoter regions. Depending on the fusion sequence (AF4, AF5, LAF4, AF9, ENL, AF10), MLL-X fusions may recruit the endogenous AF4 complex that contains P-TEFb and the histone methyltransferases DOT1L, NSD1, and CARM1. This activates gene transcription and results in enhanced epigenetic signatures (H3K79me2/3). The C-terminal portion retains CREBBP and MOF binding capacity, as well as the SET domain. In some cases (AF4, AF5, LAF4), the N-terminal fused protein sequences allow binding to P-TEFb and directly to the largest subunit of RNA polymerase II to enhance the process of transcriptional elongation. In addition, the fused protein sequences still bind NSD1 and DOT1L.
Proposed model for the oncogenic conversion of MLL fusions. (A) The physiologic situation of MLL functions. Taspase1-cleaved MLL is assembled into the holo-complex and binds to target promoter regions. This occurs via the N-terminally bound MEN1/LEDGF protein complex that allows binding to many transcription factors. The PHD domain is able to read histone core particles, whereas the SET domain allows it to write epigenetic signatures (H3K4me2/3). Associated CREBP and MOF are able to acetylate nucleosomes. CYP33 allows switching into the repressor mode by enabling the docking of a Polycomb group complex composed of BMI1, HPC2, CtBP, and several HDACs. This enables the removal of acetyl groups from nucleosomes or transcription factors to shut down gene transcription. (B) In the case of a chromosomal translocation, the intrinsic regulatory mechanism of MLL becomes destroyed. The disrupted MLL portions are fused to protein sequences deriving from a large amount of different partner genes (n > 80). The N-terminal portion of MLL retains the ability to bind MEN1 and LEDGF, and thus, to bind to target promoter regions. Depending on the fusion sequence (AF4, AF5, LAF4, AF9, ENL, AF10), MLL-X fusions may recruit the endogenous AF4 complex that contains P-TEFb and the histone methyltransferases DOT1L, NSD1, and CARM1. This activates gene transcription and results in enhanced epigenetic signatures (H3K79me2/3). The C-terminal portion retains CREBBP and MOF binding capacity, as well as the SET domain. In some cases (AF4, AF5, LAF4), the N-terminal fused protein sequences allow binding to P-TEFb and directly to the largest subunit of RNA polymerase II to enhance the process of transcriptional elongation. In addition, the fused protein sequences still bind NSD1 and DOT1L.
Near the center of the MLL protein is the plant homeodomain (PHD) domain. This region is composed by PHD1-3 subdomain, a bromodomain (BD), and PHD4 subdomain. The PHD domain exhibits 2 normal PHD subdomain structures: PHD1/2 and PHD3/4. The PHD1-3 subdomains are followed by a BD necessary to stabilize the structure of the PHD3/4 domain and has no histone-acetyl reading function in the MLL protein. The PHD3/4 subdomain is required to read H3K4me2/3 signatures within the chromatin. However, when the PHD3/4 subdomain binds to CYP33/PPIE, a conformational change is catalyzed.54,55 As long as PHD3/4 subdomain is docked via a single protein helix to BD, it exhibits its reader function for nucleosomal H3K4 methylation signatures. Isomerization via CYP33/PPIE allows disconnection of PHD3/4 subdomain from the BD and interaction with the BMI1/HPC2/HDAC1-2/CtBP complex that then becomes enabled to bind to the methyl-DNA binding domain (MBD). Binding of MLL to this polycomb-group complex converts MLL into a transcriptional repressor. This defines the CYP33/PPIE isomerase as a molecular switch that triggers the MLL complex in 2 different modes of action: transcriptional activator or repressor. Nothing is known about the precise details of this molecular switch mechanism, but it is likely that it depends on the promoter context. This MLL switch is responsible for the effects on gene transcription. For instance, when Mll-knockout cells are transcriptionally profiled and compared with their isogenic wild-type cells, 66% of the differentially expressed genes become upregulated and only 33% are downregulated in the knockout cells.56 Therefore, genetic insults affecting bona fide functions of MLL may underlie leukemia initiation (Figure 1A). Compromising a domain that is responsible to perform binary decisions linked to gene transcription and epigenetic reading-writing may well explain the profound biological effect.
What does actually happen when a chromosomal translocation occurs at the MLL gene locus? Chromosomal rearrangements usually separate the MBD from the PHD domain (Figure 1B), thereby destroying the intrinsic control mechanism of the MLL protein. Even the binding of CYP33 to the PHD domain is impaired, at least when the chromosomal breakpoint localizes within MLL intron 11.57 Consequently, chromatin reading and writing functions now become independent of each other, and both portions of MLL become constitutively active, regardless of their fused protein sequences. The MLL-X fusions still bind via MEN1/LEDGF to chromatin and transcription factors in promoter regions and the PAF complex but are disabled to exert any inhibitory function. The reciprocal X-MLL fusion proteins retain the PHD3/4 chromatin reader domain, the CREBBP/MOF binding domain, and the SET domain complex. If CYP33/PPIE binds to the PHD domain of X-MLL fusions, the binding of the Polycomb complex is disabled due to the absence of MBD. This was nicely demonstrated by experiments where the PHD domain was fused to existing MLL-X fusion proteins, being sufficient to eliminate the oncogenic properties of those artificial MLL fusions, because the repressing functions are now exerted by the fused PHD domain via binding to the BMI-1 repressor complex.58,59 This model proposes that the biological functions deriving from the N- and C-terminal fused protein sequences exert only an accessory gain of function such as changing the protein interactome.
Etiology and pathogenesis of t(4;11)+ B-ALL
Epidemiologic and genetic studies have suggested that infant MLL-r leukemia may result from transplacental exposures during the gestation to DNA topoisomerase-II inhibitors, such as bioflavonoids.60,61 Bioflavonoid-rich dietary habits of the pregnant mothers have been linked to in utero MLL breaks.62,63 However, the wide exposure of the population to bioflavonoids contrasts with the low incidence of this infant leukemia, suggesting that the probability that DNA topoisomerase-II inhibitors hit a candidate leukemia-initiating cell (LIC) is extremely low. Importantly, etoposide, a DNA topoisomerase-II inhibitor that is used widespread in chemotherapy cocktails, can induce MLL-r in different cell types61 : in embryonic stem cells (ESCs),64,65 fetal liver-derived CD34+ hematopoietic stem cells (HSCs)66 and cord blood (CB)-derived CD34+ HSCs. Clinically, ∼5% to 10% of etoposide-treated patients develop a therapy-related leukemia characterized by the presence of MLL-r.14
The driving genetic alterations underlying infant leukemias originate prenatally during embryonic-fetal development.67 Seminal studies in identical monozygotic twins with concordant leukemias first demonstrated the presence of a unique and common MLL-r in the leukemic cells from both siblings, which unequivocally shows that the leukemogenic event arises in utero in one of the twins and then is propagated through blood circulation within the single, monochorionic shared placenta.68 The concordance rate for this leukemia in both twins is close to ∼100%. MA4 fusion has also been found and expressed in BM stromal cells (BMSCs) from t(4;11)+ B-ALL infants, suggesting that the MLL fusion may arise in a very early prehematopoietic/mesodermal precursor.69 Large-scale CB screenings also demonstrated that chromosomal translocations can be generated during fetal development.70 Leukemia fusion genes were present in CB from healthy newborns, but overt leukemia only occurred in 1% of these newborns, suggesting that additional mutations are needed. In nontwinned infant ALL, the prenatal origin of MA4 was demonstrated by a retrospective screening of blood taken at birth on neonatal blood spots from children with B-ALL.71 Altogether, these studies established that t(4;11) arises in utero during fetal hematopoiesis.
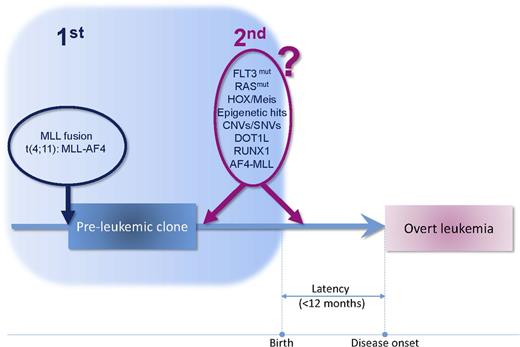
The extremely short latency of MA4+ infant B-ALL questions whether secondary mutations are required at all to develop overt leukemia. The 2-hit model postulates that cancer results from the accumulation of DNA mutations. In the case of pediatric B-ALL, the 2-hit hypothesis postulates that the prenatal chromosomal translocation is followed by a secondary prenatal or early postnatal genetic mutation. Because infant B-ALL has a shorter natural history than common B-cell precursor ALL (cALL), it is plausible that all the necessary genetic alterations occur prenatally (Figure 2).72
Two-hit cancer model in infant t(4;11)+ B-ALL. MA4 fusion is the first and driver oncogenic event. The very short latency of the disease indicates that secondary cooperating hits, if required, are expected to arise prenatally or very early after birth.
Two-hit cancer model in infant t(4;11)+ B-ALL. MA4 fusion is the first and driver oncogenic event. The very short latency of the disease indicates that secondary cooperating hits, if required, are expected to arise prenatally or very early after birth.
Although extensive efforts have been directed to decipher secondary genetic alterations in MA4+ B-ALL, clinical and experimental data remain controversial. Tamai et al showed that ectopic expression of MA4 in murine HSCs is sufficient to cause B-ALL in mice and that cooperating KRAS mutations accelerate leukemic transformation.73 However, RAS mutations are subclonal, only present in ∼25% of patients, and are partially lost at relapse, suggesting they are not tumor drivers.39,74 Preliminary data from our laboratory reveal that activated KRAS cooperates with MA4 to promote CNS infiltration and extramedullary engraftment (spleen and peripheral blood) of CB-CD34+ HSPCs but is not sufficient to initiate leukemia.
The FLT3 receptor is required for normal lymphopoiesis and is expressed in almost all AML/ALLs. Gene expression profiling showed that FLT3 is highly expressed in MLL-r B-ALL,75 leading to the characterization of FLT3 mutations as potential secondary cooperating events. However, the occurrence of FLT3 mutations in MA4+ pro-B ALL remains controversial. Some studies have shown that MLL-r B-ALL harbors FLT3 mutations in 3% to 21% of cases,76-78 whereas others have shown that FLT3 mutations are not present in MLL-r B-ALL.21,40,79-81 Given that FLT3 is consistently highly expressed in MLL-r B-ALL, increased transcriptional expression of FLT3 rather than activating mutations could represent an important hallmark of MLL-r B-ALL.21,40 We recently analyzed the prognostic significance of FLT3 mutations and expression in MA4+ and MLL-germline ALL21 and found that MA4+ B-ALL exhibited higher FLT3 expression levels than normal BM, supporting that aberrantly increased transcription of FLT3, rather than activating FLT3 mutations, contributes to the pathogenesis of MA4+ B-ALL. This is supported by studies reporting FLT3 as a direct transcriptional target of MA4.82 Similar to that reported for RAS mutations,39 high FLT3 expression is an independent prognostic factor in MA4+ B-ALL patients (but not in pediatric cALL) and is associated with lower overall survival and EFS. Unfortunately, owing to the low incidence of MLL-r infant leukemias, no clear correlation between FLT3 levels and specific MLL fusions could be established.
Increasing evidence points to AF4-MLL, the reciprocal product of MA4, as a driver oncogenic event in t(4;11)+ B-ALL.83 AF4-MLL–transduced mouse HSCs developed pro-B-ALL, whereas cotransduction with MA4 and AF4-MLL resulted in mixed lineage leukemia, suggesting that the expression of the AF4-MLL protein may induce B-ALL even in the absence of MA4. Studies from Wilkinson et al demonstrated that RUNX1 is directly activated by MA4 and the RUNX1 protein interacts with the AF4-MLL chimeric protein, suggesting how these onco-proteins could cooperate at the molecular level.84
Dysregulated immune responses to common infections have also emerged as cooperating events underlying the etiology of childhood leukemia.72 Delayed exposure of children to common pathogens may cause exacerbated T-cell responses when these infections occur later on in life.85,86 This untimely and excessive inflammatory response abolishes normal hematopoiesis promoting selective expansion of a preleukemic clone, resulting in stochastic or microenvironment-derived cooperating drivers toward overt leukemia.87,88 The delayed infection hypothesis may contribute to the etiology of pediatric leukemias with a longer latency, but it is unlikely it underlies the etiology of MA4+ infant B-ALL diagnosed during early life or even at birth, because newborns have not had natural time for exposures to infection.
Genetics of t(4;11)+ B-ALL
Whether MA4 is sufficient for leukemogenesis or requires cooperating mutations is still somehow an open debate. Genome-wide sequencing analysis revealed genetic alterations in genes involved in B-cell development/differentiation in pediatric cALL leukemias.89 In contrast, Bardini et al described a stable genome for MA4+ infant B-ALL patients as demonstrated by the absence of copy number alterations analyzed by single nucleotide polymorphism arrays.80,81 Shortly after, Dobbins et al performed WGS of leukemic blasts at diagnosis from 3 infant MA4+ B-ALLs, and no recurrent somatic mutations were found, and the mutational landscape of patients was largely silent.90 Very recently, the whole-genome mutational landscape of MLL-r B-ALLs has been revisited in a larger patient cohort as part of the Pediatric Cancer Genome Project at St. Jude Children’s Research Hospital.25 This seminal work demonstrates that infant MLL-r B-ALL has one of the lowest frequencies of somatic mutations of any sequenced cancer, with the predominant leukemic clone carrying 1.3 nonsilent mutations. The only alterations recurrently (47% of cases) detected were mutations in the kinase-phosphatidylinositol 3-kinase-RAS pathway, and these mutations were subclonal. In contrast, noninfant MLL-r B-ALL patients displayed on average 6.5 nonsilent mutations per case, mainly affecting epigenetic regulators. Importantly, noninherited germline mutations acquired during hematopoietic development cannot be ruled out as oncogenic drivers because they have not been analyzed in the available WGS studies.
The silent mutational landscape observed in infant MLL-r B-ALL coupled to the higher number of mutations emerging in pediatric (noninfants) MLL-r B-ALL supports the developmental origin of infant MLL-r B-ALL and establishes that very few genetic alterations are required for overt leukemia. This silent mutational landscape argues that the MA4 may function as a single big hit sufficient to induce overt, short latency, and aggressive B-ALL.25 This contrasts with the 2-hit model widely proposed for other childhood leukemias. For instance, TEL-AML1–expressing t(12;21) pre-B-ALL requires additional postnatal cooperating oncogenic hits such as deletion of the germline TEL allele91 or RAG-mediated deletions.92 If MA4 is the sole initiating event, the MA4-mediated transformation would then rely on alternative epigenetic cooperating lesions occurring on a critical developmentally early window of stem cell vulnerability, explaining the low prevalence of this entity. Interestingly, Chuk et al93 reported the clearance of preleukemic MLL-AF4 cells from a healthy child. Similarly, Maia et al94 described a child where the MLL fusion was detected at birth, but there was a latency of 6 years before overt leukemia was observed. These reports present evidence against the 1-hit hypothesis (specifically an MLL-ENL fusion) by proposing the existence of MLL-r preleukemic clones that eventually disappear in the absence of cooperating proliferation/survival-promoting secondary oncogenic hits.
Mutational and epigenetic mechanisms are expected to affect gene expression, allelic imbalance, and key components of the spliceosome with the subsequent generation of alternative splicing variants commonly linked to cancer.95 DNA-Seq allows identification of potentially oncogenic silent and nonsilent single nucleotide variants and is also informative in dissecting intraclonal mutation heterogeneity.96 However, to date, RNA-Seq and whole genome epigenetic studies have not been performed on this leukemia to unravel whether epigenetic rather than genetic alterations are the missing cooperating oncogenic hits. Therefore, a multilayer-omics analysis coupled to computational integration of genome-wide DNA- and RNA-Seq and genome-wide DNA methylation of infant MA4+ B-ALL will provide a unique molecular-genetic signature of the disease onset/evolution at the genome, epigenome, and transcriptome level.97,98
Epigenetics of t(4;11)+ B-ALL
The silent mutational landscape found in infant MA4+ B-ALL does not explain the rapid onset and aggressive evolution of the disease.24 MLL is an H3K4 histone methyltransferase,10,11 and leukemia transformation by MLL fusions requires H3 lysine 79 (H3K79) methyltransferase DOT1L, which is recruited to the MLL fusion transcriptional complex53,99 (Figure 1). Epigenetic mechanisms including DNA methylation and histone modification by methylation or acetylation tightly regulate gene expression during early mammalian development and hematopoiesis.100,101 Thus, we envision a key contribution of epigenetic remodeling in the pathogenesis of this B-ALL.53 However, little information is available on epigenetic dysregulation of MA4+ B-ALL. Armstrong et al demonstrated that H3K79 methylation profiles define both murine and human MA4+ B-ALL.98,102 They showed that MA4+ B-ALLs could be distinguished from other ALLs by their H3K79 profiles. Suppression of the DOT1L inhibited expression of MA4 target genes, demonstrating that H3K79 methylation is a distinguishing feature of MA4+ B-ALLs and is key for maintenance of MA4-driven gene expression.82,103
Ikawa et al104 reported dense methylation of regulatory regions of the CD10 gene promoter, suggesting that methylated transcription factor binding sites contribute to CD10 silencing as epigenetic mechanisms underlying the pro-B (CD10 neg) phenotype. In cancer cells, enhanced promoter methylation is typically accompanied by global loss of methylation in nonpromoter regions of the genome. However, Stumpel et al reported that MLL-r infant B-ALL cells display a global hypermethylated genomic state, both at promoter and nonpromoter regions.105 Because global hypomethylation usually leads to genomic instability linked to cancer development, this study might explain the global genomic stability/silent mutational landscape found in MA4+ infant B-ALLs and the remarkable sensitivity of MLL-r cells to demethylating agents.106
Further studies from Stumpel et al focused on genome-wide cytosine guanine dinucleotide island methylation and promoter methylation.107 These studies revealed that different subsets of B-ALL and different types of MLL translocations are associated with distinct patterns of DNA methylation, and the degree of DNA methylation influences clinical outcome, identifying subgroups of MLL-r infant B-ALL patients that may particularly benefit from therapeutic strategies based on demethylating drugs.107,108 Cutting edge epigenetic whole-genome approaches will shortly be applied to MA4+ B-ALLs, providing the overall genome methylation picture that should be integrated with the available whole-genome DNA-Seq data to gain insights on the genomic and epigenetic aberrations underlying MA4+ B-ALLs.
Available disease models for t(4;11)+ B-ALL
No experimental model has thus far faithfully recapitulated MA4+ pro-B-ALL latency/phenotype (Table 2). Chen et al produced MA4 knock-in mice by homologous recombination in ESCs, but these mice developed mixed lymphoid/myeloid hyperplasia and B-cell lymphomas.109 Similarly, Metzler et al used the invertor conditional technology to create a mouse model in which a floxed AF4 cDNA was knocked into MLL in an opposite orientation to transcription. Then, Rag1, Lck, and CD19-Cre expression were used to drive MA4 expression in B/T progenitors, T cells, or B cells, respectively. These mice developed mature B-cell neoplasias. Although the latency varied slightly depending on the Cre line used, in all cases it was >300 days.110 Also, Kristov et al created a mouse in which conditional expression of MA4 using interferon-inducible Mx1-Cre line resulted in cALL or AML.98 Finally, Tamai and Inokuchi established a MA4 transgenic mice, but the phenotype obtained was lymphoblastic leukemia or lymphoma111 (Table 2).
Summary of current mouse models for MA4+ B-ALL
| Strategy . | Phenotype* . | Latency† . | Tissue-specific Cre . | Reference . |
|---|---|---|---|---|
| Constitutive Mll-AF4 knock-in mice | Myeloproliferative/follicular B leukemia | 520 days (17 months) | NA | Chen et al109 |
| Mll-AF4 invertor mice | B-cell lineage neoplasias | 317-466 days | Rag-Cre | Metzler et al110 |
| 416-472 days | Lck-Cre | |||
| 460-475 days | CD19-Cre | |||
| Conditional Mll-AF4 knock-in mice | B-precursor ALL and AML | 131 days | Mx1-Cre | Krivtsov et al98 |
| Transplant of AF4-MLL-transduced murine HSPCs | AF4-MLL: pro-B ALL (63%); B/T biphenotypic (37%) | AF4-MLL: 233 days | NA | Bursen et al83 |
| Double: 266 days | ||||
| Double: B/T biphenotypic (67%); pro-B ALL (33%) | ||||
| MLL-AF4 transgenic mice | Lymphoblastic leukemia or lymphoma | 12 months | NA | Tamai et al111 |
| Strategy . | Phenotype* . | Latency† . | Tissue-specific Cre . | Reference . |
|---|---|---|---|---|
| Constitutive Mll-AF4 knock-in mice | Myeloproliferative/follicular B leukemia | 520 days (17 months) | NA | Chen et al109 |
| Mll-AF4 invertor mice | B-cell lineage neoplasias | 317-466 days | Rag-Cre | Metzler et al110 |
| 416-472 days | Lck-Cre | |||
| 460-475 days | CD19-Cre | |||
| Conditional Mll-AF4 knock-in mice | B-precursor ALL and AML | 131 days | Mx1-Cre | Krivtsov et al98 |
| Transplant of AF4-MLL-transduced murine HSPCs | AF4-MLL: pro-B ALL (63%); B/T biphenotypic (37%) | AF4-MLL: 233 days | NA | Bursen et al83 |
| Double: 266 days | ||||
| Double: B/T biphenotypic (67%); pro-B ALL (33%) | ||||
| MLL-AF4 transgenic mice | Lymphoblastic leukemia or lymphoma | 12 months | NA | Tamai et al111 |
NA, not applicable.
Disease phenotype observed differs from pro-B ALL (CD10−).
Latency, defined as time to leukemia development, is always very protracted.
Summary of current human models for MA4+ B-ALL
| Ontogeny stage . | Cell type . | Phenotype . | Leukemic cooperation with FLT3/RAS* mutants . | Reference . |
|---|---|---|---|---|
| Fetal-HSPC* | NA | — | NA | — |
| Embryonic HSPC | hESC-derived hematopoietic cells | Enhances early hemato-endothelial specification | No | 117 |
| Skew toward endothelial vs hematopoietic fate | ||||
| 118 | ||||
| Neonatal HSPC | CB-derived CD34+ HSPCs | Proliferation coupled to survival advantage | No | 116 |
| Enhanced engraftment and clonogenic potential | ||||
| 119 |
| Ontogeny stage . | Cell type . | Phenotype . | Leukemic cooperation with FLT3/RAS* mutants . | Reference . |
|---|---|---|---|---|
| Fetal-HSPC* | NA | — | NA | — |
| Embryonic HSPC | hESC-derived hematopoietic cells | Enhances early hemato-endothelial specification | No | 117 |
| Skew toward endothelial vs hematopoietic fate | ||||
| 118 | ||||
| Neonatal HSPC | CB-derived CD34+ HSPCs | Proliferation coupled to survival advantage | No | 116 |
| Enhanced engraftment and clonogenic potential | ||||
| 119 |
Unpublished preliminary data.
Bursen et al83 showed that retroviral expression of the reciprocal protein AF4-MLL in murine HSCs followed by transplantation induced pro-B ALL in transplanted mice with a latency of 233 days. A pro-B phenotype was not observed in 100% cases, and B/T biphenotypic acute leukemia (in the AF4-MLL group) and mixed lineage leukemia cases (in the MA4+AF4-MLL group) were also detected. Leukemia development was faster when MA4- and AF4-MLL–cotransduced cells were transplanted (Table 2). However, transplantation of MA4-transduced HSCs did not result in leukemia. Therefore, AF4-MLL seems capable of inducing ALL in mice without the requirement of MA4, suggesting that the reciprocal protein is the initiating event of MA4+ leukemia.83 However, controversy about the role of AF4-MLL as an oncogenic initiating event exists because AF4-MLL is not expressed in one third of the patients112 and by studies on t(4;11) cell lines showing that they display addiction to MA4 but not to AF4-MLL.113,114 However, a transient silent interfering RNA-mediated knockdown of AF4-MLL was not sufficient to downregulate the AF4-MLL fusion protein, likely due to the stability of the AF4-MLL protein. In addition, AF4-MLL was shown to cooperate with RUNX1 to mediate the leukemogenic process.84,115 Together, these data suggest that one cannot draw definitive conclusions from these conflicting reports and that further studies using mouse models and primary patient samples should address to what extent AF4-MLL drives leukemia initiation or maintenance.
Other modeling attempts have used prenatal/neonatal human stem cells. Montes et al explored the effect of MA4 expression in 2 types of human stem cells: CB-derived CD34+ HSPCs and human embryonic stem cells (hESCs). In vivo, MA4 increased the in vivo repopulation ability of CB-derived CD34+ progenitors and in vitro MA4 increased the clonogenic potential and the proliferation of CD34+ HSPCs. However, MA4 did not induce leukemogenesis.116 Similarly, MA4 enhanced the specification of hemogenic endothelium from hESCs but strongly impaired further hematopoietic commitment, being also insufficient for leukemogenesis in this context.117 Bueno et al also found that the cooperation between MA4 and FLT3 in hESCs and CD34+ HSPCs did not result in leukemia. In hESCs, enforced expression of FLT3-TKD/FLT3-WT abolished hematopoietic specification,118 whereas it conveyed a transient overexpansion but did not suffice to immortalize/transform MA4-expressing CB-CD34+ HSPCs119 (Table 2).
Cell of origin for t(4;11)+ B-ALL
The nature of the prenatal cell initially transformed by MA4 in utero is unknown. Fluorescence in situ hybridization studies indicated that the fusion gene is present in the human primitive CD34+CD19− cells.120 Furthermore, MA4+ pro-B ALL manifests with aberrant bi-phenotypic blasts that coexpress both lymphoid and myeloid markers, with MA4 fusion being present in both lymphoid and myeloid lineages, suggesting that it may originate from immature lympho-myeloid stem/progenitor cells (HSPCs). In line with genetic evidence showing that MA4 originates in utero, Greaves and Wiemels hypothesized that this leukemia could originate from lymphoid-monocytic progenitors active in the fetal liver.67 Alternatively, because BMSCs from primary MA4+ pro-B infant patients harbor and express the MA4 fusion gene, we suggested that MA4 might arise in a population of mesodermal prehematopoietic precursors.69 This observation was restricted to MA4 because other leukemic fusion genes were absent in BMSCs. However, a similar study by Shalapour et al detected MLL-ENL and TEL-AML1 in the BMSC fraction. A variable fraction of translocated-positive BM-MSCs was found among patients when using immune fluorescence in situ hybridization.121 The association between blasts and stroma is not completely elucidated, and further investigation is required to better understand the leukemia relapse origin and the role of the microenviroment in this leukemia.
Although the identity of the cell of origin has not been addressed much in the murine models, it has been investigated in the human system using ontogenically early human stem cells such as hESC-blood derivatives and CB-HSPCs. Studies from our laboratory addressed whether these populations constitute target cells for leukemia initiation.116-119 Lentiviral-mediated expression of MA4 failed to transform either hESC blood derivatives or CB-derived CD34+ HSPCs.116,117 Other sources of developmentally early stem cells which might be potential cells of origin are fetal liver and aorta-gonad-mesonephros (AGM) cells. Human fetal liver tissue could be obtained from voluntary interruption of pregnancy, but whether human fetal liver-derived CD34+ HSPCs represent the cell of origin in this leukemia has yet to be addressed. Although conceptually the AGM region could represent the candidate mesodermal prehematopoietic precursor and therefore a possible cell of origin, no AGM-derived leukemia has been reported to date either in the human or the mouse setting. Given that genetic hallmarks in pediatric B-ALL occur in the fetus at which time B-1 B-cell progenitor numbers are high, mouse B1 progenitors have been proposed as another candidate cell-of-origin population. In fact, Montecino-Rodriguez et al showed that B-cell receptor-Abelson nonreceptor tyrosine kinase (ABL)+ B-ALL can initiate in fetal B-1 progenitors. They showed that mice transplanted with BCR-ABL–transduced fetal liver or BM B-1 progenitors became moribund more rapidly than recipients of BCR-ABL–transduced B-2 progenitors, suggesting B-1 progenitors as the target cell for disease initiation.122
Conversely, there are no data about the phenotype of the LIC. The LIC may be distinct from the cell in which MA4 has a preleukemic impact and, this in turn, may also differ from the cell in which MA4 first arises. Two recent studies have assessed the LICs in MA4+ pro-B ALL patient samples using in vivo xenotransplantation models. Bardini et al showed that all the LIC potential is within the CD19+ fraction,29 and Aoki et al similarly showed that both CD34+CD38+CD19+ and CD34−CD19+ cells contained LICs.123 Interestingly, the data of Aoki et al showed that, in MLL-AF9+ leukemia, the LICs were CD34−CD19+. It also remains unknown whether this infant MA4+ B-ALL follows a hierarchical or stochastic cancer model.124 Worth mentioning, Bardini et al29 established that MA4+ infant ALL is composed of a branching subclonal architecture at diagnosis. Some MA4+ clones appear to be quiescent at diagnosis but reactivate and dominate on serial transplantation into immunodeficient mice, whereas other dominant clones at diagnosis become more quiescent, suggesting a dynamic competition between actively proliferating and quiescent subclones. They showed using paired diagnostic and relapse samples that relapses often occur from subclones already present but more quiescent at diagnosis. In sum, much information remains to be gained in both the mouse and human setting about the nature of the embryonic/fetal cell in which MA4 arises and/or exerts its transformation potential, as well as the functional LIC, which fuels leukemia maintenance/progression and therapy relapse. Identification of the cell of origin, LICs, and the subclonal architecture and dynamics in MLL+ infant leukemia should lead to improved therapeutic strategies and provide key insights for the therapy resistance and frequent relapses observed in this group of poor prognosis ALL.
Acknowledgments
This work was supported by European Research Council grant ERC-2014-CoG-646903 (to P.M.), Instituto de Salud Carlos III/Fondo Europeo de Desarrollo Regional (FEDER) Grant PI14/01191 (to C.B.), Ministerio de Economía y Competitividad (MINECO) grant SAF2013-43065 (to P.M.), The Spanish Association Against Cancer (to P.M. and C.B.), Marie Curie Career Integration grant FP7-PEOPLE-2013-CIG-631171 (to A.S.-P.), The Fundación Inocente Inocente, and The Generalitat de Catalunya grant SGR330 (to P.M.). C.B. and C.P. are supported by Miguel Servet II Contract CPII13/00011 and Ayudas Predoctorales de formación en investigación en salud (PFIS) Scholarship FI12/00468, respectively. P.M. also acknowledges financial support from The Obra Social La Caixa-Fundació Josep Carreras.
This review is dedicated to all the infants diagnosed with MLL leukemia who have participated in our research over the last years.
Authorship
Contribution: All authors contributed to review preparation by gathering, reviewing and interpreting literature, and assisting with the writing process; R.W.S. and R.M. wrote parts of the review; and A.S.-P. and P.M. conceived the review, wrote the review, and put the text and figures together.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alejandra Sanjuan-Pla, Josep Carreras Leukemia Research Institute, School of Medicine, University of Barcelona, Casanova 143, 08036 Barcelona, Spain; e-mail: asanjuan@carrerasresearch.org; or Pablo Menéndez, Josep Carreras Leukemia Research Institute, School of Medicine, University of Barcelona, Casanova 143, 08036 Barcelona, Spain; e-mail: pmenendez@carrerasresearch.org.