Abstract
The DNA-binding zinc finger transcription factors Gfi1 and Gfi1b were discovered more than 20 years ago and are recognized today as major regulators of both early hematopoiesis and hematopoietic stem cells. Both proteins function as transcriptional repressors by recruiting histone-modifying enzymes to promoters and enhancers of target genes. The establishment of Gfi1 and Gfi1b reporter mice made it possible to visualize their cell type–specific expression and to understand their function in hematopoietic lineages. We now know that Gfi1 is primarily important in myeloid and lymphoid differentiation, whereas Gfi1b is crucial for the generation of red blood cells and platelets. Several rare hematologic diseases are associated with acquired or inheritable mutations in the GFI1 and GFI1B genes. Certain patients with severe congenital neutropenia carry mutations in the GFI1 gene that lead to the disruption of the C-terminal zinc finger domains. Other mutations have been found in the GFI1B gene in families with inherited bleeding disorders. In addition, the Gfi1 locus is frequently found to be a proviral integration site in retrovirus-induced lymphomagenesis, and new, emerging data suggest a role of Gfi1 in human leukemia and lymphoma, underlining the role of both factors not only in normal hematopoiesis, but also in a wide spectrum of human blood diseases.
Biochemical functions of Gfi1 and Gfi1b
Domain structure and DNA recognition motif
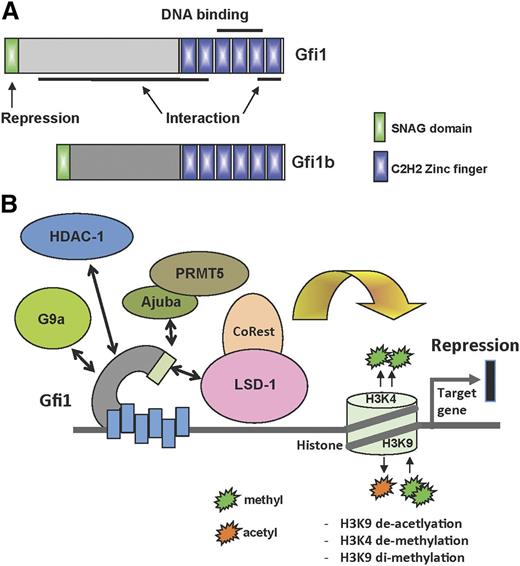
Gfi1 and Gfi1b have 3 identifiable domains: an N-terminal repressor domain 20 amino acid in length called the “SNAG” (SNAIL/GFI1) domain, a C-terminal DNA-binding domain with 6 highly conserved C2H2-type zinc fingers, and a less well-characterized middle region separating the SNAG and zinc finger domains (Figure 1A).
Structure and function of Gfi1 and Gfi1b. (A) Schematic structure of Gfi1 and Gfi1b with their respective domains. (B) Depiction of the different Gfi1 interaction partners and their ability to modify histones at Gfi1 target gene loci.
Structure and function of Gfi1 and Gfi1b. (A) Schematic structure of Gfi1 and Gfi1b with their respective domains. (B) Depiction of the different Gfi1 interaction partners and their ability to modify histones at Gfi1 target gene loci.
Whereas the amino acid sequences of the N- and C-terminal domains are very highly conserved between Gfi1 and Gfi1b, the middle part differs completely between the 2 proteins.1-6 It is not known whether this divergent middle domain performs a specific function in each of the 2 proteins or whether its role is to provide structural flexibility, enabling the interaction of these factors with other proteins (Figure 1A). Interactions between Gfi1 or Gfi1b and other proteins have been shown to require zinc fingers 1, 2, and 6, whereas zinc fingers 3 to 5 bind to a consensus DNA recognition motif with an AATC core sequence (taAATCac(t/a)gca)7,8 (Figure 1A). Although there is no evidence that Gfi1 and Gfi1b also act as transcriptional activators in mice, the Gfi1 homolog in Drosophila, called Senseless (Sens), exhibits a dual role of both activator and repressor.9 Similar to its vertebrate homologs, Sens can function as a transcriptional repressor when bound to DNA as suggested by mutational analysis of its zinc fingers. However, Sens can also bind to bHLH proneural proteins via its core zinc finger domains and can then function as a coactivator of the genes induced by proneural proteins.
Regulation of chromatin structure through histone modification
Based on their biochemical function as transcriptional repressors and recruiters of histone modifiers, both Gfi1 and Gfi1b can be considered as epigenetic regulators that modify chromatin structure. Gfi1 and Gfi1b recruit histone methyl transferases such as G9A, histone demethylases such as LSD1, and histone deacetylases (HDACs) to promoters of target genes and possibly also to other regions (Figure 1B). Both Gfi1 and Gf1b modify chromatin structure mostly to repress transcription.10-12 It is not known, however, whether Gfi1 proteins interact with these histone methyl transferases, demethylases, and deacetylases at the same time or sequentially. Neither do we have any information on whether the interaction between Gfi1, G9a, LSD1, and HDACs is mutually exclusive or only occurs in certain cell types or only acts on specific target genes. In addition, because only LSD1, but neither G9a nor HDACs, interacts with Gfi1 via its SNAG domain, the role of this domain, which is required and indispensable for the repressor activity of both Gfi1 proteins, still remains to be entirely understood. Disrupting the SNAG domain by a “knockin” mutation, which exchanges the second amino acid (proline) for an alanine, reproduces the full Gfi1 knockout phenotype in mice, confirming that the interaction with LSD1 is critical for the function of Gfi1.13 Another protein called Ajuba, which is an LIM (Lin11, Isl-1, and Mec-3) domain-containing protein, has also been found in a multiprotein complex with Gfi1 and HDAC1-3, with the Ajuba LIM domains directly interacting with Gfi1.14 In this study, the interaction between Gfi1 and Ajuba seems to be independent of the SNAG domain but was shown to be important for Gfi1 autoregulation. However, another study showed that Ajuba can recruit PRMT5 via its LIM domain to the SNAG domain of Snail, a Gfi1-related factor15 and points to the possibility that Gfi1, like Snail, may recruit an Ajuba-Prmt5 complex to target genes and mediate arginine methylation indicating that the participation of Gfi1 in multiprotein complexes that regulate chromatin structure and function provides a large range of potential tissue-specific and temporal regulatory possibilities.
Expression of Gfi1 and Gfi1b during normal hematopoiesis
Differential expression in hematopoietic stem cells (HSCs) and precursors
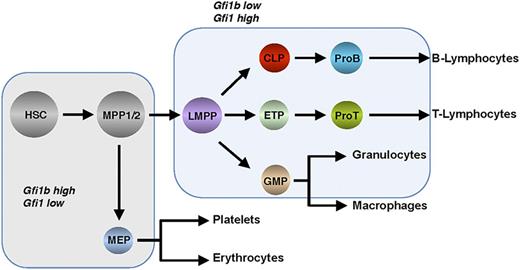
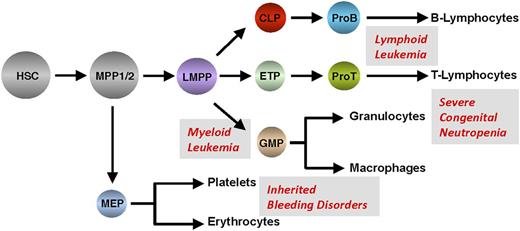
Hematopoiesis occurs in the adult bone marrow and mediates the formation of all blood and immune cells (Figure 2). Through a precisely controlled and perpetual process of self- renewal, proliferation, and differentiation, all of the hematopoietic lineages develop from HSCs.16-19 Transcription factors play a major role during hematopoiesis and represent a very distinct layer of regulation, and the zinc finger proteins Gfi1 and Gfi1b exemplify such regulatory factors20-24 (Figure 1A). The generation of mice transgenic for Gfi1 and Gfi1b promoter sequences linked to reporter genes has made it possible to analyze the expression patterns of both genes during hematopoiesis25,26 (Figure 2). Such experiments with murine cells revealed that both Gfi1 and Gfi1b are expressed in HSCs and in MPP1 and MPP2, which have lost the self-renewal capacity of HSCs but remain multipotent and thus can differentiate into all mature cell types found in the blood27-29 (Figure 2). Gfi1b expression is much higher than Gfi1 expression in the earliest HSC compartment, and its expression is downregulated upon differentiation to MPP1, MPP2, and LMPPs, whereas Gfi1 expression is progressively upregulated in these progenitors (Figure 2).29 In addition, Gfi1 is expressed in lymphoid precursor cells such as CLPs and ETPs that settle the thymus and, subsequently, in the early stages of T- and B-cell development,25,27,30,31 but also in GMPs and in monocytes and granulocytes.25
Hierarchical structure of hematopoiesis depicting the stepwise differentiation from hematopoietic stem and precursor cells into effector cells of the main lineages. Shaded fields indicate Gfi1 and Gfi1b expression levels. CLP, common lymphoid progenitor; ETP, early thymic progenitor; GMP, granulocyte-monocyte progenitor; LMPP, lymphoid primed multipotent progenitor; MEP, megakaryocyte-erythroid progenitor; MPP, multipotent progenitor.
Hierarchical structure of hematopoiesis depicting the stepwise differentiation from hematopoietic stem and precursor cells into effector cells of the main lineages. Shaded fields indicate Gfi1 and Gfi1b expression levels. CLP, common lymphoid progenitor; ETP, early thymic progenitor; GMP, granulocyte-monocyte progenitor; LMPP, lymphoid primed multipotent progenitor; MEP, megakaryocyte-erythroid progenitor; MPP, multipotent progenitor.
In contrast, expression of Gfi1b is absent in CLPs or ETPs and in GMPs, as well as in subsequent maturation stages of most of the cells of both the lymphoid and myeloid compartments. Rather, Gfi1b expression is highest in megakaryocyte-erythrocyte progenitors and during erythroid and megakaryocytic maturation,26,32 where Gfi1 is not readily detected (although a recent report suggests that Gfi1 can also affect erythroid development33 ). B-cell progenitors express both Gfi1 and Gfi1b,26 but resting peripheral B and T cells do not express readily detectable levels of either Gfi1 or Gfi1b, but Gfi1 levels rise upon antigenic activation in both types of lymphoid cells,25,34 Gfi1 being particularly important for T helper 2 cells.35-37 Finally, Gfi1 expression has been detected in dendritic cells, and although preliminary data show that it is required for cytokine signaling in this compartment, little is known about the function of Gfi1 or Gfi1b in these cells to date.38,39
Target genes and gene expression programs
A chromatin immunoprecipitation sequencing analysis with murine leukemic cells transduced with a virus directing expression of an MLL-ENL oncofusion protein allowed the identification of Gfi1 target genes.40 Previously discovered direct Gfi1 target genes, such as Hoxa9, Pbx1, Meis 1,41 and M-CSFR,42 were confirmed to be occupied by Gfi1 at promoter sites, and additional target genes were found or confirmed such as Id2, PU.1, or IL-6Ralpha.40 Overall, the analysis showed that thousands of genes were occupied by Gfi1 within 2 kb (up or downstream) of promoters.40 The significance of Gfi1 occupation outside promoters, which was also observed with this data set, remains to be elucidated. A similar chromatin immunoprecipitation sequencing analysis was done for Gfi1b using the murine HPC7 stem cell line and revealed that >2000 genes were found occupied by Gfi1b at promoters, but also demonstrated that Gfi1b was found at >6000 intergenic and intragenic locations.43 Typical Gfi1b target genes include BclxL, Socs1, Socs3, Cdkn1a, Gata3, Meis1, and Rag1/2.4,39,44-48
Although Gfi1 and Gfi1b bind to sequences in the vicinity to promoter elements, both factors have also been shown to regulate gene expression by binding to distal enhancer elements as in the case of HSCs and mast cells where Gfi1b binds to an enhancer element at −83 kb of the GATA2 gene.49 Similarly, Gfi1b can bind to the B cell–specific Erag enhancer, which regulates the recombination activating genes Rag1/2.50 In the latter case, Gfi1b binds to a site 5′ of the enhancer, which results in epigenetic changes at the Rag locus and the downregulation of its expression.50 By contrast, an activation of Gfi1 and Gfi1b by distal enhancers can occur in medulloblastoma as a result of structural variation, a process called “enhancer hijacking,”51 and a similar mechanism could well be at play in hematologic malignancies.
Genome-wide messenger RNA expression profiles generated from Gfi1- or Gfi1b-deficient cells revealed a role of Gfi1 both in early progenitor cells such as LMPPs and in early T-cell differentiation, but also in early myeloid precursors such as GMPs and in more mature myeloid cells.52-54 Similar analyses with murine Gfi1b-deficient HSCs, megakaryocytes, or erythroid cells demonstrated that Gfi1b helps to establish gene expression programs necessary for the development of both erythroid and megakaryocytic lineages29,32,55 and confirms the complementary roles of Gfi1 and Gfi1b in hematopoiesis already suspected by their differential, cell type–specific expression pattern.
Biological functions in normal hematopoiesis
Overlapping and distinct functions of Gfi1 and Gfi1b
Gfi1 and Gfi1b exert some overlapping and redundant functions during hematopoiesis that could conceivably be explained by similarities in their domain structures and expression patterns. Indeed, replacement of the Gfi1 coding sequence by Gfi1b using gene targeting in mice resulted in almost normal hematopoiesis proving that Gfi1b can largely substitute for Gfi1 in blood cell formation.13 This is likely explained by the highly conserved SNAG and zinc finger domains of both proteins but also suggests that the nonconserved middle domain in Gfi1 and Gfi1b performs an equivalent function in both proteins, at least during hematopoiesis. Gfi1 knockout mice reach adulthood, but Gfi1 ablation affects HSCs in their self-renewal ability and control of cell cycle progression,27 most likely because of a higher sensitivity to stress or DNA damage.56 In addition, Gfi1-deficient mice display defects in B- and T-cell development that may be at least partially explained by an imbalance between Gfi1 and the Ets transcription factor PU.1, which are connected in a regulatory network35,57,58 (see “Function in regulatory circuits during cell lineage determination”). It has also been reported that Gfi1 and PU.1 can directly interact.42
Among the most remarkable features of Gfi1 knockout animals is the expansion of GMPs and a strong accumulation of myelomonocytic precursors accompanied by an almost complete absence of neutrophil granulocytes. One of the consequences of this developmental arrest is that Gfi1 knockout mice display severe neutropenia and die quickly after exposure to bacterial infections.59-63 Moreover, myeloid precursors from constitutive Gfi1-deficient mice cannot differentiate into granulocytes, even after treatment with either granulocyte colony-stimulating factor (G-CSF) or granulocyte macrophage–CSF. However, granulocyte macrophage–CSF is sufficient to generate mature macrophages from Gfi1-deficient precursors.54 The developmental block toward granulopoiesis is intrinsic to the hematopoietic lineage because irradiated congenic mice transplanted with Gfi1-deficient bone marrow show the same lack of granulocyte differentiation,27 suggesting that Gfi1 controls the commitment of progenitors such as GMPs for granulo-monocytic differentiation.54,60
Both Gfi1 and Gfi1b are expressed in HSCs, and gene knockout experiments clearly indicate that Gfi1 restricts proliferation of these cells and is required for their self-renewal capacity.27,28 Although Gfi1-deficient HSCs are more numerous in gene-deficient mice, they are more sensitive to DNA damage indicating that Gfi1 exerts an important mechanisms of protection in these cells.56 In contrast, deletion of Gfi1b leads to a strong expansion of HSCs in bone marrow and in peripheral blood.29 In addition, Gfi1b-deficient HSCs maintain their self-renewal capacity and have an enhanced ability to reach the bloodstream; however, the concomitant ablation of both Gfi1 and Gfi1b is incompatible with HSC survival in mice.29
Function in regulatory circuits during cell lineage determination
A more precise role of Gfi1 in myeloid differentiation has been described in a study from Laslo and colleagues,57,58 which shows that PU.1 expression levels determine whether macrophage or neutrophil cell fate is favored in bipotential myeloid precursors. More specifically, Laslo and colleagues have demonstrated that the transcription factors Egr-1 and Egr-2 both regulate macrophage cell fate by activating the expression of macrophage genes and by repressing neutrophil-specific genes.58 Egr-1,2 can counteract Gfi1, which is required for neutrophil differentiation and achieves this by repressing macrophage-specific genes. This Egr-1/2/Gfi1 regulatory network determines macrophage vs neutrophil cell fate in the presence of lineage determinants such as PU.1 and CCAAT/enhancer binding protein α (C/EBPα). In this model, Egr-1,2/Gfi1 represent a so-called counterregulatory switch necessary to resolve mixed lineage gene expression patterns to regulate cell fate determination.57,58 This series of elegant experiments was used in a mathematical model to propose a new regulatory circuit.58 Recently, another regulatory model of myeloid differentiation has been proposed that is based on the interaction of 11 transcription factors, 1 of which is Gfi1. In this study, the differentiation of HSCs into 4 myeloid cell types has been taken into account, and a literature-derived gene regulatory network was designed that, in addition to Gfi1, includes the factors GATA1, GATA2, FOG1, EKLF, Fli-1, SCL, C/EBPα, PU.1, cJun, and Egr/Nab.64
Gfi1 is expressed in early hematopoietic progenitors such as MPPs that maintain both lymphoid and myeloid potential, and PU.1 is proposed to function in a dose-dependent manner to regulate B-lymphoid vs macrophage cell fates in a regulatory network that potentially also involves the Id proteins, which inhibit E2A activity and as a consequence block the potential of MPPs to develop along the B-lymphoid lineage.65 Because it has been shown that Gfi1 can repress Id genes,66 it is conceivable that Gfi1 is required to activate E2A to promote B-cell differentiation. Findings that Gfi1-deficient mice show defects in B-cell development that can be rescued by the ablation of PU.1 are consistent with such a regulatory circuit. In this model, a reduced concentration of PU.1, which promotes B-lymphoid development, is achieved by Ikaros and Gfi1, which both restrict PU.1 expression while promoting the expression of B-lymphoid genes.35 Hence, another regulatory network that controls lymphoid vs myeloid cell fate determination was proposed that is similar to the Egr1,2/Gfi1 circuit with the exception that the transcription factor Ikaros replaces C/EBPα as a primary determinant.35
Although less data are available, it is likely that Gfi1b also participates in regulatory networks that determine lineage fate in multipotent progenitors, in particular in those that control megakaryocyte and erythrocyte differentiation. The constitutive genetic ablation of Gfi1b in mice arrests embryonic development at approximately embryonic day 15.5. Two independent studies agree that the embryonic lethality in Gfi1b knockout mice is most likely because of defective erythropoiesis32,67 and also suggest that Gfi1b is important for megakaryopoiesis and platelet formation. In addition, adult conditional knockout mice carrying floxed alleles and transgenes for doxycycline- or pIpC-inducible Cre expression reach similar conclusions about the effect of Gf11b deletion for erythropoiesis and megakaryopoiesis,29,55 because the loss of Gfi1b in both models was characterized by a failure to produce Gfi1b-deficient red blood cells and platelets.29,55 In direct comparison with this phenotype, the ablation of Gfi1 affects erythroid differentiation much less and thus allows embryos to progress to term and the offspring to reach adulthood, suggesting that the differing functions of the 2 proteins appear to be primarily attributable to differences in their temporal and cell-specific expression and not differences in their primary sequence.
Gene expression data from a large set of single primary blood stem and progenitor cells have recently validated 2 putative regulatory interactions predicted from bioinformatic analysis, namely, that Gfi1 directly represses Gata2 expression, whereas Gata2 activates Gfi1b, suggesting that Gata2, Gfi1, and Gfi1b form a regulatory circuit that modulates the antagonism between Gfi1 and Gfi1B that had previously been established.49,68 Moreover, Gata2 inhibits lymphopoiesis and is downregulated along with Gfi1b, whereas Gfi1 is upregulated in progenitors that maintain both lymphoid and myeloid potential. This suggests that direct downregulation of Gata2 and Gfi1b by Gfi1 may represent a key event during the specification of early lymphoid cells.49
Lymphoma and leukemia
T-cell lymphoma and acute T-cell lymphoid leukemia (T-ALL)
A retroviral tagging experiment using Moloney murine leukemia virus (MMLV) that was designed to find genes conferring interleukin-2 independence to T cells led to the discovery of the rat Gfi1 gene as a proviral insertion site.1 A few years later, the MMLV infection of transgenic mice predisposed to T-cell lymphoma by constitutive expression of Myc or Pim oncogenes in lymphoid cells was used to identify new genes that could cooperate with Myc or Pim-1 in malignant transformation. In these experiments, the Gfi1 gene turned out to be one of the most frequent insertion sites selected in T-cell lymphomas that arose in infected Myc- or Pim-1-transgenic mice.2,3,6,69 Because the proviral insertions were associated with a high-level expression of Gfi1 in these tumors, Gfi1 was suspected to function as a cooperating partner of Myc or Pim-1 in tumorigenesis. That Gfi1 indeed bears such an oncogenic potential was confirmed by the rapid induction of malignant T-cell lymphoma in Myc- or Pim-1-transgenic mice crossed to mice with targeted T-cell–specific overexpression of Gfi1.2 For Gfi1b, such unequivocal evidence for activity as a dominant oncogene has not been found to date. However, an analysis of MMLV-induced B-cell lymphomas in Eµ-Myc-transgenic mice has shown that Gfi1b can also be selected as a proviral insertion site in tumors, albeit at lower frequencies than Gfi1,70 suggesting that depending on the cellular context and cooperating factors, both genes can act as dominant oncogenes when overexpressed.
Whether genes identified as proviral insertion sites in a retroviral tagging experiment such as Gfi1 would be relevant for human T-cell lymphomagenesis or T-cell leukemia remains an open question. However, the finding that the oncogenes c-Myc and Notch, which are very frequent targets of proviral insertion,70 are also activated in human T-cell leukemia either by chromosomal translocation or by mutations raises the possibility that expression of human GFI1 could also play a role in T-cell leukemogenesis. Comparison of gene expression profiles from T-ALL patients characterized by NOTCH1 mutation status and NOTCH1 target gene expression with those diagnosed of early T-cell precursor ALL showed that ETP-ALL patients had low levels of GFI1 expression compared with those with a NOTCH1 signature, where GFI1 expression was higher.52 This suggested that GFI1 overexpression contributes to the process of malignant transformation in subsets of human T-ALL. Notably, during experimental T-cell tumorigenesis in mice, Gfi1 expression is maintained at high levels in the preleukemic phase but is downregulated upon development of a full-blown leukemia, suggesting that Gfi1 exerts a more complex function in tumorigenesis than can be inferred from a simple overexpression experiment.71 Supporting this view, T-ALL patients who display a relatively lower level of GFI1 expression in blast cells at presentation relapsed at a higher frequency after therapy than patients who exhibited a higher level of GFI1 in the blast cells at initial diagnosis.72
Experiments with conditionally deficient mice have shown that Gfi1 is required for efficient T-cell tumorigenesis, regardless of whether malignant transformation was initiated retrovirally, by chemical carcinogenesis, or by Notch1 activation.52 This effect could also be shown for human cells because inhibition of GFI1 in primary human T-ALL inhibited their expansion in immune-deficient mice. More specifically, this study provided evidence that Gfi1 is an “oncorequisite” factor required for T-cell lymphomagenesis by limiting the ability of p53 to induce apoptosis during malignant transformation; that is, loss of Gfi1 led to reactivation of p53 in T-cell tumor cells and induced cell death. Recent work from Huser and colleagues73 also suggested that data from retrovirally induced cancer models have wide implications for the genetics of human lymphomas. They conclude that a wide range of genes can accomplish the final step in this type of lymphomagenesis with the common end point of growth factor–independent proliferation.73 They also suggest that the action of a small network of genes that includes Gfi1 is sufficient to overcome mechanisms such p53 activation that would otherwise inhibit malignant transformation.73
Acute myeloid leukemia (AML)
According to the World Health Organization classification, patients with at least 20% blasts in blood or bone marrow and, in addition, patients with typical clonal, recurring cytogenetic abnormalities such as t(8;21) are diagnosed with AML.74 It has been suggested that AML can be divided into 2 distinctive subgroups: AML that follows myelodysplastic syndrome (MDS) or a disease with similar features and AML that arises de novo.74-76 Moreover, AML can also emerge as a secondary cancer as a side effect of an initial nonrelated anticancer chemotherapy or radiation treatment.74 Although direct evidence that the loss of Gfi1 expression is linked to AML has not yet been documented, Gfi1-deficient mice show a block in myeloid differentiation, an accumulation of myelo-monocytic cells, and an expansion of GMPs, which are myeloid progenitors.41,56 Other studies show that loss of Gfi1 accelerates the development of a fatal myeloproliferative syndrome (MPS) in mice either in cooperation with an activated K-Ras gene or in the presence of constitutive Bcl-2 expression.41,56 The only potential connection between GFI1 and AML is the finding that a human variant GFI1 allele, in which a coding single nucleotide polymorphism (rs34631763) causes a serine to arginine exchange at amino acid position 36,77 occurs with a frequency of 0.04 in healthy white individuals but shows an increased frequency in AML patients. Hence, carriers of this variant allele have a 1.6-fold increased risk to develop AML.77 The presence of 1 GFI136N allele alone is insufficient to induce AML in mice but induces the expansion of GMPs and accelerates a K-Ras-induced myeloproliferative disease.40 How GFI136N differs functionally from the more common GFI136S is not known and requires further investigation. Initial experiments indicate that GFI136N has a different subnuclear localization than GFI136S and may act differently in the presence of the oncofusion protein AML-Eto in regulating target genes.77 GFI136N is less efficient in inducing histone deacetylation and demethylation at the Hoxa9 locus than the more common GFI136S form.40
Inherited hematologic diseases
Severe congenital neutropenia (SCN)
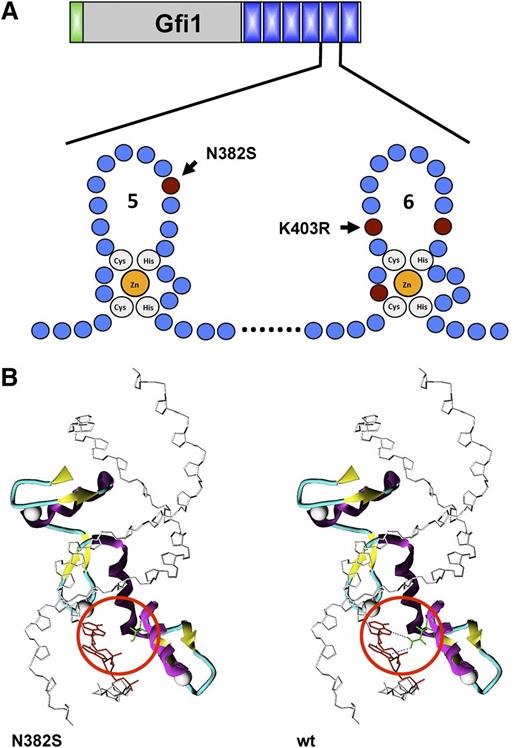
SCN is characterized by the almost complete lack of neutrophils, a condition that leads to recurrent infections and in many patients ultimately to MDS and myeloid leukemia. The observation that Gfi1 knockout mice lack neutrophil granulocytes had prompted a search for germ line mutations in the GFI1 gene in human patients with various forms of neutropenia. Although very rare, mutations in the GFI1 gene associated with hereditary neutropenia have been found in 2 families.59 One family was found in which 2 members suffered from SCN and the affected members carried heterozygous germ line mutations altering the amino acid sequence and disrupting the integrity of either the fifth or sixth zinc finger (N382S and K403R) (Figure 3A-B). It is of interest to note that the neutropenia in the Gfi1-deficient mice similar to the rare SCN patients with GFI1 mutations cannot be alleviated with G-CSF administration,78,79 whereas the vast majority of SCN patients, that is, the most common genetic subtypes (HAX1, ELANE), respond well to G-CSF therapy.80
Effect of mutations on the structure of GFI1. (A) Schematic structure of zing fingers 5 and 6 of Gfi1 and localization of the most common mutations found in families with SCN. The affected amino acids are indicated. (B) Consequence of the N382S mutation on the tertiary structure of zinc finger 5 in the human GFI1 protein: in the wild-type form, Asn-382 forms 2 hydrogen bonds with the DNA strand, which is lost in the GFI1 variant carrying the N382S mutation (red circle).
Effect of mutations on the structure of GFI1. (A) Schematic structure of zing fingers 5 and 6 of Gfi1 and localization of the most common mutations found in families with SCN. The affected amino acids are indicated. (B) Consequence of the N382S mutation on the tertiary structure of zinc finger 5 in the human GFI1 protein: in the wild-type form, Asn-382 forms 2 hydrogen bonds with the DNA strand, which is lost in the GFI1 variant carrying the N382S mutation (red circle).
Experiments suggest that the GFI1N382S mutant acts in a dominant negative manner because it still binds to components of the transcriptional machinery but lacks DNA binding. The second GFI1 mutant (K403R mutation) maintains DNA binding but probably fails to associate with accessory factors, similar to GATA1 mutants that cause thrombocytopenia. Experiments by Zarebski and colleagues demonstrated this experimentally in mice for the N382S mutation.78 Another study from the Netherlands also reported a mutation in a single patient that generated an N382S allele.81 The authors reported that this patient, similar to the 2 patients of the first family with GFI1 mutations,59 showed high numbers of circulating monocytes, which again would be consistent with the observations made in Gfi1 knockout mice. Three additional GFI1 mutations were subsequently detected in patients with SCN (R412X, L400F, and P107A) that all affected the C-terminal part of GFI1 and very likely had the same effect as the 2 originally reported mutations78,81-85 (Figures 4 and 5). Because SCN is associated at least in a subset of patients with progression to MDS and AML, the question arises whether Gfi1knockout mice would show features similar to these diseases later in life. However, the observation of Gfi1 knockout mice over many years has not revealed emergence of those diseases. One reason for this may be the fact that Gfi1-deficient bone marrow cells have a high propensity to undergo cell death56 and may be eliminated before transformation to a leukemic clone can take place, but further studies are required to clarify this issue. Alternatively, Gfi1 may not be involved in those SCN patients that progress to MDS and AML.
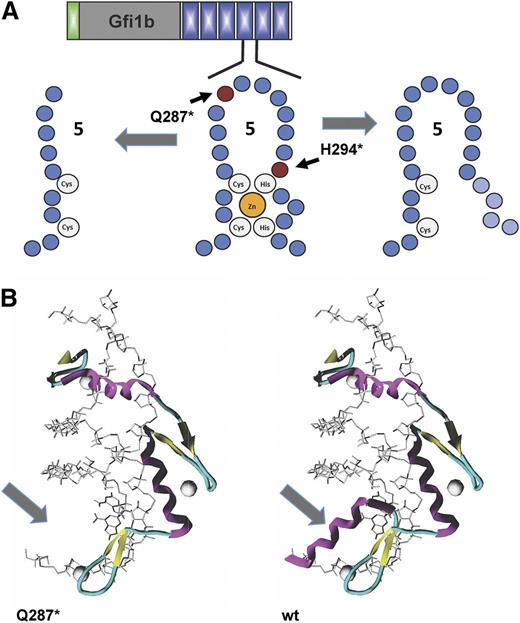
Effect of mutations on the structure of GFI1b. (A) Schematic structure of zinc finger 5 of Gfi1b and localization of the 2 mutations (Q287* and H294fs) found in families with inherited “bleeding disorder platelet-type, 17” (BDPLT17; OMIM 187900) or “Gfi1b-related thrombocytopenia” (GFI1B-RT). (B) The missense Q287* mutation introduces a premature stop codon in GFI1B causing a truncation and loss of the α helix strand in the third zinc finger domain in GFI1b (arrow).
Effect of mutations on the structure of GFI1b. (A) Schematic structure of zinc finger 5 of Gfi1b and localization of the 2 mutations (Q287* and H294fs) found in families with inherited “bleeding disorder platelet-type, 17” (BDPLT17; OMIM 187900) or “Gfi1b-related thrombocytopenia” (GFI1B-RT). (B) The missense Q287* mutation introduces a premature stop codon in GFI1B causing a truncation and loss of the α helix strand in the third zinc finger domain in GFI1b (arrow).
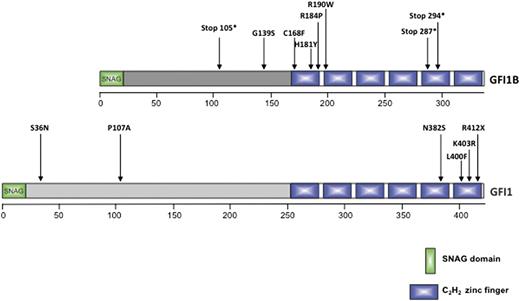
Positions of known mutations in the genes for GFI1 and GFI1B. Shown is a summary of the consequences of the known inherited and congenital mutations in the human GFIB gene (upper) and the human GFI1 gene (lower) for their respective protein sequences. All mutations in the GFI1B gene are associated with platelet disorders, and the mutations in the GFI1 gene are associated with SCN.
Positions of known mutations in the genes for GFI1 and GFI1B. Shown is a summary of the consequences of the known inherited and congenital mutations in the human GFIB gene (upper) and the human GFI1 gene (lower) for their respective protein sequences. All mutations in the GFI1B gene are associated with platelet disorders, and the mutations in the GFI1 gene are associated with SCN.
Platelet deficiencies and bleeding disorders
Whereas the loss of Gfi1 has a clear detrimental effect on the formation of neutrophils, the ablation of Gfi1b causes a rapid loss of platelets29,55 in mice. Very recently, mutations in the GFI1B gene have been described in 2 families with platelet-related bleeding disorders,86,87 illustrating the high clinical relevance of understanding GFI1B’s molecular function in thrombopoiesis. In the study performed by Stevenson and colleagues,86 a novel single nucleotide insertion in the GFI1B gene of patients with inherited bleeding disorders causes a frame-shift mutation affecting amino acid H294 (H294fs). This mutation disrupts the integrity of the fifth zinc finger and eliminates the coding sequence for the sixth zinc finger domain (Figure 4A). The mutated GFI1B protein is unable to bind DNA and loses its capacity to act as a transcriptional repressor. All patients affected by this mutation present with moderate thrombocytopenia, have large platelets, but also exhibit a red cell deficiency called anisopoikilocytosis (ie, red blood cells of unequal size).
In addition, Monteferrario and coworkers87 detected a nonsense mutation in the GFI1B gene of patients with gray platelet syndrome, characterized by low numbers of larger than normal platelets that appear gray in microscopic examination, likely caused by the lack of α granules in the platelets of these patients. The mutation found by this group introduces a premature stop codon at amino acid 287 (Q287*), which leads to a truncated form of GFI1B that lacks the 44 carboxy-terminal amino acids (Figure 4A-B). The mutated allele still expresses normal levels of messenger RNA, and the truncated GFI1B protein was found to be inactive as a repressor of established target genes. Although this mutation is very similar to the one found by Stevenson and colleagues with regard to the biochemical consequences, the affected patients in the study by Monteferrario et al study only showed aberrations in the megakaryocytic, but not in the erythroid, lineage. Since the report of these original discoveries, a number of additional mutations in the GFI1B gene associated with platelet disorders have been found88 (Figure 5).
The findings from these 2 studies with patients suffering from thrombocytopenia support conclusions about Gfi1’s biological role drawn from the analysis of Gfi1b-deficient mice. The most recent characterization of Gfi1b conditionally deficient mice showed that megakaryocytic lineage cells can develop from Gfi1b-deficient progenitors but that this development is arrested at the promegakaryocyte stage, after nuclear polyploidization, but before cytoplasmic maturation.55 Such an arrest at the promegakaryocyte stage is not seen with the ablation of other factors, and it has been proposed that Gfi1b marks a stage of thrombopoiesis where, despite a nuclear maturation, the cytoplasm remains immature.55 However, this and all previous reports on Gfi1b-deficient mice show that both erythropoiesis and platelet formation is affected. Interestingly, heterozygous mice carrying 1 intact allele of Gfi1b are entirely normal, supporting the notion that the mutated forms of GFI1B found in the patient studies mentioned previously act as dominant negative alleles, similar to the aberrant GFI1 proteins found in patients with SCN.59
Summary
Experiments with transgenic or gene-deficient mice and with cell lines overexpressing Gfi1 or mutant versions of Gfi1 have enabled the discovery of important roles of both factors in hematopoiesis and blood cell differentiation. This knowledge has helped to clarify the biochemical function of both factors and to understand how mutations or perturbed expression of Gfi1 and Gfi1b are implicated in specific hematologic diseases in patients that range from congenital neutropenia to inherited bleeding disorders to leukemia and lymphoma (Figure 6). Now, more than 20 years after the initial discovery of Gfi1 and Gfi1b, we have learned so much about their functions that a much clearer picture emerges that underlines their importance in gene regulatory networks that control blood cell formation.
Summary of hematologic diseases associated with perturbed expression or mutations of GFI1 or GFI1B.
Summary of hematologic diseases associated with perturbed expression or mutations of GFI1 or GFI1B.
Acknowledgments
This work was supported by grants from the Canadian Institute for Health Research, the Canadian Foundation for Innovation, and a Canada Research Chair (Tier 1) (T.M.); a Max Eder Grant from Deutsche Krebshilfe, Germany (C.K.); and the National Institutes of Health National Institute on Drug Abuse (grant DA004443, held by Peter Schiller, Institut de recherches cliniques de Montréal) (B.W.).
Authorship
Contribution: T.M., L.V., B.W., and C.K. wrote different parts of the review; T.M. made the figures; B.W. produced material for the figures; and T.M. edited and corrected the text and prepared the final version.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tarik Möröy, Institut de recherches cliniques de Montréal, 110 Avenue des Pins Ouest, Montréal, QC, Canada H2W 1R7; e-mail: tarik.moroy@ircm.qc.ca.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal