Abstract
Purpose: Although abnormal DNA methylation is not limited to promoter CpG islands, studies to date have focused on identifying prognostic methylation markers in gene promoters. The current study aimed at identifying methylation markers for newly-diagnosed AML with normal cytogenetics (AML-NK) using a genome-wide approach.
Patients and Methods: Pretreatment samples were from patients treated on SWOG studies S8600, S9333, S9031, S9500, S9918, S0106, and S0112 from 1986 to 2009. We combined comprehensive high-throughput array-based relative methylation analysis (CHARM) with the Cox proportional hazard model controlling for known AML prognostic factors, to identify 654 survival associated methylation regions (SAMRs) in a phase I discovery cohort (n=72). Top ranking SAMRs were then tested using quantitative bisulfite pyro-sequencing and, if validated, studied further in two additional cohorts: a phase II model building cohort (n=65) and a phase III validation cohort (n=65). In addition, an external cohort from the TCGA AML study (LAML) was used for further validation (n=93).
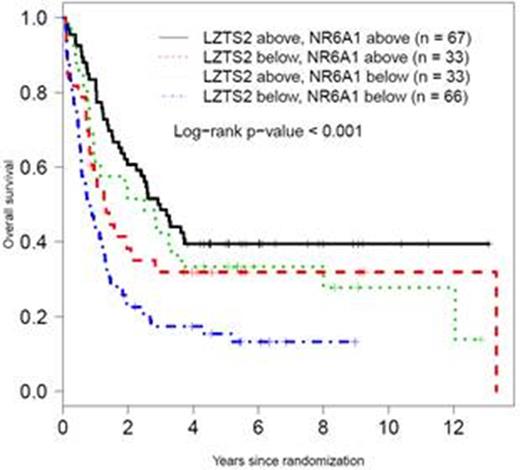
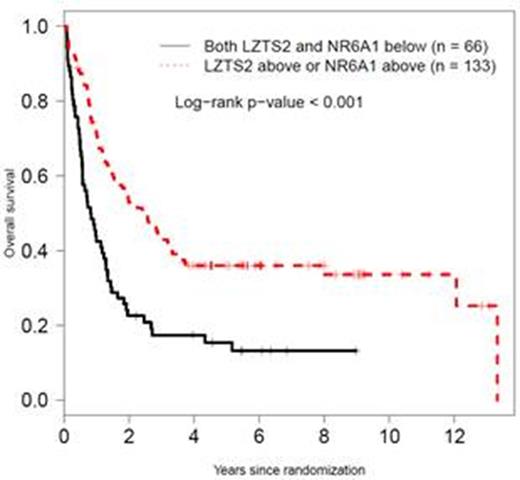
Results: Two SAMRs, located in promoter CpG island shore of LZTS2 and NR6A1, respectively, were identified. Multivariate analyses considering age, performance status, white blood count, and platelet count, showed independent prognostic significance of LZTS2 in the phase II model building cohort (P = 0.0073) and a trend of significance in the phase III validation cohort (P = 0.076), with hypomethylation associated with shorter survival. When FLT3 -ITD status was considered in addition to the above factors, hypomethylation of NR6A1 was shown as an independent marker of shorter survival in both the 2nd and 3rd cohorts (P = 0.02 and 0.03, respectively). Kaplan-Meier analyses of SWOG patients from all three phases further demonstrated that hypomethylation of either LZTS2 or NR6A1 was associated with worse overall survival of AML-NK patients (P < 0.001). The prognosis was verified in AML-NK patients from the TCGA acute myeloid leukemia (LAML) cohort. Multivariate analysis showed that methylation values below median at both markers predicted significantly worse overall survival for both cohorts (SWOG cohort, HR 1.89, P <0.001; TCGA-LAML, HR 2.08, P = 0.006). The impact of the methylation was independent of the FLT3 -ITD status, with C-statistics of 0.71 for both studies. Both LZTS2 and NR6A1 function have been associated with cancer biology. LZTS2 encodes the leucine zipper putative tumor suppressor 2 protein. The over-expression of LZTS2 protein was shown to inhibit cancer cell growth in vitro. The NR6A1 gene encodes a nuclear receptor superfamily member. Increased NR6A1 expression was reported to associate with disease progression and aggressiveness in prostate cancer. The significance of NR6A1 in hematological malignancies has not been described.
Conclusion: We identified two DNA methylation markers that are prognostic for overall survival of AML-NK patients. They warrant further validation in prospective clinical trials.
This study was supported by HOPE Foundation Development Fund and by NIH/NCI National Clinical Trials Network (NCTN) grants CA180888 and CA180819.
Methylation values (below or above median) of LZTS2 and NR6A1 survival associated methylation regions (SAMRs) are associated with overall survival in SWOG AML patients with normal karyotype.
Methylation values (below or above median) of LZTS2 and NR6A1 survival associated methylation regions (SAMRs) are associated with overall survival in SWOG AML patients with normal karyotype.
Yan:EpigenDx, Inc.: Employment. Erba:Sunesis;Pfizer; Daiichi Sankyo; Ariad: Consultancy; Millennium/Takeda; Celator; Astellas: Research Funding; Seattle Genetics; Amgen: Consultancy, Research Funding; Novartis; Incyte; Celgene: Consultancy, Patents & Royalties; GlycoMimetics; Janssen: Other: Data Safety & Monitoring Committees. Fang:Affymetrix: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal