Abstract
Deletion of the 9q21.32 locus is observed in some patients with AML, suggesting an undescribed tumor suppressor resides in this region. HNRNPK is an attractive candidate, as it is one of six genes mapped to this region and is thought to drive AML progression when mutated. Mechanistically, hnRNP K functions as a DNA and RNA binding protein that transcriptionally and translationally governs gene expression. Together, these findings suggest hnRNP K may serve as a currently uncharacterized tumor suppressor and that its haploinsufficiency plays a pivotal role in AML progression.
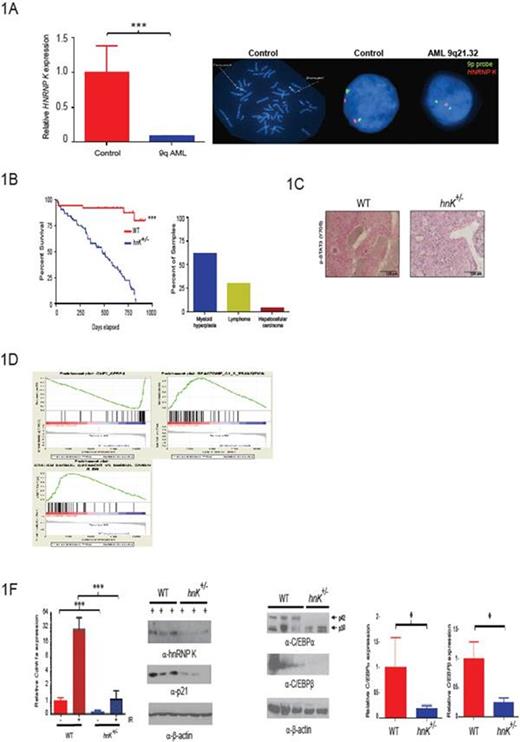
To interrogate a potential relationship between HNRNPK haploinsufficiency and AML, we performed fluorescence in situ hybridization (FISH) using bone marrow aspirates from newly diagnosed AML patients. These analyses revealed the HNRNPK gene is specifically lost in a subset of AML patients and results in a significant decrease in HNRNP K expression in CD34+ cells from patients harboring this deletion (Fig. 1A).
To directly examine the potential tumor-suppressive activities of hnRNP K in vivo, we generated a haploinsufficient hnRNP K (hnRNP K+/-) mouse model. hnRNP K haploinsufficiency resulted in reduced survival and hematologic neoplasms with marked genomic instability (Fig. 1B). Critically, hnRNP K+/- hematopoietic stem progenitor cells (HSPCs) were transplantable and had the capacity to develop hematopoietic neoplasms in recipient mice.
To examine the mechanism driving these tumor phenotypes, we analyzed the cytokine profile of hnRNP K+/- mice. This examination revealed that several critical pro-proliferation and myeloid differentiation cytokines were significantly overexpressed (e.g.; IL-3, IL-6, GM-CSF, and G-CSF), resulting in activation of the JAK-STAT pathway (Fig. 1C). Next, we analyzed the proliferation potential of hnRNP K+/- HSPCs and mouse embryo fibroblast (MEFs) using cell based studies. Here, we observed a significant increase in the number of colony forming units in HSPCs and proliferation potential in both HSPC and MEFs between cells from hnRNP K+/- and wild type mice. Critical to the proliferative advantaged observed in hnRNP K+/- cells, activation of the p53/p21 pathway was significantly reduced in hnRNP K+/- MEFs following exposure to ionizing radiation.
To more fully explore the molecular mechanisms responsible for these phenotypes, we performed whole-transcriptome analyses. Pathway analysis revealed that hnRNP K haploinsufficiency resulted in the attenuation of critical p53- and C/EBP pathways (Fig. 1D). We next validated expression changes in critical genes known to govern tumorigenesis, myeloid differentiation, and proliferation. Here, we observed a significantly down regulation in C/EBPa, C/EBPβ, and p21 at both the transcript and protein level (Fig. 1E-F). To assess the relationship between hnRNP K expression and regulation of these genes, we performed Chromatin Immunoprecipitation (ChIP) and RNA immunoprecipitation (RIP) analyses. ChIP analysis revealed the transcriptional activities of hnRNP K through its direct interaction with the promoter of these genes. Critically, reduced hnRNP K expression significantly diminished these interactions, resulting in their reduced expression. Interestingly, RIP analyses revealed that hnRNP K may also specifically regulate the expression of the tumor suppressive p42 isoform of C/EBPα through its translation regulation of the C/EBPα transcript.
Together, these results suggest hnRNP K is a bona fide tumor suppressor and that its loss contributes to the development of hematologic malignancies. Given its critical regulation of the p53 pathway, drug therapies targeting the p53 pathway may serve as efficacious treatment options for patients with 9q deletions. To address this, we are currently examining the efficacy of Mdm2-p53 antagonist (e.g.; Nutlin-3 and AMG-232) using primary patient samples and primary cells from hnRNP K+/- mice with hematologic malignancies.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal