Abstract
CoMMpass [NCT0145429], a study by the Multiple Myeloma Research Foundation (MMRF), collects longitudinal data of newly diagnosed patients' responses to treatment in the context of their genetic and genomic profiles. The rapidly shifting treatment landscape highlights the importance of a deeper understanding of drug response pathways in order to enable better drug targeting and guide drug development. CoMMpass Interim Analysis 7 (IA7) dataset provides extensive genetic and genomic data on a population of almost 800 enrolled patients. Taking advantage of the rich dataset, we apply REFS™ state-of-the-art Bayesian causal inference engine to reverse-engineer the molecular pathways that most likely affect treatment outcomes in the CoMMpass population and to assess their significance in treatment response.
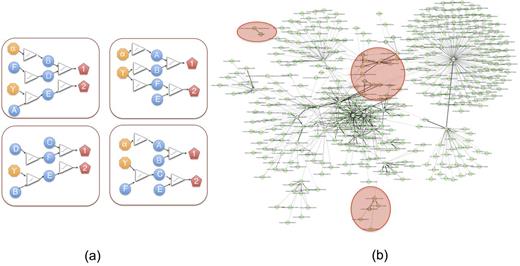
The CoMMpass IA7 dataset, after reducing to patients with reasonably complete clinical and molecular data, comprises 452 patients. We aggregated genetic variables into gene region burden scores, drop mRNA and miRNA variables with excessive zero-inflation, and collate a final model data frame of 28200 variables by 452 patients. We then performed causal modeling constrained only by a minimal set of biological considerations but otherwise entirely de novo. The objective of the modeling is to explain the variability in the dataset and, in particular, in the outcomes, by a set of models that are all consistent with the observed disease biology. Such model ensemble captures uncertainty in inference and highlights similarities among the models, allowing us to distinguish confident predictions from incidental ones. For this study, we constructed an ensemble of 256 models (Figure 1). We investigated our model ensemble by the means of systematic perturbations to model variables, observing effects from such perturbations upon treatment outcomes, an approach that we have successfully applied elsewhere. Thus we are able to assess a prospective effect upon an outcome arising from an in vitro intervention, an adjuvant drug treatment, or an imposition of an enrollment criterion upon a clinical study.
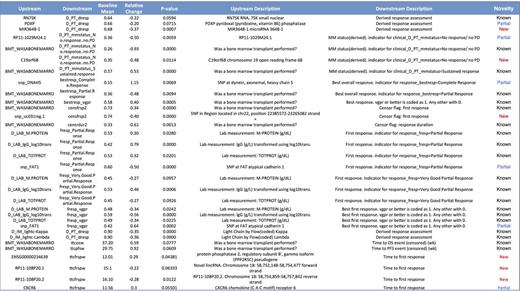
Assessing the effects of these in silico perturbations upon clinical outcomes, we conclude that key drivers of clinical outcomes, as defined by outcomes' sensitivity to perturbation, fall into broad categories of known response drivers, drivers in known pathways, and potential novel biology, or false positives. All three groups are broadly represented among top results (Table 1). The broad range of known disease modifiers, biomarkers, and drivers, such as stem cell transplant, Ig light chain, or RN7SK, identified as such by the REFS™ model de novo, leads us to have a higher confidence in the importance of the other predicted drivers of clinical outcomes, whether partially known (e.g., DNAH5, FAT1) or novel (e.g., mir3648-1, C19orf68). Furthermore, it will be possible to use future CoMMpass Interim Analyses to validate the significance of the predicted novel drivers and to help improve the quality of the causal model, promoting the discovery of further drivers of clinical outcomes.
(a) A REFS model consists of an ensemble of causal networks (b) Consensus (average) causal network topology for the CoMMpass IA7 model, with key outcome variables highlighted
(a) A REFS model consists of an ensemble of causal networks (b) Consensus (average) causal network topology for the CoMMpass IA7 model, with key outcome variables highlighted
Gruber:GNS Healthcare: Employment. Hayete:GNS Healthcare: Employment. Keats:Translational Genomic Research Institute: Employment. Karl:GNS Healthcare: Employment. Lonial:Millennium: Consultancy, Research Funding; Celgene: Consultancy, Research Funding; Onyx: Consultancy, Research Funding; Janssen: Consultancy, Research Funding; Novartis: Consultancy, Research Funding; Bristol-Myers Squibb: Consultancy, Research Funding. Khalil:GNS Healthcare: Employment.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal