Abstract
Our understanding of familial predisposition to lymphoma (collectively defined as non-Hodgkin lymphoma [NHL], Hodgkin lymphoma [HL], and chronic lymphocytic leukemia [CLL]) outside of rare hereditary syndromes has progressed rapidly during the last decade. First-degree relatives of NHL, HL, and CLL patients have an ∼1.7-fold, 3.1-fold, and 8.5-fold elevated risk of developing NHL, HL, and CLL, respectively. These familial risks are elevated for multiple lymphoma subtypes and do not appear to be confounded by nongenetic risk factors, suggesting at least some shared genetic etiology across the lymphoma subtypes. However, a family history of a specific subtype is most strongly associated with risk for that subtype, supporting subtype-specific genetic factors. Although candidate gene studies have had limited success in identifying susceptibility loci, genome-wide association studies (GWAS) have successfully identified 67 single nucleotide polymorphisms from 41 loci, predominately associated with specific subtypes. In general, these GWAS-discovered loci are common (minor allele frequency >5%), have small effect sizes (odds ratios, 0.60-2.0), and are of largely unknown function. The relatively low incidence of lymphoma, modest familial risk, and the lack of a screening test and associated intervention, all argue against active clinical surveillance for lymphoma in affected families at this time.
Introduction
Lymphomas, defined as non-Hodgkin (NHL), Hodgkin (HL), and chronic lymphocytic leukemia (CLL)/small lymphocytic lymphoma, are the most common hematologic malignancies in western countries, and combined there are an estimated 95 520 newly diagnosed cases each year in the United States.1 Although there has been a long history of case reports of familial clustering of lymphomas and leukemias, it has only been relatively recently that these malignancies were considered to have an important inherited genetic component outside of very rare hereditary cancer syndromes.2 In 2001, the World Health Organization introduced an updated classification system for lymphomas based on the Revised European American Lymphoma classification,3 which became the international gold standard.4 This classification provided the first biologically based, integrated framework for consistently defining lymphoma subtypes, thereby greatly facilitating research on this heterogeneous group of diseases.
Building from prior reviews,5-11 we focus on the strongest data addressing familial predisposition (including twin, case-control, and registry-based studies) and germline susceptibility loci (including linkage and genetic-association studies) for lymphoma, and put these findings into clinical context. One emerging theme on the etiology of lymphoma is that there is both commonality and heterogeneity for risk factors by subtype,12 and thus we consider this issue as well in the context of familial predisposition and genetic risk factors.
Evidence for familial predisposition
Twin studies
If the concordance rate of a phenotype in monozygotic twins (who share all genes) is higher than the concordance rate for dizygotic twins (who share on average half of their genes), then there is evidence for a genetic component. In a study of 44 788 pairs of twins from Scandinavia,13 there was an excess of concordant monozygotic twins compared with dizygotic twins for leukemia, and the heritability was estimated to be 21% (95% confidence interval [CI], 0-0.54); these results have been attributed to CLL, as acute lymphoblastic and myeloid leukemia have shown minimal evidence of familial clustering.14 There were insufficient cases to estimate heritability for NHL or HL. In a twin study of lymphomas,15 there was a 100-fold higher risk of HL in monozygotic twins of patients with HL compared to background rates (standardized incidence ratio = 99; 95% CI, 48-182), whereas there was no excess risk in dizygotic twins; in contrast, there was a 23-fold higher risk of NHL in monozygotic twins of patients with NHL and a 14-fold higher risk in dizygotic twins, suggesting a stronger role for shared environment for NHL.
Familial aggregation
We summarize the strongest results across different study designs that evaluate the extent that family history of lymphoma is associated with risk of developing lymphoma, including case-control, cohort, and registry-based studies. We note that none of these study designs can definitively establish an inherited genetic contribution to risk of lymphoma, as these approaches are unable to distinguish the role of shared genetics from a shared environment. Family size itself may also be associated with lymphoma risk, which can introduce bias in estimating the association of familial aggregation with lymphoma risk (Table 1).
Risk of lymphoma subtypes by family history of selected cancers in first degree relatives
| . | . | . | Family history of lymphoma subtype in first-degree relative* . | |||||
|---|---|---|---|---|---|---|---|---|
| Outcome . | Study Design . | References . | NHL . | HL . | CLL/Leukemia . | DLBCL . | FL . | LPL/WM . |
| NHL | Case-control | 12 | 1.8 (1.5-2.1) | 1.7 (1.2-2.3) | 1.5 (1.3-1.8) | — | — | — |
| Registry | 22 | 1.7 (1.4-2.2) | 1.4 (1.0-2.0) | 1.3 (0.9-1.9) | — | — | — | |
| CLL | Case-control | 12 | 1.9 (1.4-2.6) | 1.3 (0.6-2.6) | 2.4 (1.9-3.1) | — | — | — |
| Registry | 23 | 1.9 (1.5-2.3) | 1.5 (1.0-2.3) | 8.5 (6.1-12) | 1.0 (0.4-2.5) | 1.6 (0.9-2.8) | 4.0 (2.0-8.2) | |
| DLBCL | Case-control | 12 | 1.8 (1.5-2.3) | 2.1 (1.4-3.2) | 1.2 (0.9-1.5) | — | — | — |
| Registry | 24 | — | 2.4 (P < .05) | — | 9.8 (3.1-31) | No increase† | — | |
| FL | Case-control | 12 | 2.0 (1.6-2.5) | 1.5 (0.9-2.4) | 1.0 (0.7-1.3) | — | — | — |
| Registry | 24 | — | 1.4 (P > .05) | 1.8 (1.0-3.3) | No increase† | 4.0 (1.6-9.5) | — | |
| LPL/WM | Case-control | 12 | 1.2 (0.5-2.8) | 2.2 (0.5-9.4) | 2.2 (1.2-4.0) | — | — | — |
| Registry | 25 | 3.0 (2.0-4.4) | 0.8 (0.3-2.2) | 3.4 (1.7-6.6) | — | — | 20 (4.1-98) | |
| MZL | Case-control | 12 | 1.7 (1.1-2.5) | 2.7 (1.4-5.5) | 1.7 (1.2-2.4) | — | — | — |
| MCL | Case-control | 12 | 2.0 (1.1-3.3) | 1.5 (0.5-5.0) | 2.0 (1.2-3.2) | — | — | — |
| PTCL | Case-control | 12 | 1.7 (0.9-3.1) | 0.9 (0.1-4.4) | 1.8 (1.1-3.1) | — | — | — |
| Registry | 24 | — | No increase† | No increase† | No increase† | No increase† | — | |
| HL | Case-control | 26 | 3.3 (1.3-8.0) | 3.3 (0.5-22) | 6.3 (1.3-30) | — | — | — |
| Registry | 24,27 | 1.3 (0.9-1.8) | 3.1 (1.8-5.3) | 2.1 (1.2-3.8) | 2.0 (1.1-4.0) | 1.4 (P > .05) | — | |
| . | . | . | Family history of lymphoma subtype in first-degree relative* . | |||||
|---|---|---|---|---|---|---|---|---|
| Outcome . | Study Design . | References . | NHL . | HL . | CLL/Leukemia . | DLBCL . | FL . | LPL/WM . |
| NHL | Case-control | 12 | 1.8 (1.5-2.1) | 1.7 (1.2-2.3) | 1.5 (1.3-1.8) | — | — | — |
| Registry | 22 | 1.7 (1.4-2.2) | 1.4 (1.0-2.0) | 1.3 (0.9-1.9) | — | — | — | |
| CLL | Case-control | 12 | 1.9 (1.4-2.6) | 1.3 (0.6-2.6) | 2.4 (1.9-3.1) | — | — | — |
| Registry | 23 | 1.9 (1.5-2.3) | 1.5 (1.0-2.3) | 8.5 (6.1-12) | 1.0 (0.4-2.5) | 1.6 (0.9-2.8) | 4.0 (2.0-8.2) | |
| DLBCL | Case-control | 12 | 1.8 (1.5-2.3) | 2.1 (1.4-3.2) | 1.2 (0.9-1.5) | — | — | — |
| Registry | 24 | — | 2.4 (P < .05) | — | 9.8 (3.1-31) | No increase† | — | |
| FL | Case-control | 12 | 2.0 (1.6-2.5) | 1.5 (0.9-2.4) | 1.0 (0.7-1.3) | — | — | — |
| Registry | 24 | — | 1.4 (P > .05) | 1.8 (1.0-3.3) | No increase† | 4.0 (1.6-9.5) | — | |
| LPL/WM | Case-control | 12 | 1.2 (0.5-2.8) | 2.2 (0.5-9.4) | 2.2 (1.2-4.0) | — | — | — |
| Registry | 25 | 3.0 (2.0-4.4) | 0.8 (0.3-2.2) | 3.4 (1.7-6.6) | — | — | 20 (4.1-98) | |
| MZL | Case-control | 12 | 1.7 (1.1-2.5) | 2.7 (1.4-5.5) | 1.7 (1.2-2.4) | — | — | — |
| MCL | Case-control | 12 | 2.0 (1.1-3.3) | 1.5 (0.5-5.0) | 2.0 (1.2-3.2) | — | — | — |
| PTCL | Case-control | 12 | 1.7 (0.9-3.1) | 0.9 (0.1-4.4) | 1.8 (1.1-3.1) | — | — | — |
| Registry | 24 | — | No increase† | No increase† | No increase† | No increase† | — | |
| HL | Case-control | 26 | 3.3 (1.3-8.0) | 3.3 (0.5-22) | 6.3 (1.3-30) | — | — | — |
| Registry | 24,27 | 1.3 (0.9-1.8) | 3.1 (1.8-5.3) | 2.1 (1.2-3.8) | 2.0 (1.1-4.0) | 1.4 (P > .05) | — | |
DLBCL, diffuse large B-cell lymphoma; FL, follicular lymphoma; LPL, lymphoplasmacytic lymphoma; MCL, mantle cell lymphoma; MZL, marginal zone lymphoma; PTCL, peripheral T-cell lymphoma; WM, Waldenstrom macroglobulinemia.
ORs (for case-control design) or RR (for registry design) and 95% CI.
Estimate not reported.
Case-control studies.
In case-control studies, the prevalence of a family history is compared in case patients to that of controls using an odds ratio (OR) to quantify the magnitude of risk. The largest study to date is a pooled analysis of 17 471 NHL cases and 23 096 controls from 20 case-control studies in the International Lymphoma Epidemiology Consortium,12 which found an 1.8-fold increased risk of NHL (OR = 1.8, 95% CI, 1.5-2.1) for those with a first-degree (blood) relative with NHL; there was also elevated NHL risk for individuals who reported a first-degree relative with HL (OR = 1.7, 95% CI, 1.2-2.3) or leukemia (OR = 1.5, 95% CI, 1.3-1.8), suggesting susceptibility across these lymphomas.
Further evaluation of NHL subtypes in Table 1 reveals that risk of CLL was only slightly stronger for a family history of leukemia (OR = 2.4) than for NHL (OR = 1.9). In contrast, risk of DLBCL and FL were more strongly associated with a family history of NHL (ORs, 1.8-2.0) than for leukemia (ORs = 1.0-1.2), whereas MZL, MCL, and PTCL showed similar risks for either type of family history (ORs = 1.7-2.0). A family history of HL was associated with increased risk of DLBCL (OR = 2.1) and MZL (OR = 2.7), but was not significantly associated with the risk of CLL, FL, LPL/WM, MCL, or PTCL, although ORs were above 1.0 for all subtypes except PTCL. In a comprehensive analysis of all subtypes simultaneously, there was no statistically significant heterogeneity across risk of most common NHL subtypes for either a family history of NHL (PHomogeneity = 0.52) or HL (PHomogeneity = 0.47). In contrast, there was strong evidence for heterogeneity for a family history of leukemia (PHomogeneity = 3.9 × 10−5), with family history of leukemia most strongly associated with risk of CLL, LPL/WM, MCL, and PTCL. Of note, the associations for family history of NHL with risk of NHL12 or specific NHL subtypes (eg, DLBCL,16 FL,17 CLL,18 MZL,19 LPL/WM,20 and PTCL21 ) remained unchanged after adjusting for extensive subtype-specific risk factors, suggesting that the association of family history may be predominately driven by shared genetics over a shared environment (Table 1).12,22-27
Although the International Lymphoma Epidemiology Consortium did not report pooled results for risk of HL, a large case-control study conducted in Scandinavia26 reported an elevated risk of HL with a family history of HL (OR = 3.3), NHL (OR = 3.3), and CLL (OR = 6.3). Some smaller studies have reported larger ORs for risk of HL with a family history of HL.28,29
These data provide strong evidence for familial predisposition to lymphoma. However, the case-control study design is susceptible to several types of bias, particularly selection and reporting bias. The former bias can occur when there are systematic differences in how cases and controls are enrolled, most commonly due to exclusion of more aggressive cases (who die before they can be enrolled into a study) and how controls are selected (ie, controls who are not representative of the underlying population that generated the cases due to selection factors or participation rates). The main concern with reporting bias is that cases and controls can differentially report a family history. In a study from Scandinavia that compared self-report to cancer registry data,30 specificity of reporting a hematologic malignancy was very high for both cases (98%) and controls (99%), whereas sensitivity was much lower at 60% for cases and only 38% for controls. This led to inflated ORs (up to 30%) based on self-reported family history data.
Cohort studies.
Prospective cohort studies overcome many of the limitations of case-control studies, but there are few cohorts that have had detailed data on lymphoma in family members or a sufficient number of lymphoma outcomes to assess risk of specific NHL subtypes. In a national cohort study of 3.5 million people in Sweden born between 1973 and 2008, family history of HL in a parent or sibling was associated with a 7.2 and 8.8-fold higher risk of childhood/young adult HL, respectively,31 whereas another study reported a sixfold higher risk for siblings.32 In a cohort study of over 120 000 female teachers in California,33 a history of lymphoma in a first-degree relative was associated with a 1.7-fold higher risk of B-cell NHL (relative risk [RR] =1.74; 95% CI, 1.16-2.60) based on 478 cases; data on risk for NHL subtypes were not available. The latter finding was highly consistent with pooled case-control data (Table 1) and suggests a lack of major biases at least for the overall NHL association.
Registry-based studies.
Another major approach to evaluate familial aggregation is to link population-based family registry data with cancer registry data to determine the excess risk of cancer in people with a family history of cancer. Advantages of this approach include population-based assessment, which minimizes selection bias and enhances generalizability, and validation of cancer diagnoses through the use of cancer registries, which eliminates reporting bias. Based on data from the Utah Population Database and the Utah Cancer Registry,34 the risk of NHL was increased 1.7-fold in first-degree relatives of a proband with NHL (familial RR = 1.68; 95% CI, 1.04-2.48) and the risk of lymphocytic leukemia was greater than fivefold in first-degree relatives of a proband with lymphocytic leukemia (familial RR = 5.69; 95% CI, 2.58-10.0). In contrast, the risk of HL was only elevated 1.3-fold in first-degree relatives of a HL proband (familial RR = 1.27; 95% CI, 0.12-3.65), although power for this estimate was low (only 2 exposed cases). Using updated data and a different analytic approach that estimates the Genealogical Index of Familiality,14 excess relatedness was observed for NHL, HL, and CLL. For CLL and NHL, but not HL, the excess relatedness was observed for both distant and overall relatedness. Distant relatedness is due to distant relatives and may be interpreted as providing evidence that familial clustering is more likely due to shared genetic vs shared environmental contribution, as the latter would be lower in distant relationships.
The most comprehensive data available on familial aggregation by lymphoma subtypes has been published using registry data from Sweden and Denmark (summarized in Table 1). This approach compares the cancer experience in first-degree relatives of lymphoma patients with the cancer experience in relatives of matched population controls. First-degree relatives of HL patients had a 3.1-fold increase in risk of HL (95% CI, 1.8-5.3), whereas risk of HL was not associated with a family history of NHL (RR = 1.3; 95% CI, 0.9-1.8) but was associated with a family history of CLL (RR = 2.1; 95% CI, 1.2-3.8).27 In other registry-based studies, the risk of HL in first-degree relatives of HL patients has ranged from 1.2 to 5.8.35-37 Across studies, the risk of HL is stronger for HL in siblings than in parents.27,31,35,36
First-degree relatives of cases with NHL had a 1.7-fold higher risk of developing NHL (95% CI, 1.4-2.2), whereas the risk of NHL was weaker and not statistically significant for first-degree relatives with HL (RR = 1.4; 95% CI, 1.0-2.0) or CLL (RR = 1.3; 95% CI, 0.9-1.9).22 First-degree relatives of CLL patients had an 8.5-fold increased risk of CLL (RR = 8.5; 95% CI, 6.1-12), whereas the risk of CLL was also increased with a first-degree relative with NHL (RR = 1.9; 95% CI, 1.5-2.3) or HL (RR = 1.5; 95% CI, 1.0-2.3).23 It is notable that most of the risk estimates from the population-based registry studies in Table 1 were very similar or only modestly weaker than the estimates from the pooled case-control studies, again suggesting that there was only modest bias in estimates from case-control studies. The most prominent exception is for a family history of CLL, which showed a much stronger association in the registry studies compared with that of case-control studies. This may be in part due to the confusion of patients reporting a CLL as a leukemia or lymphoma.
The registry studies have also been able to evaluate risk for more detailed lymphoma subtypes. One striking finding is the clustering of risk by NHL subtype. For example, first-degree relatives of DLBCL cases had a 9.8-fold increased risk of DLBCL,24 first-degree relatives of FL had a fourfold increased risk of FL,24 and first-degree relatives of LPL/WM had a 20-fold increased risk of LPL/WM.25 In contrast, the risk of a different subtype was much weaker, and notably, relatives of DLBCL patients were not at increased risk of FL and relatives of FL patients were not at increased risk of DLBCL.24 There is very limited data on PTCL, and registry data suggests no increased risk among first-degree relatives with HL, CLL, DLBCL, or FL.24
Summary.
Multiple lines of data suggest that a family history of lymphoma is associated with an increased risk of lymphoma, familial risk is elevated for multiple lymphoma subtypes, and familial risk does not seem to be confounded by nongenetic risk factors, although there are likely unidentified risk factors and clustering of known (and unknown) risk factors within families that are difficult to exclude. This suggests at least some shared genetic etiology across the lymphoma subtypes. However, because a family history of a specific lymphoma subtype is also most strongly associated with a risk for that specific lymphoma, genetic factors are also likely to be unique to a subtype.
Genetic risk factors
We now review studies that show not only clear evidence of a genetic contribution to lymphoma risk, but also provide chromosomal locations that are associated with risk.
Linkage studies
Linkage studies use multicase families or sib pairs to screen the genome in an unbiased manner to identify chromosomal regions that show excessive sharing of inherited alleles among affected individuals. These regions can then be interrogated for causal variants using a variety of approaches, most commonly fine-mapping using dense genotyping, or sequencing. The expectation is to identify highly penetrant variants of modest to large effect size, although these variants are generally rare or very rare in the general population. Linkage studies in HL have identified both HLA class I (for Epstein-Barr virus [EBV]+) and class II (for EBV−) risk, and protective alleles and haplotypes.10,11 Beyond HLA, linkage studies in CLL,38 HL,39 and WM40 have not definitively identified genes with large effects, and there are no published studies in FL, DLBCL, or other NHL subtypes. For CLL, significant linkage was identified at 2q21.2, which contains the chemokine receptor (CXCR4) gene and for which rare coding mutations have been identified.41 The lack of strong findings for these linkage studies may be due to small sample sizes, but also raises the hypothesis that multiple, low-to-moderate risk variants that are common in the population, defined as minor allele frequency (MAF) >5%, may be more relevant in lymphoma etiology than single, highly penetrant variants that are very rare, which is referred to as the common-disease, common-variant hypothesis.42
Genetic association studies
With the advent of high-throughput and relatively inexpensive genotyping technologies, case-control studies (also commonly called association studies in the genetics literature) of sequence variation in germline DNA have become a predominant study design in genetic epidemiology.43 This design is a very efficient strategy to identify low penetrance alleles relative to linkage studies, which are underpowered for this task.44 The most common type of genetic variation in the human genome is the single nucleotide polymorphism (SNP), which is a single base-pair change in the DNA sequence. In this setting, the SNP allele or genotype frequencies in cases (patients) are compared with that of unrelated controls (who do not have the phenotype of interest) using an OR. When the genetic model (eg, dominant vs recessive) is not known a priori, the OR is typically modeled as “per risk allele” (ie, ordinal test of 0, 1, or 2 risk alleles). Although other genetic variations are of interest, including rare variants (<5% frequency), insertion/deletions, block substitutions, inversions, translocations and copy number alterations, these have not been studied as extensively.45 Two major types of association studies are candidate gene and genome-wide association studies (GWAS).
Candidate gene studies
The choice of a candidate gene has been mainly driven by a priori biologic knowledge of lymphoma and diseases associated with lymphoma (eg, infectious or autoimmune), or results identified in other cancers. Candidate gene studies have included pathways related to immune function, cell cycle/proliferation, apoptosis, DNA repair, and carcinogen metabolism. Early studies tended to evaluate a small number of genes (ie, <5) and were generally restricted to 1 or 2 SNPs within a gene. These SNPs often had some evidence for their functionality based on laboratory data or anticipated changes in protein coding or gene activity (eg, changes in promotor function). As genotyping technologies increased in throughput and decreased in cost, more SNPs within genes and more genes (often grouped into pathways) were assessed. Also, the International Haplotype Map46 and later the 1000 Genomes47 projects, which catalog human genetic variation, became available as a reference and allowed “tagging” of genes and gene regions to take advantage of linkage disequilibrium (LD) to efficiently cover all of the common genetic variations for more comprehensive genotyping studies.43
Although many studies of candidate genes have been published7-9,11,48 , most findings have failed to replicate likely due to study design, bias from population stratification (ie, confounding by race or ethnicity), small sample size (low power), uncontrolled multiple testing (leading to false-positive associations), and unrealistic expectations in our ability to choose variants and genes.49 The most robust findings have been for an LTA-TNF haplotype with DLBCL (P = 2.93 × 10−8)50,51 ; an SNP (rs3789068) in the proapoptotic BCL2L11 gene and risk for B-cell NHL (OR = 1.21; P = 2.21 × 10−11)52 ; SNPs in CASP8/CASP10 and risk of CLL53 ; an SNP (rs3132453) in PRRC2A in HLA class III and risk of B-cell NHL (OR = 0.68; P = 1.07 × 10−9)52 ; and certain HLA alleles in class I (including HLA-A*01 and *02) with EBV+ HL and class II (including HLA-DRB1) with EBV− HL.11
GWAS
In contrast to candidate gene/pathway studies, GWAS uses dense microarrays with a large number of SNPs (commonly 250 000 to 750 000 or more) spread across all chromosomes to identify genetic markers associated with case-control status.54 Although SNPs on these platforms have generally focused on common variants (MAF ≥5%), more recent arrays are enriching for rarer variants (MAF <5%). GWAS is considered agnostic (“hypothesis-free”) as all loci are considered equally. Given the large number of statistical tests involved, a stringent level of evidence (currently P < 5 × 10−8) and replication across multiple independent studies are required to declare an association as “genome-wide significant.” An advantage of having a large number of typed SNPs is that any underlying difference in population structure between cases and controls can be identified and controlled to ensure that confounding by race/ethnicity does not bias the results (Table 2).55-75
GWAS-discovered loci for lymphoma
| Subtype . | Locus . | SNP . | Nearest Gene . | RAF* (controls) . | OR† . | P . | Reference . |
|---|---|---|---|---|---|---|---|
| CLL | 2p22.2 | rs3770745 | QPCT, PRKD3 | 0.22 | 1.24 | 1.68 × 10−8 | 55 |
| CLL | 2q13 | rs13401811 | ACOXL, BCL2L11 | 0.81 | 1.41 | 2.08 × 10−18 | 55 |
| CLL | 2q13 | rs17483466 | ACOXL, BCL2L11 | 0.20 | 1.39 | 2.36 × 10−10 | 56 |
| CLL | 2q33.1 | rs3769825 | CASP10/CASP8 | 0.45 | 1.19 | 2.50 × 10−9 | 55 |
| CLL | 2q37.1 | rs13397985 | SP140, SP110 | 0.19 | 1.41 | 5.40 × 10−10 | 56 |
| CLL | 2q37.3 | rs757978 | FARP2 | 0.11 | 1.39 | 2.11 × 10−9 | 57 |
| CLL | 3q26.2 | rs10936599 | MYNN | 0.75 | 1.26 | 1.74 × 10−9 | 58 |
| CLL | 4q25 | rs898518 | LEF1 | 0.59 | 1.20 | 4.24 × 10−10 | 55 |
| CLL | 4q26 | rs6858698 | CAMK2D | 0.16 | 1.31 | 3.07 × 10−9 | 58 |
| CLL | 5p15.33 | rs10069690 | TERT | 0.25 | 1.20 | 1.12 × 10−10 | 58 |
| CLL | 6p21.31 | rs210142 | BAK1 | 0.70 | 1.40 | 9.47 × 10−16 | 59 |
| CLL | 6p21.32 | rs9273363 | HLA-DQB1 | 0.27 | 1.24 | 2.24 × 10−10 | 55 |
| CLL | 6p21.32 | rs674313 | HLA-DRB5 | 0.26 | 1.69 | 6.92 × 10−9 | 60 |
| CLL | 6p25.3 | rs872071 | IRF4 | 0.54 | 1.54 | 1.91 × 10−20 | 56 |
| CLL | 6q25.2 | rs2236256 | IPCEF1 | 0.44 | 1.23 | 1.50 × 10−10 | 58 |
| CLL | 7q31.33 | rs17246404 | POT1 | 0.71 | 1.22 | 3.40 × 10−8 | 58 |
| CLL | 8q22.3 | rs2511714 | ODF1 | 0.41 | 1.16 | 2.90 × 10−9 | 58 |
| CLL | 8q24.21 | rs2456449 | CASC19 | 0.36 | 1.26 | 7.84 × 10−10 | 57 |
| CLL | 9p21.3 | rs1679013 | CDKN2B-AS1 | 0.52 | 1.19 | 1.27 × 10−8 | 55 |
| CLL | 10q23.31 | rs4406737 | ACTA2, FAS | 0.57 | 1.27 | 1.22 × 10−14 | 55 |
| CLL | 11p15.5 | rs7944004 | C11orf21 | 0.49 | 1.20 | 2.15 × 10−10 | 55 |
| CLL | 11q24.1 | rs735665 | GRAMD1B | 0.21 | 1.45 | 3.78 × 10−12 | 56 |
| CLL | 12q24.13 | rs10735079 | OAS3 | 0.36 | 1.18 | 2.34 × 10−8 | 61 |
| CLL | 15q15.1 | rs8024033 | BMF | 0.51 | 1.22 | 2.71 × 10−10 | 55 |
| CLL | 15q21.3 | rs7169431 | RFX7 | 0.08 | 1.36 | 4.74 × 10−7‡ | 57 |
| CLL | 15q23 | rs7176508 | RPLP1 | 0.37 | 1.37 | 4.54 × 10−12 | 56 |
| CLL | 16q24.1 | rs305061 | IRF8 | 0.66 | 1.22 | 3.60 × 10−7‡ | 57 |
| CLL | 16q24.1 | rs2292982 | IRF8 | 0.34 | 0.65 | 6.48 × 10−9 | 60 |
| CLL | 18q21.32 | rs4368253 | PMAIP1 | 0.69 | 1.19 | 2.51 × 10−8 | 55 |
| CLL | 18q21.33 | rs4987855 | BCL2 | 0.91 | 1.47 | 2.66 × 10−12 | 55 |
| CLL | 18q21.33 | rs4987852 | BCL2 | 0.06 | 1.41 | 7.76 × 10−11 | 55 |
| CLL | 19q13.32 | rs11083846 | PRKD2 | 0.22 | 1.35 | 3.96 × 10−9 | 56 |
| FL | 3q28 | rs6444305 | LPP | 0.27 | 1.21 | 1.10 × 10−10 | 62 |
| FL | 6p21.32 | rs10484561 | MHC class II | 0.13 | 1.95 | 1.12 × 10−29 | 63 |
| FL | 6p21.32 | rs2647012 | HLA-DQB1 | 0.44 | 0.64 | 2.00 × 10−21 | 64 |
| FL | 6p21.32 | HLA-DRβ1 Glu-28 | 0.30 | 1.86 | 7.89 × 10−69 | 62 | |
| FL | 6p21.32 | rs17203612 | HLA-DRA | 0.49 | 1.44 | 4.59 × 10−16 | 62 |
| FL | 6p21.33 | rs3130437 | HLA-C | 0.62 | 1.23 | 8.23 × 10−9 | 62 |
| FL | 6p21.33 | rs6457327 | C6orf15 et al (STG) | 0.38 | 0.59 | 4.70 × 10−11 | 65 |
| FL | 8q24.21 | rs13254990 | PVT1 | 0.32 | 1.18 | 1.06 × 10−8 | 62 |
| FL | 11q23.3 | rs4938573 | CXCR5 | 0.20 | 1.34 | 5.79 × 10−20 | 62 |
| FL | 11q24.3 | rs4937362 | ETS1 | 0.46 | 1.19 | 6.76 × 10−11 | 62 |
| FL | 18q21.33 | rs17749561 | BCL2 | 0.91 | 1.34 | 8.28 × 10−10 | 62 |
| DLBCL | 2p23.3 | rs79480871 | NCOA1 | 0.076 | 1.34 | 4.23 × 10−8 | 66 |
| DLBCL | 3q27 | rs6773854 | BCL6/LPP | 0.22 | 1.47 | 1.14 × 10−11 | 67 |
| DLBCL | 6p21.33 | rs2523607 | HLA-B | 0.12 | 1.32 | 2.40 × 10−10 | 66 |
| DLBCL | 6p25.3 | rs116446171 | EXOC2 | 0.019 | 2.20 | 2.33 × 10−21 | 66 |
| DLBCL | 8q24.21 | rs13255292 | PVT1 | 0.32 | 1.22 | 9.98 × 10−13 | 66 |
| DLBCL | 8q24.21 | rs4733601 | PVT1 | 0.48 | 1.18 | 3.63 × 10−11 | 66 |
| MZL | 6p21.32 | rs9461741 | BTNL2 | 0.018 | 2.66 | 3.95 × 10−15 | 68 |
| MZL | 6p21.33 | rs2922994 | HLA-B | 0.11 | 1.64 | 2.43 × 10−9 | 68 |
| HL | 2p16.1 | rs1432295 | REL | 0.40 | 1.22 | 1.91 × 10−8 | 69 |
| HL | 3p24.1 | rs3806624 | EOMES | 0.45 | 1.26 | 1.14 × 10−12 | 70 |
| HL (EBV−) | 5q31 | rs20541 | IL13 | 0.18 | 1.53 | 5.40 × 10−9 | 71 |
| HL | 6p21 | rs2248462 | MICB | 0.22 | 0.61 | 1.30 × 10−13 | 71 |
| HL | 6p21 | rs2395185 | HLA-DRA | 0.33 | 0.56 | 8.30 × 10−25 | 71 |
| HL (EBV+) | 6p21 | rs2734986 | HLA-A | 0.18 | 2.45 | 1.20 × 10−15 | 71 |
| HL (EBV+) | 6p21 | rs6904029 | HCG9 | 0.30 | 0.46 | 5.50 × 10−10 | 71 |
| HL | 6p21.32 | rs2281389 | HLA-DPB1 | 0.83 | 1.73 | 6.31 × 10−13 | 72 |
| HL | 6p21.32 | rs6903608 | HLA-DRA | 0.27 | 1.70 | 2.84 × 10−50 | 69 |
| HL (NS) | 6p21.32 | rs2858870 | HLA-DRB1 | 0.13 | 0.40 | 5.61 × 10−9 | 73 |
| HL (NS) | 6p21.32 | rs204999 | PRRT1 | 0.27 | 0.60 | 2.34 × 10−8 | 73 |
| HL | 6q23.3 | rs7745098 | HBS1L, MYB | 0.48 | 1.21 | 3.42 × 10−9 | 70 |
| HL | 8q24.21 | rs2019960 | PVT1 | 0.23 | 1.33 | 1.26 × 10−13 | 69 |
| HL | 10p14 | rs501764 | GATA3 | 0.19 | 1.25 | 7.05 × 10−8 | 69 |
| HL | 19p13.3 | rs1860661 | TCF3 | 0.41 | 0.81 | 3.50 × 10−10 | 74 |
| LYM | 11q12.1 | rs12289961 | LPXN | 0.28 | 1.29 | 3.89 × 10−8 | 75 |
| Subtype . | Locus . | SNP . | Nearest Gene . | RAF* (controls) . | OR† . | P . | Reference . |
|---|---|---|---|---|---|---|---|
| CLL | 2p22.2 | rs3770745 | QPCT, PRKD3 | 0.22 | 1.24 | 1.68 × 10−8 | 55 |
| CLL | 2q13 | rs13401811 | ACOXL, BCL2L11 | 0.81 | 1.41 | 2.08 × 10−18 | 55 |
| CLL | 2q13 | rs17483466 | ACOXL, BCL2L11 | 0.20 | 1.39 | 2.36 × 10−10 | 56 |
| CLL | 2q33.1 | rs3769825 | CASP10/CASP8 | 0.45 | 1.19 | 2.50 × 10−9 | 55 |
| CLL | 2q37.1 | rs13397985 | SP140, SP110 | 0.19 | 1.41 | 5.40 × 10−10 | 56 |
| CLL | 2q37.3 | rs757978 | FARP2 | 0.11 | 1.39 | 2.11 × 10−9 | 57 |
| CLL | 3q26.2 | rs10936599 | MYNN | 0.75 | 1.26 | 1.74 × 10−9 | 58 |
| CLL | 4q25 | rs898518 | LEF1 | 0.59 | 1.20 | 4.24 × 10−10 | 55 |
| CLL | 4q26 | rs6858698 | CAMK2D | 0.16 | 1.31 | 3.07 × 10−9 | 58 |
| CLL | 5p15.33 | rs10069690 | TERT | 0.25 | 1.20 | 1.12 × 10−10 | 58 |
| CLL | 6p21.31 | rs210142 | BAK1 | 0.70 | 1.40 | 9.47 × 10−16 | 59 |
| CLL | 6p21.32 | rs9273363 | HLA-DQB1 | 0.27 | 1.24 | 2.24 × 10−10 | 55 |
| CLL | 6p21.32 | rs674313 | HLA-DRB5 | 0.26 | 1.69 | 6.92 × 10−9 | 60 |
| CLL | 6p25.3 | rs872071 | IRF4 | 0.54 | 1.54 | 1.91 × 10−20 | 56 |
| CLL | 6q25.2 | rs2236256 | IPCEF1 | 0.44 | 1.23 | 1.50 × 10−10 | 58 |
| CLL | 7q31.33 | rs17246404 | POT1 | 0.71 | 1.22 | 3.40 × 10−8 | 58 |
| CLL | 8q22.3 | rs2511714 | ODF1 | 0.41 | 1.16 | 2.90 × 10−9 | 58 |
| CLL | 8q24.21 | rs2456449 | CASC19 | 0.36 | 1.26 | 7.84 × 10−10 | 57 |
| CLL | 9p21.3 | rs1679013 | CDKN2B-AS1 | 0.52 | 1.19 | 1.27 × 10−8 | 55 |
| CLL | 10q23.31 | rs4406737 | ACTA2, FAS | 0.57 | 1.27 | 1.22 × 10−14 | 55 |
| CLL | 11p15.5 | rs7944004 | C11orf21 | 0.49 | 1.20 | 2.15 × 10−10 | 55 |
| CLL | 11q24.1 | rs735665 | GRAMD1B | 0.21 | 1.45 | 3.78 × 10−12 | 56 |
| CLL | 12q24.13 | rs10735079 | OAS3 | 0.36 | 1.18 | 2.34 × 10−8 | 61 |
| CLL | 15q15.1 | rs8024033 | BMF | 0.51 | 1.22 | 2.71 × 10−10 | 55 |
| CLL | 15q21.3 | rs7169431 | RFX7 | 0.08 | 1.36 | 4.74 × 10−7‡ | 57 |
| CLL | 15q23 | rs7176508 | RPLP1 | 0.37 | 1.37 | 4.54 × 10−12 | 56 |
| CLL | 16q24.1 | rs305061 | IRF8 | 0.66 | 1.22 | 3.60 × 10−7‡ | 57 |
| CLL | 16q24.1 | rs2292982 | IRF8 | 0.34 | 0.65 | 6.48 × 10−9 | 60 |
| CLL | 18q21.32 | rs4368253 | PMAIP1 | 0.69 | 1.19 | 2.51 × 10−8 | 55 |
| CLL | 18q21.33 | rs4987855 | BCL2 | 0.91 | 1.47 | 2.66 × 10−12 | 55 |
| CLL | 18q21.33 | rs4987852 | BCL2 | 0.06 | 1.41 | 7.76 × 10−11 | 55 |
| CLL | 19q13.32 | rs11083846 | PRKD2 | 0.22 | 1.35 | 3.96 × 10−9 | 56 |
| FL | 3q28 | rs6444305 | LPP | 0.27 | 1.21 | 1.10 × 10−10 | 62 |
| FL | 6p21.32 | rs10484561 | MHC class II | 0.13 | 1.95 | 1.12 × 10−29 | 63 |
| FL | 6p21.32 | rs2647012 | HLA-DQB1 | 0.44 | 0.64 | 2.00 × 10−21 | 64 |
| FL | 6p21.32 | HLA-DRβ1 Glu-28 | 0.30 | 1.86 | 7.89 × 10−69 | 62 | |
| FL | 6p21.32 | rs17203612 | HLA-DRA | 0.49 | 1.44 | 4.59 × 10−16 | 62 |
| FL | 6p21.33 | rs3130437 | HLA-C | 0.62 | 1.23 | 8.23 × 10−9 | 62 |
| FL | 6p21.33 | rs6457327 | C6orf15 et al (STG) | 0.38 | 0.59 | 4.70 × 10−11 | 65 |
| FL | 8q24.21 | rs13254990 | PVT1 | 0.32 | 1.18 | 1.06 × 10−8 | 62 |
| FL | 11q23.3 | rs4938573 | CXCR5 | 0.20 | 1.34 | 5.79 × 10−20 | 62 |
| FL | 11q24.3 | rs4937362 | ETS1 | 0.46 | 1.19 | 6.76 × 10−11 | 62 |
| FL | 18q21.33 | rs17749561 | BCL2 | 0.91 | 1.34 | 8.28 × 10−10 | 62 |
| DLBCL | 2p23.3 | rs79480871 | NCOA1 | 0.076 | 1.34 | 4.23 × 10−8 | 66 |
| DLBCL | 3q27 | rs6773854 | BCL6/LPP | 0.22 | 1.47 | 1.14 × 10−11 | 67 |
| DLBCL | 6p21.33 | rs2523607 | HLA-B | 0.12 | 1.32 | 2.40 × 10−10 | 66 |
| DLBCL | 6p25.3 | rs116446171 | EXOC2 | 0.019 | 2.20 | 2.33 × 10−21 | 66 |
| DLBCL | 8q24.21 | rs13255292 | PVT1 | 0.32 | 1.22 | 9.98 × 10−13 | 66 |
| DLBCL | 8q24.21 | rs4733601 | PVT1 | 0.48 | 1.18 | 3.63 × 10−11 | 66 |
| MZL | 6p21.32 | rs9461741 | BTNL2 | 0.018 | 2.66 | 3.95 × 10−15 | 68 |
| MZL | 6p21.33 | rs2922994 | HLA-B | 0.11 | 1.64 | 2.43 × 10−9 | 68 |
| HL | 2p16.1 | rs1432295 | REL | 0.40 | 1.22 | 1.91 × 10−8 | 69 |
| HL | 3p24.1 | rs3806624 | EOMES | 0.45 | 1.26 | 1.14 × 10−12 | 70 |
| HL (EBV−) | 5q31 | rs20541 | IL13 | 0.18 | 1.53 | 5.40 × 10−9 | 71 |
| HL | 6p21 | rs2248462 | MICB | 0.22 | 0.61 | 1.30 × 10−13 | 71 |
| HL | 6p21 | rs2395185 | HLA-DRA | 0.33 | 0.56 | 8.30 × 10−25 | 71 |
| HL (EBV+) | 6p21 | rs2734986 | HLA-A | 0.18 | 2.45 | 1.20 × 10−15 | 71 |
| HL (EBV+) | 6p21 | rs6904029 | HCG9 | 0.30 | 0.46 | 5.50 × 10−10 | 71 |
| HL | 6p21.32 | rs2281389 | HLA-DPB1 | 0.83 | 1.73 | 6.31 × 10−13 | 72 |
| HL | 6p21.32 | rs6903608 | HLA-DRA | 0.27 | 1.70 | 2.84 × 10−50 | 69 |
| HL (NS) | 6p21.32 | rs2858870 | HLA-DRB1 | 0.13 | 0.40 | 5.61 × 10−9 | 73 |
| HL (NS) | 6p21.32 | rs204999 | PRRT1 | 0.27 | 0.60 | 2.34 × 10−8 | 73 |
| HL | 6q23.3 | rs7745098 | HBS1L, MYB | 0.48 | 1.21 | 3.42 × 10−9 | 70 |
| HL | 8q24.21 | rs2019960 | PVT1 | 0.23 | 1.33 | 1.26 × 10−13 | 69 |
| HL | 10p14 | rs501764 | GATA3 | 0.19 | 1.25 | 7.05 × 10−8 | 69 |
| HL | 19p13.3 | rs1860661 | TCF3 | 0.41 | 0.81 | 3.50 × 10−10 | 74 |
| LYM | 11q12.1 | rs12289961 | LPXN | 0.28 | 1.29 | 3.89 × 10−8 | 75 |
LYM, lymphoma; NS, nodular sclerosis.
Risk AF among controls.
OR (per allele).
Considered genome-wide significant at the time of initial publication.
CLL.
The estimated contribution of all common variations to the heritability of CLL is 46% to 59%.55,76 The first GWAS in a lymphoid malignancy was conducted for CLL56 and to date, GWAS analyses55,57-61 have identified 32 SNPs from 28 loci for CLL, which accounts for ∼19% of familial risk of CLL.58 Many of the established SNPs are near or in genes plausibly linked to CLL, including genes involved in apoptosis (including FAS, PMAIP1, BAK1, BCL2, BCL2L11, BMF, and CASP8/CASP10), telomere function (POT1, TERT, and TERC), transcription factors important in B-cell differentiation (IRF8, LEF1, PRKD3, and SP140), and B-cell receptor activation (IRF3 and HLA-DQA1). Notably, there has been little evidence of interaction among these SNPs, suggesting independent effects. None of the SNPs have individually shown a strong relationship with age at diagnosis, although cases diagnosed at a younger age tended to carry a greater number of risk alleles,58 supporting the hypothesis that early onset CLL is enriched for genetic susceptibility.
In an East Asian population, GWAS-discovered SNPs for CLL near IRF4 (rs872071), SP140 (rs13397985), and ACOXL (rs17483466) were associated with CLL risk (nominal P < .05), with a suggestive association with GRAMD1B (rs735665).77 The MAFs of these SNPs were much lower than in populations of European descent, supporting the hypothesis that the lower prevalence of CLL genetic risk factors might explain part of the lower incidence of CLL in East Asian populations.
FL.
Three early GWA studies based on small discovery sets (<400 cases) identified loci at 6p21.3365 and 6p21.3263,64 in the major histocompatibility complex (MHC) associated with FL. In a meta-analysis of those studies plus a new GWAS of over 2100 cases, the HLA region showed overwhelming association with FL, with 8104 SNPs achieving genome-wide significance. A top SNP from this region, rs12195582, reached P = 5.35 × 10−100 after additional validation.62 HLA alleles and amino acids (AA) were imputed and the top signal mapped to four-linked DRβ1 multi-allelic AA at positions at 11, 13, 28, and 30, suggesting an important role for DRβ1 peptide presentation in FL.62 Additional independent signals were also identified in HLA class II (rs17203612) and class I (rs3130437, near HLA-C); after accounting for all of these signals, no other previously identified SNPs from the MHC achieved genome-wide significance. Outside of the HLA region, 5 novel loci have been identified including 11q23.3 (near CXCR5), 11q24.3 (near ETS1), 3q28 (in LPP), 18q21.33 (near BCL2), and 8q24 (near PVT1).62 These genes are linked to B-cell biology making them plausible in the etiology of FL.
HL.
Classical HL (cHL) makes up ∼95% of HL, and cHL compromises several subtypes: in young children and older adults, mixed cellularity HL (typically EBV+) predominates, whereas in adolescents and young adults, nodular sclerosing HL (typically EBV−) predominates.78 Five GWAS analyses have been published in HL,69-71,73,74 and the strongest findings have been for SNPs mapping to HLA class II69,71,73 in close proximity to HLA-DRA and HLA-DRB1, regions previously linked to HL by HLA-typing studies.79,80 The 6p21.32 locus marked by rs6903608 (near HLA-DRA) was associated with cHL overall, and more specifically to EBV− cHL,69,71 early onset,69,72 and young adult nodular sclerosing HL73 (largely EBV−). Additional GWAS signals at 6p21 have been identified in HLA class I,71 with statistically independent associations for rs2248462 (near MICB) for all cHL (irrespective of EBV status); and rs2734986 (3′ untranslated region of HLA-G and near HLA-A) and rs6904029 (near HCG9) for EBV+ cHL. These results confirm earlier studies linking HLA-A*01 and *02 to EBV+ cHL,81-83 and support a role for class I but not class II genes in EBV+ HL. Using SNPs to impute classic HLA alleles, two independent signals in the HLA class II region (rs6903608 and rs2281389) were linked to early onset HL, but no specific classical HLA alleles from this region were significant after conditioning on these two SNPs.72 The class II SNP rs6903608 was estimated to account for ∼6% of the familial risk in HL.72
Outside of the MHC region, GWAS-discovered loci for HL include 2p16.169 (near REL), 10p1469 (near GATA3), 8q24.2169 (telomeric to PVT1 and near MYC), 5q3171 (a nonsynonymous SNP in IL13), 3p24.170 (5′ to EOMES), 6q23.370 (intergenic to HBS1L and MYB), and 19p13.374 (in intron 2 of TCF3), with only the 2p16.1 and 5q31 loci showing stronger associations with EBV (negative) status. Genes from these non-HLA regions are involved in hematopoiesis and immunoregulation, making them plausible susceptibility loci for cHL. HLA and non–HLA-linked loci appear to be independent, and non-HLA loci were estimated to account for ∼7% of the familial risk in HL.70
DLBCL.
In a GWAS conducted in an East Asian population, a locus at 3q27 (near BCL6 and LPP) was identified,67 although this could not be replicated in independent studies of East Asian84 or European ancestry.66 In a large GWAS of European ancestry,66 novel loci identified included 6p25.3 (EXOC2), 6p21.33 (HLA-B), 2p23.3 (NCOA1), and 8q24.21 (near PVT1 and MYC); the strongest finding after imputing HLA alleles and AA was with HLA-B*08:01, although this could not be statistically distinguished from the HLA-B SNP due to high LD. The latter study also estimated that common SNPs, including but not limited to the GWAS-discovered loci, explained ∼16% of the variance in DLBCL risk overall.
Three of the five GWAS-discovered SNPs for DLBCL in Europeans were significantly associated with DLBCL in an East Asian population,84 including EXOC2 (OR = 2.04; P = 3.9 × 10−10), PVT1 (OR = 1.34; P = 2.1 × 10−6), and HLA-B (OR = 3.05; P = .009). Overall, MAFs were similar or only modestly lower in the East Asian population for all SNPs except for one of the 8q24 SNPs, which was much rarer.
MZL.
The only GWAS of this subtype68 identified two distinct loci at 6p21.32 (intragenic to BTNL2, in HLA class II) and 6p21.33 (HLA-B, in HLA class I); these two loci were in low LD and were statistically independent of each other. There was no strong heterogeneity in these results when stratified on mucosa-associated lymphoid tissue vs non–mucosa-associated lymphoid tissue (splenic MZL and nodal MZL) subtypes, although this was based on a modest sample size. These loci are also associated with autoimmune diseases and immune response, suggesting shared biologic underpinnings with MZL.
Lymphoma.
Only one GWAS has been conducted based on all lymphomas (including HL, multiple myeloma, and T-cell cases) as the outcome in both the discovery and validation stages.75 An SNP at 11q12.1 (near LPXN) was identified, and the associations were consistent across the common subtypes. However, this locus has not been replicated in larger GWAS studies based on specific subtypes.
Summary
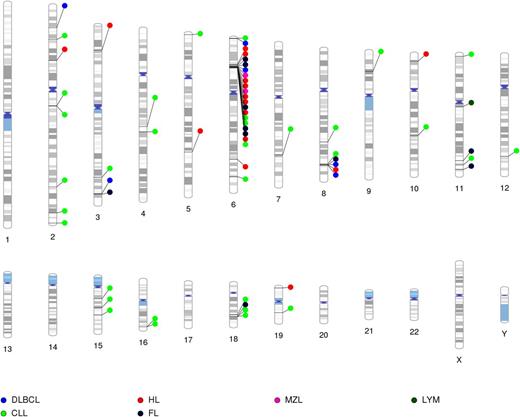
To date, GWAS have successfully identified 67 SNPs from 41 genetic loci, mainly associated with specific subtypes (Figure 1), with only two regions (ie, the HLA region and 8q24) associated with multiple lymphoma subtypes; few candidate gene loci have been replicated by GWAS. As shown in Figure 2, the established loci are common (MAF >5%) and have small effect sizes, supporting a polygenic model for susceptibility. In contrast to GWAS, candidate gene studies in lymphoma have had only minimal success, similar to other cancers.85 Linkage studies have also not been successful in identifying rare alleles causing Mendelian disease, and the evaluation of low-frequency variants with intermediate effects is still in early research phases for lymphoma, but will be challenged by sample size issues.86 The GWAS-identified SNPs that have been identified are largely of unknown function. However, a leading hypothesis related for the mechanistic role of these common SNPs is their effect on gene expression (eg, through effects on promotors or enhancers), but this effect is difficult to identify given an expected modest impact of these SNPs on gene expression and the fact that this impact could occur at any time before diagnosis.58
GWAS-discovered loci for lymphoma subtypes mapped to chromosomal location. Except for 6p21 and 8q24, there is minimal or little overlap of loci for lymphoma subtype-specific susceptibility loci. Lym, lymphoma.
GWAS-discovered loci for lymphoma subtypes mapped to chromosomal location. Except for 6p21 and 8q24, there is minimal or little overlap of loci for lymphoma subtype-specific susceptibility loci. Lym, lymphoma.
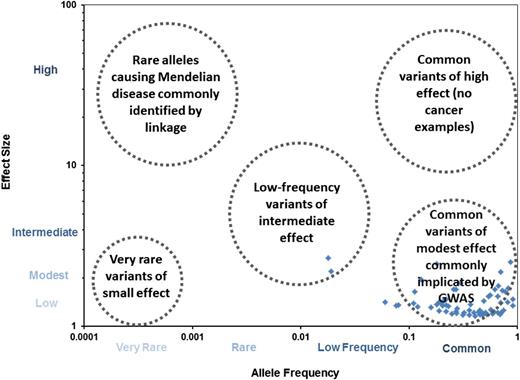
Lymphoma susceptibility loci by effect size and AF. The blue diamonds represent established lymphoma susceptibility loci plotted by AF (x-axis) vs effect size (y-axis). For lymphoma, most of the loci are common variants of low to modest effect size (mainly discovered by GWAS), although a few low-frequency variants have been identified. No rare alleles of low frequency (generally identified through linkage studies and sequencing) have been definitively linked to lymphoma. Very rare variants of low effect size are difficult to identify using current genetic approaches, whereas there are very few examples of common variants of high effect size for common diseases (and none in lymphoma).
Lymphoma susceptibility loci by effect size and AF. The blue diamonds represent established lymphoma susceptibility loci plotted by AF (x-axis) vs effect size (y-axis). For lymphoma, most of the loci are common variants of low to modest effect size (mainly discovered by GWAS), although a few low-frequency variants have been identified. No rare alleles of low frequency (generally identified through linkage studies and sequencing) have been definitively linked to lymphoma. Very rare variants of low effect size are difficult to identify using current genetic approaches, whereas there are very few examples of common variants of high effect size for common diseases (and none in lymphoma).
Practice implications
Given the estimated lifetime risk of NHL is 1 in 48 (2.1%) in the United States1 and an RR of 1.7 for the risk of NHL in a first-degree relative, then the absolute lifetime risk of NHL is 3.6% in first-degree relatives of an NHL patient. The absolute risk is even lower for specific lymphoma subtypes, which are less common. Although the absolute lifetime risk of NHL is not trivial, the relatively low incidence of lymphoma, the modest familial risk, and the lack of a screening test and associated intervention all argue against active clinical surveillance of family members of lymphoma patients at this time. One hope is that genetic risk scores, alone or in combination with other risk factors, might improve prediction ability.87 Although there are currently no validated risk scores for lymphoma, this advance is anticipated as more loci are characterized.
Future directions
Characterization of genetic susceptibility in lymphoma is rapidly evolving. It is expected that additional common variants will be discovered for the different lymphoma subtypes,88 and perhaps pan-lymphoma loci will also be identified. As new lymphoma entities and precursor lesions are defined, the evaluation of heritability and genetic susceptibility should be addressed. Additional work needs to occur in other racial and ethnic groups, particularly with contrasting lymphoma incidence rates. It is not yet clear if rare and low-frequency variants will play a major role in lymphoma susceptibility. This will be challenging to address due to phenotype heterogeneity and the need for large sample sizes for these relatively rare entities, and both family and association study designs along with bioinformatics and laboratory-based studies will all need to be integrated to achieve progress.86 Other genetic mechanisms (eg, copy number variation), epigenetics, and gene-environment interactions are additional frontiers.85 Finally, integrating somatic and germline genomics should provide additional insights into lymphoma etiology and pathogenesis,89 and hopefully provide novel insights into how to prevent and treat this malignancy.
Acknowledgments
The authors thank Dr Thomas Habermann for his critical review of the manuscript, Curtis Olswald for technical assistance, and Sondra Buehler for editorial assistance.
This work was supported by grants from the National Institutes of Health National Cancer Institute (R01 CA92153, U01 CA118444, and P50 CA97274).
Authorship
Contribution: J.R.C. and S.L.S. did the background research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: James R. Cerhan, Department of Health Sciences Research, Mayo Clinic, 200 First St SW, Rochester, MN 55905; e-mail: cerhan.james@mayo.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal