Key Points
Comprehensive study of glucocorticoid-induced osteonecrosis identifies glutamate receptor gene variants as risk factors.
Abstract
Glucocorticoids are important therapy for acute lymphoblastic leukemia (ALL) and their major adverse effect is osteonecrosis. Our goal was to identify genetic and nongenetic risk factors for osteonecrosis. We performed a genome-wide association study of single nucleotide polymorphisms (SNPs) in a discovery cohort comprising 2285 children with ALL, treated on the Children’s Oncology Group AALL0232 protocol (NCT00075725), adjusting for covariates. The minor allele at SNP rs10989692 (near the glutamate receptor GRIN3A locus) was associated with osteonecrosis (hazard ratio = 2.03; P = 3.59 × 10−7). The association was supported by 2 replication cohorts, including 361 children with ALL on St. Jude’s Total XV protocol (NCT00137111) and 309 non-ALL patients from Vanderbilt University’s BioVU repository treated with glucocorticoids (odds ratio [OR] = 1.87 and 2.26; P = .063 and .0074, respectively). In a meta-analysis, rs10989692 was also highest ranked (P = 2.68 × 10−8), and the glutamate pathway was the top ranked pathway (P = 9.8 × 10−4). Osteonecrosis-associated glutamate receptor variants were also associated with other vascular phenotypes including cerebral ischemia (OR = 1.64; P = 2.5 × 10−3), and arterial embolism and thrombosis (OR = 1.88; P = 4.2 × 10−3). In conclusion, osteonecrosis was associated with inherited variations near glutamate receptor genes. Further understanding this association may allow interventions to decrease osteonecrosis. These trials are registered at www.clinicaltrials.gov as #NCT00075725 and #NCT00137111.
Introduction
Cure rates for childhood acute lymphoblastic leukemia (ALL) have approached 90% with therapeutic advances over the last several decades.1-6 Intensification of therapy with glucocorticoids has played a crucial role in achieving these outcomes. However, one of the most common therapy-related and dose-limiting toxicities of therapy in children with ALL is glucocorticoid-induced osteonecrosis, particularly in those older than 10 years of age. The majority of symptomatic cases of osteonecrosis occur within the first 2 years of treatment,7,8 often precipitating early withdrawal of glucocorticoids from therapy for ALL.
The incidence of glucocorticoid-induced osteonecrosis varies widely.7,9 Age remains the most significant risk factor, with symptomatic osteonecrosis (defined as grades 2-4) occurring in 10% to 30% of children older than 10 years of age.7,8,10-12 Glucocorticoid-induced osteonecrosis also complicates treatment of nonmalignant conditions such as solid organ transplant and arthritis.13-15 Osteonecrosis can result in debilitation and adversely affect quality of life, often requiring surgical intervention.
In this study, we conducted the largest genome-wide association study (GWAS) to date of glucocorticoid-induced osteonecrosis, with replication cohorts including not only children treated for ALL7 but also adults and children treated with glucocorticoids for other medical conditions. Our goal was to identify germline genetic variants that predispose to glucocorticoid-induced osteonecrosis.
Methods
Subjects
The discovery cohort included children with newly diagnosed ALL with germline DNA available who were treated on the Children’s Oncology Group (COG) AALL0232 protocol (NCT00075725) for high-risk B-precursor ALL (n = 2285) (Table 1; see supplemental Figure 1 and supplemental Table 1 on the Blood Web site). Validation cohorts included children with newly diagnosed ALL treated on the St. Jude (SJ) Total XV protocol (NCT00137111) (n = 361)7 (supplemental Figure 1 and supplemental Table 2), and a separate cohort comprising children and adults treated with corticosteroids in the Vanderbilt University Medical Center Biorepository BioVU database16 (n = 309) (Table 1; supplemental Figure 1 and supplemental Table 3). Patients included in the genetic association analyses represented 80% (n = 2285 of 2868) of participants on the COG AALL0232 protocol, and 73% (n = 361 of 498) of patients on the SJ Total XV protocol (supplemental Figure 1).
Patient characteristics by cohort
| . | COG AALL0232 . | SJ Total XV . | Vanderbilt BioVU . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| . | All (n = 2285) . | Cases (n = 250) . | Controls (n = 2035) . | All (n = 361) . | Cases (n = 68) . | Controls (n = 293) . | All (n = 309) . | Cases (n = 82) . | Controls (n = 227) . |
| Ancestry classification | |||||||||
| White | 1275 | 164 (12.9%) | 1111 (87.1%) | 260 | 53 (20.4%) | 207 (79.6%) | 216 | 57 (26.4%) | 159 (73.6%) |
| Black | 139 | 4 (2.9%) | 135 (97.1%) | 55 | 6 (10.9%) | 49 (89.0%) | 54 | 15 (27.8%) | 39 (72.2%) |
| Hispanic | 601 | 58 (9.7%) | 543 (90.3%) | 25 | 6 (24%) | 19 (75%) | 14 | 4 (28.6%) | 10 (71.4%) |
| Asian | 48 | 5 (10.4%) | 43 (89.6%) | 4 | 0 (0%) | 4 (100%) | 2 | 1 (50.0%) | 1 (50.0%) |
| Other | 222 | 19 (8.6%) | 203 (91.4%) | 17 | 3 (17.6%) | 14 (82.4%) | 23 | 5 (21.7%) | 18 (78.3%) |
| Gender | |||||||||
| Male | 1244 | 120 (9.6%) | 1124 (90.4%)) | 197 | 37 (18.8%) | 160 (81.2%) | 100 | 29 (29.0%) | 71 (71.0%) |
| Female | 1041 | 130 (12.5%) | 911 (87.5%) | 164 | 31 (18.9%) | 133 (81.1%) | 209 | 53 (25.4%) | 156 (74.6%) |
| Age (y, at diagnosis) | |||||||||
| 1-10 | 817 | 20 (2.4%) | 797 (97.6%) | 270 | 28 (10.4%) | 242 (89.6%) | 13 | 2 (15.4%) | 11 (84.6%) |
| ≥10 | 1468 | 230 (15.7%) | 1238 (84.3%) | 91 | 40 (44.0%) | 51 (56%) | 296 | 80 (27.0%) | 216 (73.0%) |
| . | COG AALL0232 . | SJ Total XV . | Vanderbilt BioVU . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| . | All (n = 2285) . | Cases (n = 250) . | Controls (n = 2035) . | All (n = 361) . | Cases (n = 68) . | Controls (n = 293) . | All (n = 309) . | Cases (n = 82) . | Controls (n = 227) . |
| Ancestry classification | |||||||||
| White | 1275 | 164 (12.9%) | 1111 (87.1%) | 260 | 53 (20.4%) | 207 (79.6%) | 216 | 57 (26.4%) | 159 (73.6%) |
| Black | 139 | 4 (2.9%) | 135 (97.1%) | 55 | 6 (10.9%) | 49 (89.0%) | 54 | 15 (27.8%) | 39 (72.2%) |
| Hispanic | 601 | 58 (9.7%) | 543 (90.3%) | 25 | 6 (24%) | 19 (75%) | 14 | 4 (28.6%) | 10 (71.4%) |
| Asian | 48 | 5 (10.4%) | 43 (89.6%) | 4 | 0 (0%) | 4 (100%) | 2 | 1 (50.0%) | 1 (50.0%) |
| Other | 222 | 19 (8.6%) | 203 (91.4%) | 17 | 3 (17.6%) | 14 (82.4%) | 23 | 5 (21.7%) | 18 (78.3%) |
| Gender | |||||||||
| Male | 1244 | 120 (9.6%) | 1124 (90.4%)) | 197 | 37 (18.8%) | 160 (81.2%) | 100 | 29 (29.0%) | 71 (71.0%) |
| Female | 1041 | 130 (12.5%) | 911 (87.5%) | 164 | 31 (18.9%) | 133 (81.1%) | 209 | 53 (25.4%) | 156 (74.6%) |
| Age (y, at diagnosis) | |||||||||
| 1-10 | 817 | 20 (2.4%) | 797 (97.6%) | 270 | 28 (10.4%) | 242 (89.6%) | 13 | 2 (15.4%) | 11 (84.6%) |
| ≥10 | 1468 | 230 (15.7%) | 1238 (84.3%) | 91 | 40 (44.0%) | 51 (56%) | 296 | 80 (27.0%) | 216 (73.0%) |
Patient characteristics are listed by osteonecrosis cases and controls within each cohort. Ancestry was genomically determined as described in “Methods.”
Informed consent was obtained from patients 18 years and older, and from parents or guardians for patients under the age of 18 in accordance with the Declaration of Helsinki. The COG AALL0232 protocol was approved by the National Cancer Institute and the Institutional Review Boards (IRBs) of participating institutions. The SJ Total XV trial was approved by the SJ IRB.
The second validation cohort for this study was derived from BioVU, Vanderbilt’s repository linking DNA from remnant blood samples to de-identified electronic medical record data.17 Due to de-identification, use of this resource qualifies as nonhuman subjects research. IRB exemption was required by the institution and was obtained prior to the study. Cases of glucocorticoid-induced osteonecrosis were identified using the de-identified electronic medical records in BioVU using keyword searches, International Classification of Disease, 9th edition codes, and exclusion criteria described in supplemental Materials. Controls were selected in a 3:1 (controls:cases) ratio, and matched on age, race, gender, and primary diagnoses. All cases and controls were manually reviewed to confirm inclusion/exclusion criteria (supplemental Figure 2).
Detection of adverse events
Osteonecrosis was graded using the National Cancer Institute Common Terminology Criteria for Adverse Events, version 3.0 for SJ Total XV and version 4.0 for COG AALL0232, and categorized as absent (grade 0), asymptomatic (grade 1), moderate (grade 2), severe (grade 3), or disabling (grade 4). Patients with symptomatic grades 2-4 were considered to have osteonecrosis. For patients on AALL0232, patients with symptoms suggestive of osteonecrosis were evaluated by magnetic resonance imaging (MRI) to verify the diagnosis. In the SJ cohort, all patients were prospectively screened for osteonecrosis with serial MRI, regardless of symptoms. Details for case ascertainment in the Vanderbilt cohort are provided (supplemental Materials and methods).
Genotyping
Single nucleotide polymorphism (SNP) genotyping was performed on germline DNA from patients from the COG AALL0232 and SJ Total XV protocols using the Illumina Human Exome BeadChip v1.1 and Affymetrix GeneChip Human Mapping Array 6.0. The Illumina Exome Chip, Omni1-Quad, 1M-Duo, and Human660W-Quad BeadChip Arrays were used for the Vanderbilt BioVU cohort. For SNPs not interrogated on the arrays, imputation was performed using the 1000 Genomes Project (http://www.1000genomes.org/) database as the reference genome, leveraging linkage disequilibrium (LD) within racial ethnic groups (Northern European ancestry, West African ancestry, and other) with MaCH-Admix software (University of North Carolina) for the COG AALL0232 and SJ Total XV cohorts, and IMPUTE2 (University of Oxford, United Kingdom) for the Vanderbilt cohort.18-20 For the imputation, reference groups for imputing SNPs in patients classified as white (see below) were as follows: the European individuals in 1000 Genomes, for patients classified as black were the 1000 Genomes Africans, and the remaining patients were all individuals in the 1000 Genomes database.
Genetic ancestry race classification
Genetic ancestry for each patient was determined using STRUCTURE (version 2.2.3).21 When ancestry was assessed as a categorical variable, individuals were classified as white, black, Hispanic, and Asian based on percentage-inferred genetic ancestry as follows: >90% Northern European (CEU) were classified as white; >70% West African (YRI) were classified as black; >90% East Asian (CHB/JPT) were classified as Asian; those with Native American ancestry >10% and greater than the percentage of West African ancestry were classified as Hispanic. Patients not falling into these groups were categorized as “Other.”
Quality control
Identical quality control measures were enforced for the discovery and validation cohorts. All SNPs with a call rate <95% were excluded. SNPs with a minor allele frequency (MAF) >1% but deviating from Hardy–Weinberg equilibrium within Europeans (P < .0001) were also excluded. Otherwise, no MAF threshold was enforced.
Statistical analyses
For the discovery GWAS, SNP genotypes were compared in 250 ALL osteonecrosis cases and 2035 controls enrolled on COG AALL0232. Adjusting for gender, age, percent ancestry as a continuous variable, and treatment (see supplemental Materials and methods for details on consideration of treatment variables), association of genotypes with ON was tested with a Cox proportional hazard model for time-dependent analyses and logistic regression for time-independent analyses. For the time-independent analyses, only patients with a follow-up time of 800 days or greater from the start of therapy on COG AALL0232 were included in the analysis. Results from analyses with imputed SNPs and each independent platform were merged and rank ordered by P value. Analyses were performed using R software (version 3.0; www.r-project.org).
We excluded rare/low frequency SNPs (MAF <0.1) with a protective, negative correlation with osteonecrosis risk (odds ratio [OR] <1).
Time-independent analyses were performed using a logistic regression model in the 2 validation cohorts: 68 osteonecrosis cases and 293 controls from the SJ Total XV cohort as previously published,7 and 82 cases and 227 controls from the Vanderbilt BioVU cohort. Meta-analysis of the discovery and validation cohorts was performed using Stouffer’s Z-score method.22
Pathway and network analyses were performed to identify biological networks enriched within the top ranked SNPs for each of the cohorts using QIAGEN Ingenuity Pathway Analysis (www.qiagen.com/ingenuity). Top SNPs were selected with meta-analysis P value cutoffs < .0001 or < .001, and genes closest to SNPs based on chromosomal location were used for the pathway analyses.
The phenome-wide association studies (PheWAS) analysis was performed as described23,24 (supplemental Materials and methods) for the 137 SNPs within the glutamate receptor signaling pathway genes. We tested for their associations with 1358 phenotypes using the extant BioVU genotype data linked to electronic health records for over 10 000 individuals.
Results
Covariates included in the GWAS
The overall frequency of symptomatic grades 2-4 osteonecrosis in patients included in the entire COG AALL0232 cohort (all ages) was 10.9%. For the discovery COG AALL0232 cohort, a Cox proportional hazard model with nongenetic and clinical factors was used to identify covariates to include in the GWAS. Multivariate analysis revealed age ≥10 years (hazard ratio [HR] = 11.1; 95% CI, 6.7-18.1; P = 1.57 × 10−21), and female gender (HR = 1.39; 95% CI, 1.08-1.78; P = 9.82 × 10−3) (supplemental Table 1) to be associated with a higher risk of osteonecrosis. African genetic ancestry was associated with a decreased risk of osteonecrosis compared with individuals of European genetic ancestry (HR = 0.19; 95% CI, 0.07-0.51; P = 1.02 × 10−3). Treatment variables A and B (supplemental Materials and methods and supplemental Table 4) were also included as covariates in the GWAS.
GWAS
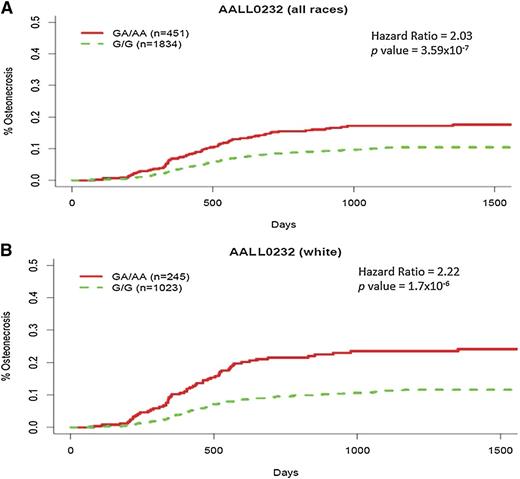
In the discovery GWAS in the COG AALL0232 cohort, adjusting for gender, age, ancestry, and treatment, a variant at 9q31.1 (rs10989692, P = 3.59 × 10−7), ∼170kb 5′ of the GRIN3A locus (glutamate [NMDA] receptor subunit 3A), had the strongest association with osteonecrosis risk in both a time-dependent and time-independent analysis (supplemental Tables 5 and 6; supplemental Figure 3). Patients with an additional A allele at rs10989692 had a higher risk of osteonecrosis (HR = 2.03; 95% CI, 1.55-2.66) (Figure 1A), with 73 of 250 cases carrying a risk allele (69 heterozygous and 4 homozygous). The correlation between rs10989692 genotypes and osteonecrosis was stronger in patients with >90% European ancestry (n = 1268; HR = 2.22; 95% CI, 1.64-2.99) (Figure 1B), risk allele frequency (RAF) in patients with and without osteonecrosis (0.173 vs 0.089; P = 1.7 × 10−6) than in patients of non-European ancestry (n = 1017; HR = 1.63; 95% CI, 0.98-2.63; P = .057) (Figure 2; supplemental Figure 4). SNP rs10989692 was further validated in the Vanderbilt cohort (OR = 2.26; P = .0074), again with a stronger association in whites (n = 216; OR = 2.68; P = .012; RAF: 0.132 in cases vs 0.06 in controls) than in those with non-European ancestries (n = 93; OR = 1.72; P = .26). The effect of rs10989692 in the SJ cohort was marginal when all patients were included (OR = 1.87; P = .063), but reached statistical significance when only patients with >90% European ancestry were considered (n = 260; OR = 2.29; P = .028; RAF: 0.123 in cases vs 0.086 in controls). The RAF was very similar in both the discovery and replication cohorts (0.106, 0.116, and 0.130 for COG, SJ, and Vanderbilt cohorts, respectively). An additional SNP (rs28584318) in high LD with rs10989692 (r2 = 1 and 0.83 in HapMap CEU and African populations, respectively) was also associated with osteonecrosis (P = 4.86 × 10−7; P = .07; P = .0073 in AALL0232, SJ, and Vanderbilt cohorts, respectively).25 We evaluated the effect of the rs10989692 SNP across different ancestral groups by analysis of variance test of the interaction term of the genotype and genetic ancestry in AALL0232. The P value for the interaction term, P = .075, suggests that there may be a difference in the effect size of this polymorphism across ancestral groups. When restricting the analysis to patients >10 years, the risk associated with rs10989692 was maintained in AALL0232 (n = 1468; HR = 2.07; 95% CI, 1.59-2.70; P = 1.44 × 10−6; RAF 0.152 in cases vs 0.0905 in controls) with a similar trend in the SJ cohort that did not reach statistical significance (n = 91; HR = 1.67; 95% CI, 0.55-5.10; P = .37; RAF 0.10 in cases vs 0.077 in controls) (supplemental Tables 7 and 8; supplemental Figure 3C), and interaction analyses between genotype and age did not show a significant interaction between GRIN3A rs10989692 genotype and age (P = .86 and P = .89) in the AALL0232 and TOTALXV cohorts, respectively.
Cumulative incidence of osteonecrosis by rs10989692 genotype in COG AALL0232. The cumulative incidence of osteonecrosis was higher in those carrying the A allele at rs10989692 (5′ of GRIN3A) in COG AALL0232 (A) in all ancestry groups combined, adjusting for ancestry (n = 2285), and (B) among whites only (n = 1268).
Cumulative incidence of osteonecrosis by rs10989692 genotype in COG AALL0232. The cumulative incidence of osteonecrosis was higher in those carrying the A allele at rs10989692 (5′ of GRIN3A) in COG AALL0232 (A) in all ancestry groups combined, adjusting for ancestry (n = 2285), and (B) among whites only (n = 1268).
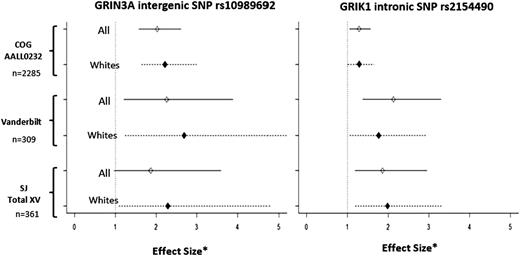
Effect size for rs10989692 and rs2154490 genotypes by cohort. Effect sizes are computed as the HR for COG AALL0232 based on time-dependent analysis, and OR for SJ Total XV and Vanderbilt BioVU based on time-independent analyses. Effect sizes for rs10989692 and rs2154490 are shown after adjusting for age, gender, treatment arm, and ancestry in COG and SJ, and after adjusting for age, gender, and ancestry in Vanderbilt BioVU. Asterisk (*) denotes effect size.
Effect size for rs10989692 and rs2154490 genotypes by cohort. Effect sizes are computed as the HR for COG AALL0232 based on time-dependent analysis, and OR for SJ Total XV and Vanderbilt BioVU based on time-independent analyses. Effect sizes for rs10989692 and rs2154490 are shown after adjusting for age, gender, treatment arm, and ancestry in COG and SJ, and after adjusting for age, gender, and ancestry in Vanderbilt BioVU. Asterisk (*) denotes effect size.
Meta-analysis
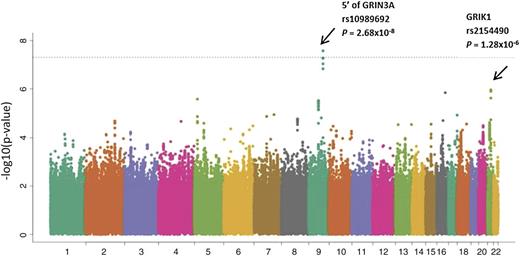
Meta-analysis was performed by combining GWAS results from both the discovery cohort and the 2 validation cohorts. The 9q31.1 locus (near GRIN3A) was the highest ranked region, including multiple SNPs in LD with rs10989692 (P = 2.68 × 10−8) (Table 2; Figure 3). After the 9q31.1 locus, the next highest ranked variant was an intronic SNP rs2154490 in glutamate receptor, ionotropic, kainate (GRIK1) on chromosome 21 (P = 1.28 × 10−6) (Table 2; Figure 3). The additional minor A allele at rs2154490 conferred a higher risk of osteonecrosis in all 3 cohorts: COG AALL0232 (HR = 1.29; P = .016), SJ Total XV (OR = 1.86; P = .0078), and Vanderbilt (OR = 2.13; P = 9.1 × 10−4) (supplemental Figure 5).
Top SNPs from meta-analysis across COG AALL0232, SJ Total XV, and Vanderbilt BioVU cohorts
| . | . | . | . | . | . | . | COG AALL0232 . | SJ Total XV . | Vanderbilt BioVU . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP ID* . | Chr . | Position (HG19)† . | Alleles‡ . | Gene§ . | SNP function . | Meta P . | RAF . | P . | HR (95%CI) . | RAF . | P . | OR (95%CI) . | RAF . | P . | OR (95%CI) . |
| rs10989692 | 9 | 104674555 | G/A | 5′ of GRIN3A | Intergenic | 2.68 × 10−8 | 0.106 | 3.59 × 10−7 | 2.03 (1.55 - 2.66) | 0.116 | 0.063 | 1.87 (0.97 - 3.60) | 0.130 | 0.0074 | 2.26 (1.24 - 4.11) |
| rs2154490 | 21 | 30915962 | G/A | GRIK1 | Intronic | 1.28 × 10−6 | 0.222 | 0.016 | 1.29 (1.05 - 1.56) | 0.233 | 0.0078 | 1.86 (1.18 - 2.94) | 0.240 | 9.1 × 10−4 | 2.13 (1.36 - 3.32) |
| rs72733993 | 5 | 18408908 | G/A | NA | Intergenic | 2.67 × 10−6 | 0.156 | 0.043 | 1.29 (1.01 - 1.64) | 0.102 | 0.0051 | 2.62 (1.34 - 5.15) | 0.119 | 9.4 × 10−4 | 2.89 (1.54 - 5.42) |
| rs11144550 | 9 | 78261548 | G/A | 5′ of PCSK5 | Intergenic | 3.08 × 10−6 | 0.137 | 2.37 × 10−5 | 1.70 (1.33 - 2.17) | 0.136 | 0.0629 | 1.71 (0.97 - 3.01) | 0.078 | 0.046 | 2.04 (1.01 - 4.11) |
| rs1536407 | 13 | 75095567 | C/A | 5′ of KLF12 | Intergenic | 4.43 × 10−6 | 0.646 | 0.0064 | 1.32 (1.08 -1.61) | 0.638 | 0.002 | 2.13 (1.32 - 3.45) | 0.660 | 0.034 | 1.59 (1.03 - 2.44) |
| rs4789693 | 17 | 80421870 | A/C | NARF | intronic | 5.73 × 10−6 | 0.274 | 0.030 | 1.25 (1.02 - 1.54) | 0.245 | 0.013 | 1.86 (1.14 - 3.05) | 0.264 | 0.0014 | 1.95 (1.29 - 2.93) |
| rs6797178 | 3 | 137253713 | G/A | 5′ of SOX14 | Intergenic | 5.74 × 10−6 | 0.390 | 0.025 | 1.24 (1.03 - 1.49) | 0.413 | 0.0023 | 2.00 (1.28 - 3.11) | 0.430 | 0.01 | 1.63 (1.12 - 2.37) |
| rs10849004 | 12 | 4292862 | T/C | 5′ of CCND2 | Intergenic | 5.75 × 10−6 | 0.819 | 0.0029 | 1.47 (1.14 - 1.89) | 0.817 | 3.1 × 10−4 | 3.45 (1.75 -6.67) | 0.791 | 0.20 | 1.39 (0.83 - 2.27) |
| rs11594258 | 10 | 79218030 | T/A | KCNMA1 | Intronic | 8.61 × 10−6 | 0.822 | 2.8 × 10−4 | 1.59 (1.23 - 2.04) | 0.867 | 0.2 | 1.52 (0.80 - 2.86) | 0.842 | 0.0052 | 2.44 (1.30 - 4.55) |
| . | . | . | . | . | . | . | COG AALL0232 . | SJ Total XV . | Vanderbilt BioVU . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP ID* . | Chr . | Position (HG19)† . | Alleles‡ . | Gene§ . | SNP function . | Meta P . | RAF . | P . | HR (95%CI) . | RAF . | P . | OR (95%CI) . | RAF . | P . | OR (95%CI) . |
| rs10989692 | 9 | 104674555 | G/A | 5′ of GRIN3A | Intergenic | 2.68 × 10−8 | 0.106 | 3.59 × 10−7 | 2.03 (1.55 - 2.66) | 0.116 | 0.063 | 1.87 (0.97 - 3.60) | 0.130 | 0.0074 | 2.26 (1.24 - 4.11) |
| rs2154490 | 21 | 30915962 | G/A | GRIK1 | Intronic | 1.28 × 10−6 | 0.222 | 0.016 | 1.29 (1.05 - 1.56) | 0.233 | 0.0078 | 1.86 (1.18 - 2.94) | 0.240 | 9.1 × 10−4 | 2.13 (1.36 - 3.32) |
| rs72733993 | 5 | 18408908 | G/A | NA | Intergenic | 2.67 × 10−6 | 0.156 | 0.043 | 1.29 (1.01 - 1.64) | 0.102 | 0.0051 | 2.62 (1.34 - 5.15) | 0.119 | 9.4 × 10−4 | 2.89 (1.54 - 5.42) |
| rs11144550 | 9 | 78261548 | G/A | 5′ of PCSK5 | Intergenic | 3.08 × 10−6 | 0.137 | 2.37 × 10−5 | 1.70 (1.33 - 2.17) | 0.136 | 0.0629 | 1.71 (0.97 - 3.01) | 0.078 | 0.046 | 2.04 (1.01 - 4.11) |
| rs1536407 | 13 | 75095567 | C/A | 5′ of KLF12 | Intergenic | 4.43 × 10−6 | 0.646 | 0.0064 | 1.32 (1.08 -1.61) | 0.638 | 0.002 | 2.13 (1.32 - 3.45) | 0.660 | 0.034 | 1.59 (1.03 - 2.44) |
| rs4789693 | 17 | 80421870 | A/C | NARF | intronic | 5.73 × 10−6 | 0.274 | 0.030 | 1.25 (1.02 - 1.54) | 0.245 | 0.013 | 1.86 (1.14 - 3.05) | 0.264 | 0.0014 | 1.95 (1.29 - 2.93) |
| rs6797178 | 3 | 137253713 | G/A | 5′ of SOX14 | Intergenic | 5.74 × 10−6 | 0.390 | 0.025 | 1.24 (1.03 - 1.49) | 0.413 | 0.0023 | 2.00 (1.28 - 3.11) | 0.430 | 0.01 | 1.63 (1.12 - 2.37) |
| rs10849004 | 12 | 4292862 | T/C | 5′ of CCND2 | Intergenic | 5.75 × 10−6 | 0.819 | 0.0029 | 1.47 (1.14 - 1.89) | 0.817 | 3.1 × 10−4 | 3.45 (1.75 -6.67) | 0.791 | 0.20 | 1.39 (0.83 - 2.27) |
| rs11594258 | 10 | 79218030 | T/A | KCNMA1 | Intronic | 8.61 × 10−6 | 0.822 | 2.8 × 10−4 | 1.59 (1.23 - 2.04) | 0.867 | 0.2 | 1.52 (0.80 - 2.86) | 0.842 | 0.0052 | 2.44 (1.30 - 4.55) |
Listed are 9 loci with meta P <1 × 10−5. Within each locus, only the SNP with the lowest P value is shown. RAF in each of the 3 cohorts; HR from time-dependent analysis in COG AALL0232 cohort; OR from time independent analysis in SJ Total XV and Vanderbilt BioVU cohorts.
CI, confidence interval.
Rs numbers according to the National Center for Biotechnology Information dbSNP release 139.
Physical location of SNPs based on human genome assembly (hg19).
The second allele listed is the risk allele.
Refseq gene located closest to the SNP within a distance of 1M base pairs.
Manhattan plot of results from meta-analysis. Inverse of log P value for SNP associations with osteonecrosis risk from meta-analysis of COG AALL0232 (n = 2285), SJ Total XV (n = 361), and Vanderbilt BioVU (n = 309), adjusting for age, gender, treatment, and ancestry group in COG AALL0232 and SJ, and adjusting for age, gender, and ancestry groups in Vanderbilt BioVU. SNPs near GRIN3A and within GRIK1 had the strongest association.
Manhattan plot of results from meta-analysis. Inverse of log P value for SNP associations with osteonecrosis risk from meta-analysis of COG AALL0232 (n = 2285), SJ Total XV (n = 361), and Vanderbilt BioVU (n = 309), adjusting for age, gender, treatment, and ancestry group in COG AALL0232 and SJ, and adjusting for age, gender, and ancestry groups in Vanderbilt BioVU. SNPs near GRIN3A and within GRIK1 had the strongest association.
The top nonsense SNP from the meta-analysis (supplemental Table 9) was within pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 (or PLEKHH1) (P = .0023), involved in phosphate binding and implicated in liver lipid homeostasis.26 Another nonsense SNP (P = .0056) was within STEAP4, a ferrireductase involved in osteoclastogenesis27 and adipocyte development.28 Additional missense loci of interest from the meta-analysis include those in ZFHX3, a gene involved in atrial fibrillation and Kawasaki disease29 (P = 1.46 × 10−6), and loci in COL22A1 (collagen, type XXII, α 1; P = .0003), involved in maintaining integrity of tissue junctions.
Pathway analysis and PheWAS
Based on the meta-analysis, there were 197 SNPs with P values < .0001, which were annotated to 64 genes (supplemental Table 10). Ingenuity Pathway Analysis using these 64 genes (supplemental Table 11) showed that the glutamate receptor-signaling pathway, including 3 genes GRIN3A, GRIK1, and GRM7, was the top canonical pathway (P = 4.8 × 10−4). Using a P value cutoff of .001, there were 433 genes, and the glutamate receptor-signaling pathway remained the top pathway, with additional genes in the pathway including GRM3, GRIK4, and GRIA1 (P = 9.8 × 10−4) (supplemental Table 11).
All SNPs with P value < .05 in or near genes within the glutamate pathway found to be significant in at least 1 of 3 cohorts were selected for analysis in a PheWAS (supplemental Materials) of the Vanderbilt University Medical Center BioVU database to explore whether these same osteonecrosis variants were associated with any of 1358 other phenotypes in the BioVU repository (supplemental Table 12). None of these associations achieved a Bonferroni significance threshold of 3.7 × 10−5. However, among the top 5 phenotypes associated with SNPs in GRIN3A were diseases of the respiratory tract (OR = 3.02; P = 3.1 × 10−4) and the long-term use of antithrombotics within the circulatory system (OR = 2.13; P = 4.7 × 10−4) (supplemental Table 13). Other phenotypes included cerebral ischemia (OR = 1.64; P = 2.5 × 10−3), and arterial embolism and thrombosis (OR = 1.88; P = 4.2 × 10−3).
Discussion
The development of osteonecrosis can result in serious debilitation and requires surgical intervention in many cases. For example, in the Children’s Cancer Group 1961 study, 143 patients (of 2056 enrolled) developed symptomatic osteonecrosis at 377 confirmed sites leading to 139 surgeries.8 Identification of risk factors for the development of osteonecrosis in patients undergoing therapy for ALL might facilitate tailoring therapy to minimize its risk.
In this study, we performed a GWAS, which identified a locus at 9q31.1 near GRIN3A as associated with osteonecrosis in both the discovery and validation cohorts, and the second highest ranked variant was in a related gene, GRIK1. The same GRIN3A variant was associated in the meta-analysis of the discovery and validation cohorts (P = 2.68 × 10−8), and the glutamate receptor pathway was the top canonical pathway based on the meta-analysis of all 3 cohorts. These findings suggest the involvement of the glutamate pathway in the pathogenesis of osteonecrosis not only in childhood ALL, but in a heterogeneous cohort of children and adults who were treated with prolonged corticosteroids for other medical conditions.30,31 Interestingly, we previously reported that polymorphisms within a third glutamate receptor gene, GRIA1, were the top genomic variants associated with asparaginase allergy in a GWAS of children with ALL in the SJ cohort,32 a finding recently independently replicated.33 We have also reported that asparaginase allergy was associated with lower systemic exposure to asparaginase,34 which in turn was associated with a lower risk of osteonecrosis in the SJ cohort.35 This prompted us to evaluate asparaginase allergy in the COG AALL0232 discovery cohort (n = 1845 with follow-up >800 days), and consistent with the SJ cohort, grades 2-4 asparaginase allergy was associated with the risk of osteonecrosis: 206 out of 1647 (12.5%) for those without asparaginase allergies had osteonecrosis compared with 12 out of 198 (6.7%) for those with asparaginase allergies (P = .0096).
Concomitant asparaginase treatment is associated with higher plasma exposure to dexamethasone.7,34 Although it is possible that the glutamate receptor pathway association was via secondary effects on asparaginase, which affects risk of osteonecrosis in ALL patients,35 this does not explain the association of the glutamate pathway within the BioVU cohort, whose patients did not receive asparaginase, and thus also implicates a mechanism with a direct effect of glutamate receptors. Multiple mechanisms can lead to osteonecrosis, with contributions to risk from therapy and host-specific factors.
Previously described biologic mechanisms by which glucocorticoids may induce osteonecrosis include thrombosis, hyperlipidemia-associated enlarged adipocytes in bone, arteriopathy, and direct toxicity to osteocytes.36-41 Prior candidate gene and genome-wide investigations have implicated polymorphisms involved in lipid homeostasis (ACP1), fibrinolysis (SERPINE1), antifolate pharmacodynamics (thymidylate synthetase), and glucocorticoid response (VDR).7,10,42,43 These variants did not replicate in the discovery cohort, possibly due to differences in therapy and methods of case/control ascertainment. Differences in therapy and in frequency of performing MRIs may also explain the different rates of osteonecrosis seen in the AALL0232 and SJ Total XV protocols. Importantly, the GRIN3A rs10989692 variant was associated with osteonecrosis in both cohorts despite these underlying protocol differences.
Glutamate receptor genetic variations have not been previously reported as risk factors for osteonecrosis. Glutamate is released by osteocytes in response to mechanical load, which opens stretch-sensitive calcium channels and causes activation of osteoblast receptors,44 and glutamate impairs endothelial barrier function.45 Glucocorticoids have been shown to induce the expression of glutamine synthetase in osteoblasts46 and hepatoma47 cells. Genetic variation in GRIN3A has been associated with the severity of vascular complications of Kawasaki disease.48 Interestingly, the disruption of the vascular supply to bone is a proximal event to glucocorticoid-induced osteonecrosis in a murine model, with or without asparaginase.41 Herein, in our PheWAS of GRIN3A SNPs, vascular phenotypes including cerebral ischemia, arterial embolism, and thrombosis trended toward association with glutamate receptor variants. Thus, glutamate and variations in glutamate receptors may contribute to a proximal vascular event that leads to an increased risk of osteonecrosis in individuals exposed to glucocorticoid therapy.
In conclusion, we hypothesize that different mechanisms of glucocorticoid-induced osteonecrosis may predominate among patients, influenced by variation in concurrent drug therapy as well as inherited genetic risk factors. To our knowledge, this is the largest genome-wide investigation of glucocorticoid-induced osteonecrosis. Our findings suggest for the first time a possible association between inherited genetic variations in glutamate receptors and the development of glucocorticoid-induced osteonecrosis in both ALL and non-ALL settings. The ability to identify genetic risk factors for osteonecrosis has implications for understanding the underlying mechanism of this common and serious adverse effect of glucocorticoids, and may have implications for modifying therapy decisions in the future.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by the National Institutes of Health, Institute of General Medical Sciences, National Cancer Institute, Human Genome Research Institute, and National Library of Medicine grants GM 92666, GM 115279, CA142665, CA 21765, CA 36401, CA98543 (COG Chair’s grant), CA98413 (COG Statistical Center), CA114766 (COG Specimen Banking), U01-HG04603, RC2-GM092618, and R01-LM010685; the Vanderbilt Clinical Translational Science Award grants UL1-TR000445, 5T32-GM007569, and LLS 6168; and the American Lebanese Syrian Associated Charities.
Authorship
Contribution: E.A.R., C.-H.P., and M.V.R. were responsible for concept and design; S.L.V.D., S.K., E.B., D.M.R., J.C.D., E.L., N.W., M.L.L., S.P.H., P.S., M.D., L.A.M., and M.V.R. collected and assembled the data; W.Y., S.E.K., T.Y.C., M.B., L.B., D.M.R., J.C.D., W.L.C., C.C., D.P., C.A.F., C.L., C.S., P.S., S.J., W.E.E., M.D., L.A.M., and M.V.R. were responsible for the analysis and interpretation of data; and all authors contributed to the writing of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mary V. Relling, Pharmaceutical Department, St. Jude Children’s Research Hospital, 262 Danny Thomas Pl, Memphis, TN 38105; e-mail: mary.relling@stjude.org.
References
Author notes
S.E.K. and W.Y. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal