Key Points
Genome editing of primary human HSPCs generates MLL leukemias that model clinical disease features and molecular pathogenesis.
Abstract
Chromosomal rearrangements involving the mixed-lineage leukemia (MLL) gene occur in primary and treatment-related leukemias and confer a poor prognosis. Studies based primarily on mouse models have substantially advanced our understanding of MLL leukemia pathogenesis, but often use supraphysiological oncogene expression with uncertain implications for human leukemia. Genome editing using site-specific nucleases provides a powerful new technology for gene modification to potentially model human disease, however, this approach has not been used to re-create acute leukemia in human cells of origin comparable to disease observed in patients. We applied transcription activator-like effector nuclease–mediated genome editing to generate endogenous MLL-AF9 and MLL-ENL oncogenes through insertional mutagenesis in primary human hematopoietic stem and progenitor cells (HSPCs) derived from human umbilical cord blood. Engineered HSPCs displayed altered in vitro growth potentials and induced acute leukemias following transplantation in immunocompromised mice at a mean latency of 16 weeks. The leukemias displayed phenotypic and morphologic similarities with patient leukemia blasts including a subset with mixed phenotype, a distinctive feature seen in clinical disease. The leukemic blasts expressed an MLL-associated transcriptional program with elevated levels of crucial MLL target genes, displayed heightened sensitivity to DOT1L inhibition, and demonstrated increased oncogenic potential ex vivo and in secondary transplant assays. Thus, genome editing to create endogenous MLL oncogenes in primary human HSPCs faithfully models acute MLL-rearranged leukemia and provides an experimental platform for prospective studies of leukemia initiation and stem cell biology in a genetic subtype of poor prognosis leukemia.
Introduction
Aberrations of the mixed-lineage leukemia (MLL) gene are present in primary and treatment-related acute leukemias of children and adults, and portend an intermediate to poor prognosis. The MLL gene encodes a DNA-binding protein that functions as a histone methyltransferase to positively regulate expression of target genes, including multiple HOX genes.1 Its epigenetic role is corrupted by fusions with over 60 different translocation partner proteins in leukemias of various lineages, including acute myeloid leukemia (AML), acute lymphoblastic leukemia (ALL), and a distinctive mixed-phenotype acute leukemia (MPAL).2-4 Despite their poor prognosis, MLL leukemias are genetically simple and appear to require very few additional driver mutations beyond the activated MLL oncogene for their pathogenesis, consistent with the short latency between MLL gene rearrangements in utero and clinical presentation of leukemia in infants.5-7 Given their genomic simplicity and short progression, MLL leukemias are particularly amenable to experimental modeling for study of their pathogenesis. However, many productive attempts to model this process are based on supraphysiological expression of MLL fusion proteins in primary mouse or human cells using retroviral vectors. Unlike human leukemias, these models maintain 2 normal copies of the MLL gene and circumvent the endogenous feedback regulation of the fusion gene. Other approaches have simulated MLL oncogenic fusions by creating knock-in mouse models using homologous recombination in embryonic stem cells.8-10 Although these studies have provided important insights, it remains uncertain whether the experimental models accurately reflect the pathology underlying the disease as it manifests in human patients.
In the past several years, new experimental techniques have been developed to edit the genome in situ for potential correction or modeling of human diseases. These approaches are based on the use of custom DNA nucleases including zinc finger nucleases,11,12 transcription activator-like effector nucleases (TALENs),13 and RNA-guided endonucleases of the clustered regularly interspaced short palindromic repeats14 that specifically cleave genome target sites to facilitate site-specific mutation or recombination. Genome editing of murine hematopoietic stem and progenitor cells (HSPCs) has been used to generate myeloid malignancy in mice15 but the approach has not been used to induce acute leukemia in human cells that serve as de novo targets for disease origination in patients. Here, we used TALENs to specifically engineer endogenous activation of 2 common MLL oncogenes, MLL-AF9 and MLL-ENL, in primary human HSPCs. Their transplantation in mice led to human leukemias that manifest the pathological and clinical attributes seen in MLL leukemia patients.2-4,16 Our study highlights the application of genome-editing tools in primary human HSPCs to activate oncogenes under the control of the endogenous promoter to faithfully model MLL-rearranged leukemias.
Methods
TALEN construction and validation
The MLL cleavage site was selected based on the most commonly found patients’ breakpoint cluster region (BCR) in the MLL gene available through GenBank using the TAL Effector Nucleotide Targeter 2.0.17,18 Three pairs of TALENs were created using the Golden Gate TALEN Assembly Method.19 Following nucleofection of the TALEN pairs, genomic DNA (gDNA) was isolated and the targeted region of interest was amplified by polymerase chain reaction (PCR) with MLL-specific primers (5′GCCTTTTAATAGTCCGTGTCT3′ and 5′TCTTTAGCTGGTTTAAACAGG3′) and analyzed using the T7 endonuclease assay as previously described.20
For sequence analysis of TALEN cut sites, gel-extracted DNA was cloned into the pCR2.1-TOPO vector with the TOPO TA Cloning kit (Invitrogen) and transformed into competent XL-1 Escherichia coli cells. For analyzing allele modification frequencies, the purified PCR products were Sanger-sequenced and each sequence chromatogram was analyzed with the online Tracking of In/dels by Decomposition (TIDE) software (available at http://tide.nki.nl). Analyses were performed using a reference sequence (green fluorescent protein [GFP] sample).21
AF9 and ENL2/7 knock-in construct design
The knock-in DNA templates contained MLL homology arms (∼700 bp) flanking the TALEN cleavage site, fusion partner complementary DNA (cDNA) sequences, an internal ribosomal entry site (IRES), a fluorescent marker gene coding NeonGreen, and a polyA tail (nucleotide sequences provided in supplemental Figure 1, see supplemental Data available at the Blood Web site).22 The constructs were synthesized commercially (GenScript USA Inc).
Cell culture, nucleofection, and retroviral transduction
K562 cells were cultured and nucleofected as previously described.20 HSPCs were isolated from fresh human umbilical cord blood (huCB) obtained from the maternity ward of Stanford Hospital (under institutional review board–approved research protocol) using Ficoll-Paque plus (GE Healthcare Life Sciences) followed by the EasySep CD34+ Human Selection kit (StemCell Technologies) to obtain enrichment >90%. Following isolation, CD34+ cells were maintained in serum-free StemSpan II media (StemCell Technologies) supplemented with cytokines (PeproTech: stem cell factor [SCF], thrombopoietin [TPO], Flt3L, interleukin-6 [IL-6], IL-3 [100 ng/mL each]; Cellagen Technology: StemRegenin1 [SR1; 0.75 μM]). The following day, CD34+ cells were nucleofected using the Lonza 4-D Nucleofector system (program EO-100). Cells were incubated at 37°C, 7.5% CO2 in serum-free media (StemSpan II) + cytokines + 20 μM Z-Vad-FMK (Enzo Life Sciences). After 48 hours, 10% filtered umbilical cord blood plasma was added.
For retroviral transductions, the MLL-AF9 cDNA was cloned into the pMSCV-ires-GFP retroviral vector using standard cloning techniques.23 Transductions were performed on retronectin-coated (10 µg/mL; Takara Bio Inc) 48-well plastic non-tissue-culture plates that were preloaded with retrovirus by centrifugation at 3000 rpm for 2 hours. CD34+ cells were transduced 2 times on the prepared plates for 4 hours each time at 32°C with freshly loaded retrovirus and transplanted into NSG mice. Explanted leukemia cells were used for further analysis.
CFC assays
CD34+ cells were nucleofected with knock-in constructs without (control) or with MLL TALENs or MLL TALENs alone (control), followed by consecutive fluorescence-activated cell sorting (FACS) for NeonGreen expression at days 2 to 4 of cell culture. On day 21 of liquid culture, cells were seeded in triplicate (10 000 cells per dish) in Methocult H4230 methylcellulose medium (StemCell Technologies) supplemented with SCF, TPO, Flt3L, IL-6, IL-3 (100 ng/mL each) and SR1 (0.75 μM). Colony-forming cell (CFC) assays were performed as previously described.20,24 Identical conditions were used for CFC assays with explanted leukemic cells from MPAL or AML mice.
Reverse transcription PCR and qPCR
MLL oncogene knock-in was assessed by genomic PCR with the LongAmp Taq PCR kit (New England BioLabs). For each PCR, 200 to 1000 ng of gDNA was used with genomic-specific primers (proof of genomic integration): MLL primer: 5′ATCCCTGTAAAACAAAAACCAAAA3′; AF9 primer: 5′TTGTCATCAGAATGCAGATCTTTC3′, ENL2 primer: 5′GTACCCCGACTCCTCTACTTTGTA3′, ENL7 primer: 5′GTAGGTGCCCTTCTTGAGGATCT3′; and for the detection of the wild-type (WT) MLL gene: 5′ACAACTTTGGATGGAAAATAAGGA3′ PCR products were visualized on agarose gel and extracted for Sanger sequencing.
Bone marrow (BM) cells were obtained from the Division of Blood and Marrow Transplantation at Stanford University (under institutional review board–approved research protocol). Mononuclear cells (MNCs) were isolated by using Ficoll-Paque plus and further enriched for B-cell subpopulations by sorting for CD10+CD19+ or CD10−CD19+ cells. RNA was then isolated from MNCs, B-cell subpopulations, or leukemic cells harvested from mice postmortem using the RNeasy Mini kit (Qiagen). RNA was used to generate cDNA using the SuperScript III First-Strand Synthesis System (Invitrogen). PCR was subsequently performed for MLL fusion transcripts, MLL primer: 5′ATCCCTGTAAAACAAAAACCAAAA3′; AF9 primer: 5′TTATAGACCTCAAAGGACCTTGTTG3′; ENL2 primer: 5′GTACCCCGACTCCTCTACTTTGTA3′; ENL7 primer: 5′GAAGTCTGAGTCTGAGCTGGAGT3′. PCR products were visualized, gel extracted, and subjected to Sanger sequencing. For detection of target genes MEIS1 (HS00180020_m1) and HOXA9 (HS00365956_m1) by quantitative PCR (qPCR), TaqMan gene expression assays were used (Life Technologies). qPCR was performed in triplicate followed by melting curve analysis in the Bio-Rad CFX384 C1000 real-time system relative to Kasumi-1 cells or leukemic BM cells. Results were normalized to the housekeeping gene ACTB.
Confocal microscopy
K562 cells were nucleofected with TALENs and the knock-in constructs followed by consecutive FACS for NeonGreen expression at days 5 and 8 of cell culture. Confocal microscopy was performed using a Zeiss LSM 710 confocal scope. A 10× objective and a GFP laser with the excitation 450 to 490 nm were used for the detection of NeonGreen-expressing cells.
DOT1L inhibitor treatment
EPZ00477 (Millipore) was prepared in stock solutions with dimethylsulfoxide (DMSO). CFC assays were performed with cell lines and leukemic cells explanted from MPAL and AML mice as described above (CFC assays) with the addition of increasing EPZ00477 concentrations. After 12 to 14 days, CFC assays were diluted and cell count, CD14, MEIS1, and HOXA9 expression was determined.
Microarray data analysis
BM cells (sorted for human CD45 [hCD45]) of leukemic mice (ALL) were used for global gene expression measurement using the Affymetrix Microarray GeneChip platform (HG-U133 Plus 2.0). The data, together with those from 70 MLL patients (ALL) and 6 control samples (same GeneChip platform) from the leukemia study group, were used for unsupervised hierarchical clustering analysis.16 The gene expression matrix from the arrays was normalized in the same way using the RMA model from Bioconductor affy package. Gene expression values were then independently filtered to remove low-variance probes using the Bioconductor package, genefilter 1.50. Finally, 25 000 probes were then subjected to unsupervised hierarchical clustering analysis using R packages, ape, and heatmap 2.0.
Results
Generation of site-specific DNA DSBs in the MLL gene using TALENs
TALENs were designed to introduce DNA breaks at a preselected site at the intersection of exon 11 and intron 11 of the MLL gene (Figure 1A). Intron 11 contains a BCR that harbors common sites of chromosomal translocations in patients with MLL-AF9 or MLL-ENL leukemias.17 To test their abilities to specifically cleave the MLL gene, pairs of TALENs were nucleofected into K562 and CD34+ cells and resultant double-strand breaks (DSBs) from the different TALEN pairs were detected using the surveyor assay (Figure 1B).25 Specific cleavage activity was further assessed by cloning and sequence analyses of target site DNA, which showed unique deletions and insertions in the repaired cutting sites of the TALENs (K562, 34.3%; CD34+, 15.7%) in the cloned fragments (Figure 1C). Additionally, we quantified allele modification frequencies using TIDE analysis (Figure 1D).21 The MLL TALEN pair most efficient at cleavage was used in subsequent experiments to target a DNA DSB at the preselected genomic site of the MLL gene in primary cells.
TALENs induce specific DNA DSBs within the MLL gene. (A) Schematic illustration of the human MLL gene shows recognition sites (bold sequences) for TALEN pairs designed to cleave within the 5′ portion of MLL intron 11. Black boxes represent respective exons of the MLL gene. (B) Gel image shows results of the T7 endonuclease assay performed on gDNA isolated from K562 or CD34+ cells nucleofected with the 3 different MLL TALEN pairs (P1, P2, P3) or control (GFP) as indicated. Digested PCR products (*) of the MLL locus represent the presence of strand mismatches resulting from indels that are generated during nonhomologous end joining (NHEJ) repair of DSBs. (C) DNA sequences of the amplified endogenous MLL locus resulting from the best TALEN pair (P3) show unique insertions or deletions resulting from NHEJ (K562 24 of 70: 34.3%; CD34+ 11 of 70: 15.7% mutation ratio). Italic letters represent insertions; underlined letters denote TALEN binding sites; red and blue text indicates MLL exon 11 and MLL intron 11, respectively. (D) Indel frequencies were measured by TIDE and compared with indel frequencies of the control sample.21 eff., efficiency.
TALENs induce specific DNA DSBs within the MLL gene. (A) Schematic illustration of the human MLL gene shows recognition sites (bold sequences) for TALEN pairs designed to cleave within the 5′ portion of MLL intron 11. Black boxes represent respective exons of the MLL gene. (B) Gel image shows results of the T7 endonuclease assay performed on gDNA isolated from K562 or CD34+ cells nucleofected with the 3 different MLL TALEN pairs (P1, P2, P3) or control (GFP) as indicated. Digested PCR products (*) of the MLL locus represent the presence of strand mismatches resulting from indels that are generated during nonhomologous end joining (NHEJ) repair of DSBs. (C) DNA sequences of the amplified endogenous MLL locus resulting from the best TALEN pair (P3) show unique insertions or deletions resulting from NHEJ (K562 24 of 70: 34.3%; CD34+ 11 of 70: 15.7% mutation ratio). Italic letters represent insertions; underlined letters denote TALEN binding sites; red and blue text indicates MLL exon 11 and MLL intron 11, respectively. (D) Indel frequencies were measured by TIDE and compared with indel frequencies of the control sample.21 eff., efficiency.
Creation of endogenous MLL oncogenes in CD34+ human cord blood cells by genome editing
To promote genome editing at the targeted cleavage site during homology-directed repair,26 exogenous DNA templates were designed with flanking sequences homologous to the nuclease target site (supplemental Figure 1). Three different templates were designed for insertion of either AF9 (exon 5-9), ENL2 (exon 2-12), or ENL7 (exon 7-12) sequences to create the respective fusion oncogenes expressed under control of the endogenous MLL promoter along with the fluorescent marker gene NeonGreen (Figure 2A).22 The AF9 construct represented the most common fusion seen in MLL-AF9 patients.17,27,28 The ENL constructs contained different lengths of the ENL cDNA accounting for the majority of MLL-ENL fusions seen in patients associated with either ALL or AML.29-31
Generation of MLL-AF9 and MLL-ENL knock-in genes by genome engineering. (A) Schematic illustration of experimental strategy to induce a DSB by TALENs followed by integration of the knock-in template in the MLL gene locus by homology-directed repair (HDR). (B) FACS profiles show fluorescence of K562 and CD34+ cells nucleofected with the knock-in template alone (gray line) or in combination with the MLL TALENs (black line) sorted on days 5 and 3, respectively. (C) Summary of NeonGreen expression in K562 (n[AF9] = 3, n [ENL2/7] = 2) and CD34+ cells (n[AF9] = 11, n[ENL2] = 7, n[ENL7] = 5) pre- and postsort. *P < .05 was considered statistically significant. Error bars indicate standard error of the mean (SEM). (D) Confocal microscopy images show NeonGreen expression in sorted K562 and control cells as indicated. Top row, cell density (brightfield); bottom row, NeonGreen expression detected by GFP excitation (450-490 nm) and ×10 objective. (E) PCR/RT-PCR was performed on gDNA and cDNA isolated from NeonGreen-positive K562 and CD34+ cells to detect integration and transcription of the construct under control of the endogenous MLL promoter (representative results shown for MLL-AF9). BGH, bovine growth hormone.
Generation of MLL-AF9 and MLL-ENL knock-in genes by genome engineering. (A) Schematic illustration of experimental strategy to induce a DSB by TALENs followed by integration of the knock-in template in the MLL gene locus by homology-directed repair (HDR). (B) FACS profiles show fluorescence of K562 and CD34+ cells nucleofected with the knock-in template alone (gray line) or in combination with the MLL TALENs (black line) sorted on days 5 and 3, respectively. (C) Summary of NeonGreen expression in K562 (n[AF9] = 3, n [ENL2/7] = 2) and CD34+ cells (n[AF9] = 11, n[ENL2] = 7, n[ENL7] = 5) pre- and postsort. *P < .05 was considered statistically significant. Error bars indicate standard error of the mean (SEM). (D) Confocal microscopy images show NeonGreen expression in sorted K562 and control cells as indicated. Top row, cell density (brightfield); bottom row, NeonGreen expression detected by GFP excitation (450-490 nm) and ×10 objective. (E) PCR/RT-PCR was performed on gDNA and cDNA isolated from NeonGreen-positive K562 and CD34+ cells to detect integration and transcription of the construct under control of the endogenous MLL promoter (representative results shown for MLL-AF9). BGH, bovine growth hormone.
Cotransfection of MLL TALENs with the respective knock-in templates resulted in insertions both in K562 cells and in primary HSPCs isolated from huCB with knock-in efficiencies of ∼22% (range, 6%-36%) and ∼9% (range, 2%-35%), respectively, based on NeonGreen expression, which could be further enriched by FACS (Figure 2B-C). NeonGreen expression in nucleofected cells was also demonstrated by confocal microscopy (Figure 2D). PCR analyses of sorted NeonGreen-expressing K562 and primary CD34+ cells confirmed correct integration and expression of the constructs (Figure 2E). These studies demonstrate that TALENs promote homology-directed insertional mutagenesis to create endogenous MLL oncogenes in cell lines and primary human HSPCs.
Insertional activation of endogenous MLL oncogenes alters the growth and survival of primary CD34+ cells
To assess the effects of activating an endogenous MLL oncogene in primary HSPCs, human CD34+ cells were nucleofected with MLL TALENs and knock-in templates or template alone (control). In liquid culture, the engineered CD34+ cells displayed a distinct survival advantage and proliferated for ∼100 to 120 days whereas control samples terminally differentiated and eventually died out after 60 to 80 days (supplemental Figure 2A-B).
CFC assays were performed to further assess the clonal expansion capacity and transformative potential of the knock-in cells. For this purpose, 10 000 sorted knock-in or control cells (harvested on day 21 of liquid culture) were plated in semisolid medium. Both populations generated colonies after 10 to 14 days, but the knock-in cells displayed a significantly higher clonogenic potential after the second round of replating as compared with control cells (supplemental Figure 3A). Knock-in cells also demonstrated more compact colony morphologies in contrast to the controls, consistent with more immature cells (supplemental Figure 3B).24 Despite their enhanced growth properties, knock-in cells were not fully transformed because further replating resulted in decreasing cell numbers consistent with the proliferative exhaustion observed in extended liquid cultures.
CD34+ cells engineered to express MLL oncogenes induce acute leukemias
CD34+ huCB cells were cotransfected with MLL TALENs and the knock-in constructs, and were then transplanted using 2 alternative approaches: unsorted nucleofected cells were either injected directly (<3 days) into NSG immunocompromised mice or cultured in vitro (3 weeks) under myeloid growth conditions prior to transplantation (Figure 3A). IV injection of uncultured CD34+ HSPCs (0.75-3.6 × 106, median 2 × 106 CD34+ cells) or cultured cells (1 × 106) induced acute leukemia within 8 to 33.5 weeks (median 16 weeks) posttransplantation (Figure 3B). MLL-AF9 knock-in cells induced ALL, MPAL, or AML, whereas the ENL knock-in cells exclusively resulted in an ALL phenotype (Table 1). Mice transplanted with control cells (nucleofected with template alone, hereafter referred to as control mice) displayed no overt pathology. Rather, periodic BM aspirations showed an initial lymphoid engraftment in the control mice, which subsequently developed additional features of myeloid hematopoiesis with evidence of terminal differentiation characterized by increased expression levels of CD38. Control mice presented with common gates for lymphocytes, monocytes, and granulocytes detected by different CD45 expression levels and side-scatter (SSC) profiles (Figure 3C first row, 16 weeks postinjection).
Induction of acute leukemias following transplantation of human CD34+ cells containing knock-in MLL oncogenes. (A) Experimental scheme depicts nucleofection of CD34+ cells and their subsequent transplantation directly into sublethally irradiated NSG recipient mice or culture for 3 weeks in vitro prior to transplantation. (B) Kaplan-Meier plot is shown for cohorts of mice transplanted with CD34+ cells transfected with templates plus TALENs (n [AF9] = 25: direct inject = 19, cultured cells = 6; n [ENL2] = 7: direct inject = 5, cultured cells = 2; n [ENL7] = 8: direct inject = 5, cultured cells = 3) or template alone (control, n = 7). P < .05 was considered statistically significant. Mice were sacrificed upon signs of illness. (C) Flow cytometry profiles show representative phenotypes of various leukemias that developed in mice transplanted with CD34+ cells (n = 17) compared with blasts from patients with MLL translocations (AML/ALL), which display comparable phenotypes. Also shown are representative profiles of BM cells from control mice (n = 2) that received CD34+ cells nucleofected with knock-in construct alone (week 16 posttransplantation) and representative analysis of knock-in cells 3 weeks after cell culture prior to transplantation. l, lymphoid; m, myeloid; (Pat.), patient.
Induction of acute leukemias following transplantation of human CD34+ cells containing knock-in MLL oncogenes. (A) Experimental scheme depicts nucleofection of CD34+ cells and their subsequent transplantation directly into sublethally irradiated NSG recipient mice or culture for 3 weeks in vitro prior to transplantation. (B) Kaplan-Meier plot is shown for cohorts of mice transplanted with CD34+ cells transfected with templates plus TALENs (n [AF9] = 25: direct inject = 19, cultured cells = 6; n [ENL2] = 7: direct inject = 5, cultured cells = 2; n [ENL7] = 8: direct inject = 5, cultured cells = 3) or template alone (control, n = 7). P < .05 was considered statistically significant. Mice were sacrificed upon signs of illness. (C) Flow cytometry profiles show representative phenotypes of various leukemias that developed in mice transplanted with CD34+ cells (n = 17) compared with blasts from patients with MLL translocations (AML/ALL), which display comparable phenotypes. Also shown are representative profiles of BM cells from control mice (n = 2) that received CD34+ cells nucleofected with knock-in construct alone (week 16 posttransplantation) and representative analysis of knock-in cells 3 weeks after cell culture prior to transplantation. l, lymphoid; m, myeloid; (Pat.), patient.
Pathological features of leukemias arising in mice transplanted with engineered hCD34+ HSPCs
| No. . | Oncogene insertion . | Cell count prenuc. ×106 . | In vitro culture, days . | Latency, weeks . | BM hCD45, % . | Phenotype . | Spleen, mg . | Secondary Tx, pos/total inj. . |
|---|---|---|---|---|---|---|---|---|
| 1 | AF9 | 1 | 21 | 9 | 75 | AML | 20 | 1/3 |
| 2 | AF9 | 1.0 | 21 | 14 | 83 | AML | 40 | 0/2 |
| 3 | AF9 | 1.5 | 1 | 33.5 | 87 | AML | 100 | nd |
| 4 | AF9 | 1.2 | 0 | 14 | 75 | MPAL | 50 | 1/1 |
| 5 | AF9 | 2.8 | 0 | 16 | 84 | MPAL | 40 | 1/1 |
| 6 | AF9 | 3.3 | 1 | 20.5 | 82 | MPAL | 40 | 2/4 |
| 7 | AF9 | 2.7 | 0 | 23 | 81 | MPAL | 20 | 1/1 |
| 8 | AF9 | 1.5 | 0 | 8.5 | 90 | ALL | 180 | nd |
| 9 | AF9 | 2.4 | 0 | 10 | 93 | ALL | 120 | 1/3 |
| 10 | AF9 | 2.2 | 2 | 10.5 | 48 | ALL | 140 | nd |
| 11 | AF9 | 1.5 | 1 | 32 | 77 | ALL | 70 | nd |
| 12 | AF9 | 2 | 2 | 29 | 72 | ALL | 110 | nd |
| 13 | ENL2 | 3 | 0 | 11.5 | 95 | ALL | 80 | 2/2 |
| 14 | ENL2 | 2.7 | 0 | 28 | 92 | ALL | 120 | nd |
| 15 | ENL7 | 3.6 | 1 | 8 | 90 | ALL | 80 | 1/2 |
| 16 | ENL7 | 2.7 | 0 | 14.5 | 95 | ALL | 170 | nd |
| 17 | ENL7 | 0.75 | 1 | 30.5 | 82 | ALL | 30 | nd |
| No. . | Oncogene insertion . | Cell count prenuc. ×106 . | In vitro culture, days . | Latency, weeks . | BM hCD45, % . | Phenotype . | Spleen, mg . | Secondary Tx, pos/total inj. . |
|---|---|---|---|---|---|---|---|---|
| 1 | AF9 | 1 | 21 | 9 | 75 | AML | 20 | 1/3 |
| 2 | AF9 | 1.0 | 21 | 14 | 83 | AML | 40 | 0/2 |
| 3 | AF9 | 1.5 | 1 | 33.5 | 87 | AML | 100 | nd |
| 4 | AF9 | 1.2 | 0 | 14 | 75 | MPAL | 50 | 1/1 |
| 5 | AF9 | 2.8 | 0 | 16 | 84 | MPAL | 40 | 1/1 |
| 6 | AF9 | 3.3 | 1 | 20.5 | 82 | MPAL | 40 | 2/4 |
| 7 | AF9 | 2.7 | 0 | 23 | 81 | MPAL | 20 | 1/1 |
| 8 | AF9 | 1.5 | 0 | 8.5 | 90 | ALL | 180 | nd |
| 9 | AF9 | 2.4 | 0 | 10 | 93 | ALL | 120 | 1/3 |
| 10 | AF9 | 2.2 | 2 | 10.5 | 48 | ALL | 140 | nd |
| 11 | AF9 | 1.5 | 1 | 32 | 77 | ALL | 70 | nd |
| 12 | AF9 | 2 | 2 | 29 | 72 | ALL | 110 | nd |
| 13 | ENL2 | 3 | 0 | 11.5 | 95 | ALL | 80 | 2/2 |
| 14 | ENL2 | 2.7 | 0 | 28 | 92 | ALL | 120 | nd |
| 15 | ENL7 | 3.6 | 1 | 8 | 90 | ALL | 80 | 1/2 |
| 16 | ENL7 | 2.7 | 0 | 14.5 | 95 | ALL | 170 | nd |
| 17 | ENL7 | 0.75 | 1 | 30.5 | 82 | ALL | 30 | nd |
inj., injected; nd, not determined; pos, positive; prenuc., prenucleofection; Tx, transplantation.
Leukemic mice transplanted with MLL oncogene knock-in cells presented with typical blasts characterized by a lower expression of hCD45 and distinct SSC characteristics. Different surface phenotypes (ALL, AML, MPAL) of blast cells showed similarities to patient samples harboring MLL translocations (Figure 3C rows 3 and 8). The majority of leukemic mice (10 of 17) directly injected with nucleofected cells developed ALL displaying a prelymphoid phenotype with CD10+/CD19+/CD38+/CD34+ and very low expression levels of mature B-cell markers CD20 and immunoglobulin M (IgM) (Figure 3C rows 6 and 7). In some cases, MPAL was observed with immunophenotypic features of AML and ALL in the same recipient mouse (Figure 3C rows 5 and 6). Two distinct blast populations were clearly distinguished by their SSC properties. The ALL cells were phenotypically similar to those in pure ALLs (Figure 3C row 7), whereas a separate blast cell subpopulation displayed a myelomonocytic phenotype (CD38+/CD33+/CD15+/CD64+/CD4+/CD117+) (Figure 3C row 5). Furthermore, coexpression of myeloid and lymphoid antigens was observed on both blast populations.
Short-term in vitro culture (3 weeks) of knock-in cells under myeloid conditions prior to transplantation resulted in AML (CD38+/CD33+/HLA DR+/CD64+/CD117+) with a median latency of ∼12 weeks (Figure 3C row 4). The cultured cells at the time of transplant and their corresponding leukemias displayed similar immature myelomonocytic phenotypes (Figure 3C second row) consistent with a pivotal role of the microenvironment for the lineage determination of MLL leukemias.32,33 Conversely, prolonged (>3 months) antecedent in vitro culture of nucleofected cells prior to transplantation resulted in no engraftment, indicating that knock-in cells did not maintain oncogenic potential in vitro consistent with their differentiation and exhaustion observed in extended in vitro cultures and serial replating CFC assays.
Genome editing of CD34+ cells models human leukemia in mice
All mice that succumbed to fatal leukemia presented with a similar disease profile that included anemia, leukocytosis, and thrombocytopenia (Figure 4A) and blasts in peripheral blood (Figure 4B). Splenomegaly was most prominent in ALL mice (Figure 4C). Histopathological examination confirmed extensive replacement of BM and infiltration of the spleen, liver, and other peripheral organs (Figure 4D) by leukemic blasts that expressed the respective oncogenes and normal WT MLL gene detected by PCR, reverse transcription PCR (RT-PCR), and western blot (Figure 4E-F). The expression level under control of the endogenous promoter was lower compared with that in AMLs induced by ectopic expression of MLL-AF9 by retroviral transduction of huCB cells.23 Cytospins showed monomorphic immature blast cells (supplemental Figure 4). Moreover, the leukemic blasts demonstrated increased expression levels of common MLL target genes (MEIS1, HOXA9) compared with human BM cells or non-MLL leukemic cell lines and similar to MLL rearranged cell lines (Mono Mac 6 or MV4-11) and patient samples harboring MLL translocations (Figure 4G). Furthermore, gene expression profiling of MLL-AF9– or MLL-ENL–expressing blast cells demonstrated a signature that closely parallels that of publicly available patient samples harboring an MLL rearrangement (Figure 4H).16 These studies establish a novel experimental model to generate MLL leukemias deriving from primary human HSPCs expressing the fusion oncogene under control of the endogenous promoter.
Pathologic and molecular features of acute leukemias induced by genome editing of the MLL oncogene. (A) Plots show results of hematologic analyses performed on control (n = 7) and leukemic mice (n = 16), respectively. (B) Representative peripheral blood smears of control and leukemic mice are shown and summarized by calculating the percentage of blast cells (n = 16). Scale bars define 10 µm. (C) Spleen size and weight are shown for 1 representative control and leukemic mice (n = 17). (D) Hematoxylin-and-eosin–stained paraffin sections demonstrate disruption of organ architecture due to tumor infiltration compared with control mice. Scale bars define 100 µm. (E) PCR/RT-PCR was performed on gDNA and cDNA of leukemia cells to detect integration and expression of the MLL oncogenes and WT MLL gene. (F) Representative western blot analysis shows WT MLLN and MLL-AF9 expression in control (CD34+ cells nucleofected with template alone) and explanted blast cells from xenotransplants induced by either retroviral transduction or under the expression of the endogenous promoter (MPAL, AML). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), loading control. Bottom, relative MLL-AF9 band intensities compared with WT MLLN. (G) Representative qPCR analyses show elevated expression levels of MLL target genes compared with non-MLL leukemic cell lines or controls (human BM or BM CD10+/−/CD19+) but similar to cell lines and patients with MLL translocations. Representative results from 8 independent experiments are shown. (H) Unsupervised hierarchical cluster analysis of 3 leukemic mice (ALL) and 70 MLL-rearranged ALL patients showing similar gene expression profiling in contrast to control samples. Each dot (A-C) represents a mouse; horizontal bars represent the mean. *P < .05 was considered statistically significant. Error bars indicate SEM. HGB, hemoglobin; PB, peripheral blood; PLT, platelet; WBC, white blood cell.
Pathologic and molecular features of acute leukemias induced by genome editing of the MLL oncogene. (A) Plots show results of hematologic analyses performed on control (n = 7) and leukemic mice (n = 16), respectively. (B) Representative peripheral blood smears of control and leukemic mice are shown and summarized by calculating the percentage of blast cells (n = 16). Scale bars define 10 µm. (C) Spleen size and weight are shown for 1 representative control and leukemic mice (n = 17). (D) Hematoxylin-and-eosin–stained paraffin sections demonstrate disruption of organ architecture due to tumor infiltration compared with control mice. Scale bars define 100 µm. (E) PCR/RT-PCR was performed on gDNA and cDNA of leukemia cells to detect integration and expression of the MLL oncogenes and WT MLL gene. (F) Representative western blot analysis shows WT MLLN and MLL-AF9 expression in control (CD34+ cells nucleofected with template alone) and explanted blast cells from xenotransplants induced by either retroviral transduction or under the expression of the endogenous promoter (MPAL, AML). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), loading control. Bottom, relative MLL-AF9 band intensities compared with WT MLLN. (G) Representative qPCR analyses show elevated expression levels of MLL target genes compared with non-MLL leukemic cell lines or controls (human BM or BM CD10+/−/CD19+) but similar to cell lines and patients with MLL translocations. Representative results from 8 independent experiments are shown. (H) Unsupervised hierarchical cluster analysis of 3 leukemic mice (ALL) and 70 MLL-rearranged ALL patients showing similar gene expression profiling in contrast to control samples. Each dot (A-C) represents a mouse; horizontal bars represent the mean. *P < .05 was considered statistically significant. Error bars indicate SEM. HGB, hemoglobin; PB, peripheral blood; PLT, platelet; WBC, white blood cell.
Leukemic cells explant, proliferate ex vivo, and serial transplant with enhanced oncogenic potential
Leukemic blast cells were explanted from MPAL and AML mice into serial CFC assays where they formed predominantly compact colonies (Figure 5A), which increased in number and total cell counts after each serial replating (Figure 5B-C). The cellular morphology and phenotypes were comparable to the AML blast portion present in leukemic mice (Figure 5D-E). Cultured cells demonstrated significantly increased expression of MLL target genes MEIS1 and HOXA9 after each replating, suggesting expansion and enrichment for leukemia-propagating cells in vitro (Figure 5F). The explanted cells displayed indefinite growth ex vivo in contrast to the limited in vitro growth potentials of fresh knock-in cells that eventually exhausted during sequential replatings. The CFCs were serially transplanted into secondary recipients, which showed an analogous pathologic phenotype as primary recipients except that the median latency of secondary disease was significantly shorter than the primary leukemias, and only the AML blast portion was present after culturing them under myeloid growth conditions in CFC assays (Figure 5G). The more robust oncogenic properties of leukemic cells vs fresh nucleofected cells suggest that activation of the MLL oncogene alone did not fully transform human HSPCs, which must acquire additional transformation events in vivo to advance to acute leukemia.
Immortalization ex vivo and increased oncogenic potential through CFC assays of MLL edited acute leukemia cells. CFC assays were performed to assess the replating efficiency of leukemic cells ex vivo in semisolid medium. Scale bars define 100 µm. (A) Images show representative morphologies of compact colonies displaying increased density after each replating. (B) Bar graph represents the mean number of colonies generated per 104 seeded cells. (C) Plot indicates cell numbers after each replating. Pooled data from 3 independent experiments. (D) Representative morphologies and phenotypes (E) are shown for colony-forming cells. Scale bar defines 10 µm. (F) Representative qPCR analyses of MLL target genes show increasing levels after each replating in CFC assays. Results from 1 of 3 independent experiments performed in triplicate. (G) Kaplan-Meier plot is shown for each cohort of animals (direct inject = 4 and secondary inject = 4). *P < .05 was considered statistically significant. Error bars indicate SEM. CFU, colony-forming unit; R, round.
Immortalization ex vivo and increased oncogenic potential through CFC assays of MLL edited acute leukemia cells. CFC assays were performed to assess the replating efficiency of leukemic cells ex vivo in semisolid medium. Scale bars define 100 µm. (A) Images show representative morphologies of compact colonies displaying increased density after each replating. (B) Bar graph represents the mean number of colonies generated per 104 seeded cells. (C) Plot indicates cell numbers after each replating. Pooled data from 3 independent experiments. (D) Representative morphologies and phenotypes (E) are shown for colony-forming cells. Scale bar defines 10 µm. (F) Representative qPCR analyses of MLL target genes show increasing levels after each replating in CFC assays. Results from 1 of 3 independent experiments performed in triplicate. (G) Kaplan-Meier plot is shown for each cohort of animals (direct inject = 4 and secondary inject = 4). *P < .05 was considered statistically significant. Error bars indicate SEM. CFU, colony-forming unit; R, round.
Gene-edited leukemias display increased sensitivity to DOT1L inhibition
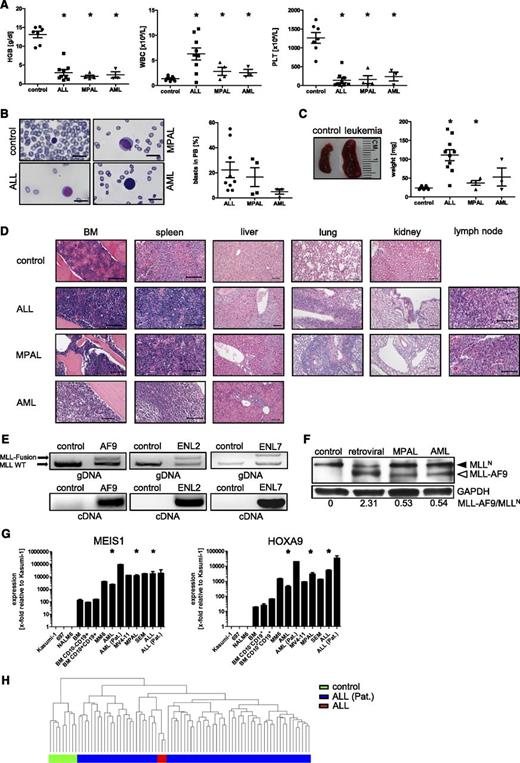
DOT1L catalyzes the methylation of H3K79, a chromatin modification that leads to enhanced expression of leukemogenic genes, including HOXA9 and MEIS1 in MLL-rearranged leukemias.34-44 Small-molecule inhibitors of DOT1L have been developed and are currently undergoing phase 1 clinical trials.45-50 We tested leukemic blast cells explanted from MPAL and AML mice for sensitivity to the DOT1L inhibitor EPZ004777 in comparison with the human MLL-rearranged leukemia cell line MV4-11 and non-MLL-rearranged K562 cells. In CFC assays, the explanted blasts and MV4-11 cells displayed high sensitivity to EPZ004777 in a dose-dependent manner characterized by a decrease of colony number and size, whereas K562 cells showed no significant differences (Figure 6A-B). EPZ004777 induced differentiation of leukemic and MV4-11 cells characterized by the induction and increased expression of CD14 (Figure 6C). The compound also inhibited expression of target genes, HOXA9 and MEIS1, in the MLL leukemic cells (Figure 6D). These data demonstrate the dependence of gene-edited leukemias on DOT1L, a signature feature of primary acute MLL-rearranged leukemias.
Edited leukemia cells display increased sensitivity to DOT1L inhibition. (A) Bar graph represents the mean number of colonies generated per 5000 or 104 seeded cells after 12 to 14 days in the presence of increasing concentrations of EPZ004777 or vehicle (DMSO). Data from 4 (MPAL) and 3 (AML) independent experiments performed in duplicate were pooled together (IC50 values: MV4-11, 10 nM; MPAL/AML, 51/11 nM; K562, >50 µM). (B) Images show representative morphologies of colonies displaying decreased density after drug treatment. Scale bars define 100 µm. (C) MV4-11 and leukemic cells (n [MPAL] = 4; n [AML] = 3) were analyzed by flow cytometry for cell surface expression of CD14 after incubation with 10 µM EPZ04777 (black line) or DMSO (gray line) in CFC assays. (D) Representative qPCR analyses of MLL target genes show decreasing levels relative to DMSO treatment. Shown are representative data from 1 of 4 (MPAL) and 3 (AML) independent experiments. *P < .05 was considered statistically significant. Error bars indicate SEM.
Edited leukemia cells display increased sensitivity to DOT1L inhibition. (A) Bar graph represents the mean number of colonies generated per 5000 or 104 seeded cells after 12 to 14 days in the presence of increasing concentrations of EPZ004777 or vehicle (DMSO). Data from 4 (MPAL) and 3 (AML) independent experiments performed in duplicate were pooled together (IC50 values: MV4-11, 10 nM; MPAL/AML, 51/11 nM; K562, >50 µM). (B) Images show representative morphologies of colonies displaying decreased density after drug treatment. Scale bars define 100 µm. (C) MV4-11 and leukemic cells (n [MPAL] = 4; n [AML] = 3) were analyzed by flow cytometry for cell surface expression of CD14 after incubation with 10 µM EPZ04777 (black line) or DMSO (gray line) in CFC assays. (D) Representative qPCR analyses of MLL target genes show decreasing levels relative to DMSO treatment. Shown are representative data from 1 of 4 (MPAL) and 3 (AML) independent experiments. *P < .05 was considered statistically significant. Error bars indicate SEM.
Discussion
Using genome-editing techniques, we generated MLL-AF9 and MLL-ENL oncogenes in primary human HSPCs based on knock-in mutations of the endogenous MLL gene to model the consequences of oncogene activation in human leukemia both in vitro and in vivo using a huCB xenotransplantation assay. Our studies demonstrate that the CD34+ fraction of huCB cells is capable of initiating leukemia in response to endogenous activated MLL-AF9 or MLL-ENL oncogenes. The productive insertion of knock-in constructs under control of the endogenous MLL promoter was confirmed by the expression of a NeonGreen marker gene by flow cytometry, confocal microscopy, and PCR. Two alternative transplantation approaches differed with respect to whether knock-in cells were subjected to antecedent culture prior to transplantation in NSG mice. Culture in vitro (3 weeks) under myeloid growth conditions prior to transplantation resulted in AML, whereas direct injection (<3 day of in vitro culture) led to the development of ALL, AML, or MPAL induced by MLL-AF9 oncogene expression, or ALL induced by MLL-ENL constructs. Although limited by overall cohort sizes, our data support and are consistent with previous reports showing that both microenvironmental cues and the fusion partner serve primary roles in lineage determination of leukemia.32,33
Although previous studies utilizing a variety of in vivo models have substantially advanced our understanding of MLL leukemia pathogenesis,32,33,51-53 they may not fully reconstitute leukemia as it manifests in human patients. For example, genetically engineered mouse models driven by germline or somatic MLL oncogene activation may be limited by species-specific differences in genetic background and requirements for leukemic transformation.8-10 Retroviral transduction/transplantation models using mouse HSPCs are widely used but may not accurately recapitulate the role of MLL fusion proteins in human diseases.23,54 Xenotransplantation studies using transduced primary human cells offer a potentially more relevant approach for modeling leukemia pathogenesis, but suffer from limitations of nonphysiological oncogene expression under control of the retroviral long terminal repeat, which is more broadly expressed and resistant to differentiation-linked downregulation compared with the endogenous MLL promoter. Furthermore, retroviral models maintain the presence of 2 wild-type MLL genes. Therefore, it remains uncertain to what extent these models mirror the naturally occurring disease in humans.
The genome-editing approach developed in our studies bypasses these limitations and provides a novel method to explore the roles of various initiating events in leukemia pathogenesis de novo in a prospective, physiologic, and faithful manner using primary human HSPCs. Notably, activation of endogenous MLL oncogenes in human HSPCs induced transient enhancement of in vitro growth potentials, but the effects were not as strong as those observed in retroviral transduction of both primary murine and human HSPCs, which display unlimited growth potentials in methylcellulose assays.23,33 Thus, previous models may actually induce more robust oncogenic readouts than the genomic aberrations that activate the endogenous MLL gene in primary human HSPCs.
huCB leukemias generated by our approach faithfully recapitulate many features of the clinical disease present in a subset of leukemia patients associated with MLL chromosomal translocations. They are consistent with the original designation of MLL as the mixed lineage leukemia oncogene.55 Moreover, the phenotype, morphology, and molecular features of the induced leukemias presented in our study are similar to patient leukemic blasts. This includes expression of an MLL-associated transcriptional program with elevated levels of crucial MLL target genes HOXA9 and MEIS1, which have well-described roles in both the induction and maintenance of MLL-fusion leukemias.56-58 Similar to clinical observations, the leukemias displayed heightened sensitivity to DOT1L inhibition consistent with aberrant recruitment and dependence on this histone methyltransferase in leukemias driven by MLL-AF9 and MLL-ENL oncogenes and could serve for drug screening of human cells.
In the present study, the development of leukemia was relatively rapid, with a median latency of 16 weeks. Although expression of MLL-AF9 or MLL-ENL is the primary genetic defect responsible for the defining characteristics of disease, MLL fusion alone was not sufficient to drive unlimited proliferation of human HSPCs in our cultures compared with the more robust growth of explanted leukemia cells consistent with the acquisition of additional genetic or epigenetic events in vivo. The rapid progression of gene-edited HSPCs to a fully developed leukemia phenotype in vivo supports the hypothesis, based on clinical and genomic observations, that MLL fusion genes require fewer cooperating oncogenic events for leukemic transformation than other fusion oncoproteins.5,7 This is consistent with MLL rearrangements that arise in utero in the majority of infants with acute leukemia, and are unique in their ability to produce overt clinical disease within a few months.59,60 Preliminary analyses suggest that our leukemias lack mutations in candidate driver genes KRAS, NRAS, FLT3, and PIK3CA7 when assessed using previously published primers.61,62 However, these studies were not exhaustive and in the future, our model system can be used to further elucidate the pivotal role of secondary mutations in MLL-rearranged leukemogenesis and provide an experimental platform to further investigate driver and passenger mutations. It promises potential insights into the early events of MLL fusion-driven leukemogenesis and allows for further prospective studies of leukemia initiation and stem cell biology.
The Affymetrix gene expression data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE68879).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dan Voytas (University of Minnesota) for generously providing the TALEN Golden Gate library, and Daniel E. Vega Salazar (Stanford Hospital) for his efforts and assistance in the collection of huCB.
This work was supported in part by grants from the National Institutes of Health, National Cancer Institute grants CA116606 and CA160384 (M.L.C.), Alex’s Lemonade Stand Foundation (M.L.C.), Hyundai Hope on Wheels (M.P.), the Dr. Mildred Scheel Stiftung (C.B., D.S.), the St. Baldrick’s Foundation (E.H.B.), and the German Research Foundation (Deutsche Forschungsgemeinschaft, ref. DU128712-1; J.D.).
Authorship
Contribution: C.B. designed and performed the research, analyzed data, and wrote the manuscript; E.H.B., D.S., J.J., C.-H.L., J.D.-A., S.H.K.W., and K.S.S. performed research and analyzed data; R.S.N. provided patient material; M.P. provided fruitful discussions and guidance; M.L.C. provided overall guidance; and all authors edited the manuscript for content.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michael L. Cleary, Lokey Stem Cell Research Building, Room G2034, 1291 Welch Rd, Stanford, CA 94305; e-mail: mcleary@stanford.edu.


![Figure 2. Generation of MLL-AF9 and MLL-ENL knock-in genes by genome engineering. (A) Schematic illustration of experimental strategy to induce a DSB by TALENs followed by integration of the knock-in template in the MLL gene locus by homology-directed repair (HDR). (B) FACS profiles show fluorescence of K562 and CD34+ cells nucleofected with the knock-in template alone (gray line) or in combination with the MLL TALENs (black line) sorted on days 5 and 3, respectively. (C) Summary of NeonGreen expression in K562 (n[AF9] = 3, n [ENL2/7] = 2) and CD34+ cells (n[AF9] = 11, n[ENL2] = 7, n[ENL7] = 5) pre- and postsort. *P < .05 was considered statistically significant. Error bars indicate standard error of the mean (SEM). (D) Confocal microscopy images show NeonGreen expression in sorted K562 and control cells as indicated. Top row, cell density (brightfield); bottom row, NeonGreen expression detected by GFP excitation (450-490 nm) and ×10 objective. (E) PCR/RT-PCR was performed on gDNA and cDNA isolated from NeonGreen-positive K562 and CD34+ cells to detect integration and transcription of the construct under control of the endogenous MLL promoter (representative results shown for MLL-AF9). BGH, bovine growth hormone.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/126/14/10.1182_blood-2015-05-646398/4/m_1683f2.jpeg?Expires=1766005018&Signature=vNPBfkKcIfD5C2rpgUgrNm6vBmrxNUTN6Jtd8E6-QYc0zeBFOOmkVHtd0y35EzSNBzrFlbjH4zXj1temJ~t05c13K-eKh9biOMjp4SR8RC7AQWe9wC8MQRkoj4dt0HHe93H1xCSs9xj8jikBMh81bCRCepsWoqCZ907aSPwEBLhCmlRSc~x9q984QzkUbJwI7X48wET2UtoiJOfSkwm6aGn95YN9z140uxJ23D5oCHHy7938lv7uZnQrYKwmTkM6AaA7EQWjFH6Mld7DxULETPzRqrAgbkJSA49vIt1895UV2zqFyUxO9EVylBuKxU~9Ti6pyY4vwPxJwmIvFFo6uw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. Induction of acute leukemias following transplantation of human CD34+ cells containing knock-in MLL oncogenes. (A) Experimental scheme depicts nucleofection of CD34+ cells and their subsequent transplantation directly into sublethally irradiated NSG recipient mice or culture for 3 weeks in vitro prior to transplantation. (B) Kaplan-Meier plot is shown for cohorts of mice transplanted with CD34+ cells transfected with templates plus TALENs (n [AF9] = 25: direct inject = 19, cultured cells = 6; n [ENL2] = 7: direct inject = 5, cultured cells = 2; n [ENL7] = 8: direct inject = 5, cultured cells = 3) or template alone (control, n = 7). P < .05 was considered statistically significant. Mice were sacrificed upon signs of illness. (C) Flow cytometry profiles show representative phenotypes of various leukemias that developed in mice transplanted with CD34+ cells (n = 17) compared with blasts from patients with MLL translocations (AML/ALL), which display comparable phenotypes. Also shown are representative profiles of BM cells from control mice (n = 2) that received CD34+ cells nucleofected with knock-in construct alone (week 16 posttransplantation) and representative analysis of knock-in cells 3 weeks after cell culture prior to transplantation. l, lymphoid; m, myeloid; (Pat.), patient.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/126/14/10.1182_blood-2015-05-646398/4/m_1683f3.jpeg?Expires=1766005018&Signature=Gjj6lg6-r2Y5id9wsv-kUEwMSSWpnxMX3kp9dBOiIm7AbcvkHc33F0t8zcilwAkY9GivviXQwHTpTb-W6oOWsRjUBlexG~tpsIEq4txXHyIumnfDCBxCTyZWMn1eNriJ2gmNtMFl079EcCmqhTyFpdPqSx7-8kLVWa5h07IQ-Uuqq8y5mIIo2mawWtJqNHYBaRF4IFVlhpK2wRIHIh03dWmD-TdoeReHGYIFdHGLqBzqLWocJt5Ap71B6d1F4emDFzcrDzSQHl2JAB6T45~IoYqU~iO8kTWOBDSXGj0R6a5Br1KMapnVCigj-dv8onGH307TFXDk5UTf1sAW57fSYw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 6. Edited leukemia cells display increased sensitivity to DOT1L inhibition. (A) Bar graph represents the mean number of colonies generated per 5000 or 104 seeded cells after 12 to 14 days in the presence of increasing concentrations of EPZ004777 or vehicle (DMSO). Data from 4 (MPAL) and 3 (AML) independent experiments performed in duplicate were pooled together (IC50 values: MV4-11, 10 nM; MPAL/AML, 51/11 nM; K562, >50 µM). (B) Images show representative morphologies of colonies displaying decreased density after drug treatment. Scale bars define 100 µm. (C) MV4-11 and leukemic cells (n [MPAL] = 4; n [AML] = 3) were analyzed by flow cytometry for cell surface expression of CD14 after incubation with 10 µM EPZ04777 (black line) or DMSO (gray line) in CFC assays. (D) Representative qPCR analyses of MLL target genes show decreasing levels relative to DMSO treatment. Shown are representative data from 1 of 4 (MPAL) and 3 (AML) independent experiments. *P < .05 was considered statistically significant. Error bars indicate SEM.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/126/14/10.1182_blood-2015-05-646398/4/m_1683f6.jpeg?Expires=1766005018&Signature=hiHJRdpktq5gUDtGn5LHTOs6Vxch6tw2tIZoo9R4HYG8KsX-kXGNubzloFAyyqQnIm-xxGIVED03BKuZUni95px~lBBKmS~sHz6pKxSQXwM4a6420KK6muqSDQACL1-tz77GDrm2jSq0yeKzyt~EVEfi~~KZ6hqiaaThad7n--qCNpgNNFJUORS5-cVQYi93S92DLqtYierkc~C65bA88e74-JFcUCp~ucaDZuV8uNuoc-LLw~Aom6Mp-NW8s1IPufbXoEuCYA9JTRjAsp9FvxfGaCpF1VRt~YyuOimXB21uBarjQSt5jxo4JOOoocmHjpZnpiwwJj31wWL05wEOmw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)