Key Points
Direct analysis of the HLA-presented peptidome identifies a distinct antigenic signature in MM.
T-cell responses for these antigens are detectable exclusively in MM patients and can be induced in vitro in response-naive patients.
Abstract
Direct analysis of HLA-presented antigens by mass spectrometry provides a comprehensive view on the antigenic landscape of different tissues/malignancies and enables the identification of novel, pathophysiologically relevant T-cell epitopes. Here, we present a systematic and comparative study of the HLA class I and II presented, nonmutant antigenome of multiple myeloma (MM). Quantification of HLA surface expression revealed elevated HLA molecule counts on malignant plasma cells compared with normal B cells, excluding relevant HLA downregulation in MM. Analyzing the presentation of established myeloma-associated T-cell antigens on the HLA ligandome level, we found a substantial proportion of antigens to be only infrequently presented on primary myelomas or to display suboptimal degrees of myeloma specificity. However, unsupervised analysis of our extensive HLA ligand data set delineated a panel of 58 highly specific myeloma-associated antigens (including multiple myeloma SET domain containing protein) which are characterized by frequent and exclusive presentation on myeloma samples. Functional characterization of these target antigens revealed peptide-specific, preexisting CD8+ T-cell responses exclusively in myeloma patients, which is indicative of pathophysiological relevance. Furthermore, in vitro priming experiments revealed that peptide-specific T-cell responses can be induced in response-naive myeloma patients. Together, our results serve to guide antigen selection for T-cell–based immunotherapy of MM.
Introduction
Antigen-specific immunotherapy holds the potential to induce clinically effective anticancer T-cell responses1,2 and might be harnessed to guide and increase the specificity of cancer immunotherapy in future combination trials.3 To this end, the exact knowledge of tumor-associated/specific T-cell epitopes is crucial. After decades of research into overexpressed tumor antigens, more recently the focus has shifted to the patient-individualized identification of mutation-derived neoantigens.4,5 The encouraging findings of these new studies6-8 have led to neoepitopes being viewed as the dominant targets of anticancer immune responses.9-11
However, analyzing the antigenome of hematologic malignancies, we have recently demonstrated that nonmutated antigens are relevant targets of spontaneous antileukemia T-cell responses.12,13 The strategy implemented in these studies differentially maps the naturally presented HLA ligandomes of hematologic cells in health and disease by mass spectrometry and was found to efficiently identify relevant tumor-associated antigens (TAAs).
Here, we translated this approach to multiple myeloma (MM), a low-grade B-cell lymphoma, characterized by the proliferation of malignant plasma cells in the bone marrow.14 Despite recent advances in treatment, including high-dose chemotherapy followed by autologous stem cell transplantation, novel immunomodulatory drugs, and proteasome inhibitors, MM remains largely incurable.15,16 This is mostly due to the persistence of minimal residual disease (MRD), which leads to high relapse rates.17,18 So far, the only established immunotherapeutic approach for MM is allogenic stem cell transplantation, which is associated with a high morbidity and mortality and remains an option for only a fraction of patients.19-21 Antigen-specific T-cell–based immunotherapy,22,23 especially in the constellation of MRD characterized by favorable effector-to-target ratios, might present an effective, low side-effect option.24
An array of myeloma-associated T-cell antigens has been described in previous studies.25-35 Most of these antigens were identified based on gene expression analysis and reverse immunology. Some of these antigens (WT1,36,37 RHAMM,38,39 hTERT,40 and Survivin40,41 ) have already found their way into clinical trials, showing promising results in terms of induction of specific T-cell responses as well as clinical responses in single patients. However, broad clinical effectiveness has not yet been achieved. These previous studies were restricted to very few HLA allotypes and single antigens/epitopes,42 limiting both the population of patients eligible for this therapeutic approach and the spectrum of inducible tumor-specific T-cell responses. Of note, recent studies demonstrated lacking degrees of tumor association for several of these tumor antigens, both on the transcriptome level43 and importantly also on the level of HLA-restricted presentation.12,13
By analyzing the antigenic landscape of MM directly on the HLA ligand level, we here provide new insights on antigenic distribution/specificity and identify a panel of novel myeloma-associated epitopes suited for antigen-specific immunotherapy.
Materials and methods
Patients, blood, and bone marrow samples
Bone marrow mononuclear cells (BMNCs) and peripheral blood mononuclear cells (PBMCs) from MM patients at the time of diagnosis or at relapse before therapy, as well as PBMCs, BMNCs and granulocytes of healthy volunteers (HVs), were isolated by density gradient centrifugation (Biocoll; Biochrom GmbH) and erythrocyte lysis (EL buffer; Qiagen). Informed consent was obtained in accordance with the Declaration of Helsinki protocol. The study was performed according to the guidelines of the local ethics committee (142/2013BO2). Patient characteristics are provided in Table 1.44 HLA typing was carried out by the Department of Hematology and Oncology, University of Tübingen, Tübingen, Germany.
Patient characteristics
| UPN . | Sex . | Age (y) . | ISS Stage . | Durie and Salmon . | Light chain restriction . | Cytogenetic risk . | Previous therapy . | HLA typing . | Experiment . |
|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 62 | 3 | 3B | λ | High | Yes | A*26, A*30, B*15, B*42 | Q; L |
| 2 | F | 55 | 2 | 1A | Non | Standard | No | A*01, A*24, B*18, B*08 | L |
| 3 | F | 69 | 2 | 3B | κ | Standard | No | A*02, A*01, B*08, B*37 | L |
| 4 | M | 72 | 3 | 3A | κ | Standard | No | A*02, A*33, B*15, B*18 | L |
| 5 | M | 54 | 2 | 3A | κ | Low | No | A*03, A*26, B*40, B*55 | L |
| 6 | M | 60 | 3 | 3B | λ | High | No | A*02, A*24, B*07, B*27 | L |
| 7 | F | 39 | 3 | 3A | κ | Standard | No | A*02, A*03, B*07, B*35 | L |
| 8 | F | 73 | 3 | 3B | λ | Unknown | No | A*24, A*25, B*39, B*40 | L |
| 9 | M | 52 | 3 | 1B | κ | Unknown | No | A*02, B*07, B*44 | L |
| 10 | M | 47 | 1 | 3A | κ | Standard | No | A*03, A*33, B*07 | L |
| 11 | F | 72 | 2 | 3A | κ | High | Yes | A*02, B*27, B*44 | Q |
| 12 | F | 68 | 2 | 3A | κ | Standard | No | n.d. | Q |
| 13 | M | 74 | 1 | 3A | κ | Standard | No | n.d. | Q |
| 14 | F | 50 | 1 | 3A | λ | Standard | No | A*01, A*68, B*08, B*35 | Q |
| 15 | M | 62 | 1 | 3A | κ | Standard | Yes | n.d. | Q |
| 16 | M | 74 | 3 | 2B | κ | Standard | Yes | n.d. | Q |
| 17 | M | 68 | 1 | 3A | κ | Unknown | Yes | A*03, A*33, B*14, B*18 | Q |
| 18 | M | 73 | 1 | 1A | κ | Standard | Yes | A*02, A*32, B*40, B*44 | Q |
| 19 | F | 70 | 1 | 1A | κ | Standard | No | n.d. | Q |
| 20 | F | 53 | 3 | 3A | κ | Standard | No | n.d. | Q |
| 21 | M | 59 | 2 | 3A | λ | Standard | Yes | n.d. | Q |
| 22 | M | 57 | 1 | 1A | κ | Standard | Yes | n.d. | Q |
| 23 | F | 65 | 3 | 1A | λ | Standard | Yes | n.d. | Q |
| 24 | M | 76 | 2 | 3A | λ | Standard | Yes | n.d. | Q |
| 25 | F | 74 | 2 | 2A | κ | Standard | Yes | A*01, A*02, B*08, B*44 | Q |
| 26 | F | 67 | 1 | 3A | λ | Standard | Yes | n.d. | Q |
| 27 | M | 50 | 1 | 1A | λ | Unkwnon | Yes | A*01, A*03, B*35, B*40 | Q |
| 28 | M | 70 | 3 | 3A | λ | Unknown | No | n.d. | Q |
| 29 | M | 75 | 1 | 3A | κ | Standard | Yes | n.d. | Q |
| UPN . | Sex . | Age (y) . | ISS Stage . | Durie and Salmon . | Light chain restriction . | Cytogenetic risk . | Previous therapy . | HLA typing . | Experiment . |
|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 62 | 3 | 3B | λ | High | Yes | A*26, A*30, B*15, B*42 | Q; L |
| 2 | F | 55 | 2 | 1A | Non | Standard | No | A*01, A*24, B*18, B*08 | L |
| 3 | F | 69 | 2 | 3B | κ | Standard | No | A*02, A*01, B*08, B*37 | L |
| 4 | M | 72 | 3 | 3A | κ | Standard | No | A*02, A*33, B*15, B*18 | L |
| 5 | M | 54 | 2 | 3A | κ | Low | No | A*03, A*26, B*40, B*55 | L |
| 6 | M | 60 | 3 | 3B | λ | High | No | A*02, A*24, B*07, B*27 | L |
| 7 | F | 39 | 3 | 3A | κ | Standard | No | A*02, A*03, B*07, B*35 | L |
| 8 | F | 73 | 3 | 3B | λ | Unknown | No | A*24, A*25, B*39, B*40 | L |
| 9 | M | 52 | 3 | 1B | κ | Unknown | No | A*02, B*07, B*44 | L |
| 10 | M | 47 | 1 | 3A | κ | Standard | No | A*03, A*33, B*07 | L |
| 11 | F | 72 | 2 | 3A | κ | High | Yes | A*02, B*27, B*44 | Q |
| 12 | F | 68 | 2 | 3A | κ | Standard | No | n.d. | Q |
| 13 | M | 74 | 1 | 3A | κ | Standard | No | n.d. | Q |
| 14 | F | 50 | 1 | 3A | λ | Standard | No | A*01, A*68, B*08, B*35 | Q |
| 15 | M | 62 | 1 | 3A | κ | Standard | Yes | n.d. | Q |
| 16 | M | 74 | 3 | 2B | κ | Standard | Yes | n.d. | Q |
| 17 | M | 68 | 1 | 3A | κ | Unknown | Yes | A*03, A*33, B*14, B*18 | Q |
| 18 | M | 73 | 1 | 1A | κ | Standard | Yes | A*02, A*32, B*40, B*44 | Q |
| 19 | F | 70 | 1 | 1A | κ | Standard | No | n.d. | Q |
| 20 | F | 53 | 3 | 3A | κ | Standard | No | n.d. | Q |
| 21 | M | 59 | 2 | 3A | λ | Standard | Yes | n.d. | Q |
| 22 | M | 57 | 1 | 1A | κ | Standard | Yes | n.d. | Q |
| 23 | F | 65 | 3 | 1A | λ | Standard | Yes | n.d. | Q |
| 24 | M | 76 | 2 | 3A | λ | Standard | Yes | n.d. | Q |
| 25 | F | 74 | 2 | 2A | κ | Standard | Yes | A*01, A*02, B*08, B*44 | Q |
| 26 | F | 67 | 1 | 3A | λ | Standard | Yes | n.d. | Q |
| 27 | M | 50 | 1 | 1A | λ | Unkwnon | Yes | A*01, A*03, B*35, B*40 | Q |
| 28 | M | 70 | 3 | 3A | λ | Unknown | No | n.d. | Q |
| 29 | M | 75 | 1 | 3A | κ | Standard | Yes | n.d. | Q |
Cytogenetic risk according to the IMWG risk stratification.44
F, female; IMWG, International Myeloma Working Group; ISS, International Staging System; L, HLA ligand isolation; M, male; n.d., not determined; Q, HLA quantification; UPN, uniform patient number; y, years.
Myeloma cell lines
For HLA ligandome analysis, myeloma cell lines (MCLs; U266, RPMI 8226, JJN3, LP-1, MM.1S) were cultured in the recommended cell media (RPMI 1640, Gibco; Iscove modified Dulbecco medium [IMDM], Lonza) supplemented with 10%/20% fetal calf serum, 100 IU/L penicillin, 100 mg/L streptomycin, and 2 mmol/L glutamine at 37°C and 5% CO2. The MCLs RPMI 8226, JJN3, MM.1S and LP-1 were obtained from the Department of Hematology and Oncology, University of Tübingen, Tübingen, Germany.
Quantification of HLA surface expression
HLA surface expression on MM patient and HV bone marrow cells including CD38+CD138+ myeloma cells/plasma cells, CD19+CD20+ B cells, CD3+ T cells, and CD34+CD38− hematopoietic progenitor cells (HPCs) were analyzed using the QIFIKIT bead–based quantitative flow cytometric assay (Dako) according to the manufacturer’s instructions as described previously.12 In brief, samples were stained with the pan-HLA class I–specific monoclonal antibody (mAb) W6/32, HLA-DR–specific mAb L243 (produced in-house) or immunoglobulin G isotype control (BioLegend), respectively. Surface marker staining was carried out with directly labeled CD138, anti-κ, anti-λ, CD19, CD20 (BioLegend) and CD38, CD3, and CD34 (BD) antibodies. 7-aminoactinomycin D (BioLegend) was added as viability marker immediately before flow cytometric analysis on a LSR Fortessa (BD Biosciences).
Isolation of HLA ligands from primary samples and MCLs
HLA class I and II molecules were isolated using standard immunoaffinity purification as described45 using the pan-HLA class I–specific mAb W6/32, the pan-HLA class II–specific mAb Tü39, and the HLA-DR–specific mAb L243 (produced in-house).
Analysis of HLA ligands by liquid chromatography–tandem mass spectrometry
HLA ligand extracts were analyzed in 5 technical replicates as described previously.13 In brief, peptide samples were separated by nanoflow high-performance liquid chromatography (RSLCnano; Thermo Fisher Scientific) using a 50 μm × 25 cm PepMap rapid separation liquid chromatography column (Thermo Fisher Scientific) and a gradient ranging from 2.4% to 32.0% acetonitrile over the course of 90 minutes. Eluting peptides were analyzed in an online-coupled LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific) using a top 5 collision-induced dissociation fragmentation method.
Database search and spectral annotation
Data processing was performed as described previously.13 In brief, the Mascot search engine (Mascot 2.2.04; Matrix Science) was implemented to search the human proteome as comprised in the Swiss-Prot database (20 279 reviewed protein sequences, September 2013) without enzymatic restriction. Potential mutated HLA ligands were searched implementing a database containing the human proteome concatenated with proteins containing single amino acid variants listed in the COSMIC database (http://cancer.sanger.ac.uk/cosmic/). Only recurrent single amino acid variants described in 2 or more samples of hematologic origin were included. Oxidized methionine was allowed as a dynamic modification. The false discovery rate (FDR) was estimated using the Percolator algorithm46 and set to 5%. Peptide lengths were limited to 8 to 12 aa for HLA class I and 12 to 25 aa for HLA class II. Protein inference was disabled, allowing for multiple protein annotations of peptides. HLA annotation was performed using SYFPEITHI47 or an extended in-house database. Experimental validation of peptide identifications and HLA annotations was performed by mass spectrometric and functional characterization of synthetic peptides for a subset of peptides.
Peptide and HLA peptide monomer synthesis
The automated peptide synthesizer EPS221 (Abimed) was used to synthesize peptides using the 9-fluorenylmethyl-oxycarbonyl/tert-butyl (Fmoc/tBu) strategy.48 Synthetic peptides were used for validation of liquid chromatography–tandem mass spectrometry (MS) identifications as well as for functional experiments. Biotinylated recombinant HLA molecules and fluorescent HLA-peptide tetramers were produced as described previously.49
Amplification of peptide-specific T cells
PBMCs from MM patients and HVs were cultured as described previously.12,13 In brief, for CD8+ T-cell stimulation, PBMCs were pulsed with 1 µg/mL per peptide and cultured for 12 days adding interleukins 4 and 7 (IL-4 and IL-7) on days 0 and 1 as well as IL-2 on days 3, 5, 7, and 9. HLA-A*02 (KLFEKVKEV)- and B*07 (KPSEKIQVL)-restricted control peptides derived from benign tissues (HV-exclusive HLA ligands) served as negative control. Peptide-stimulated PBMCs were analyzed by enzyme-linked immunospot (ELISPOT) assays on day 12. For CD4+ T-cell stimulation, culture was performed as described for CD8+ T cells except for 2 modifications: pulsing was carried out with 10 µg/mL HLA class II peptide and no IL-4 or IL-7 was added.
IFNγ ELISPOT assay
Interferon γ (IFNγ) ELISPOT assays were carried out as described previously.50 In brief, 96-well nitrocellulose plates (Millipore) were coated with 1 mg/mL IFNγ mAb (Mabtech) and incubated overnight at 4°C. Plates were blocked with 10% human serum for 2 hours at 37°C. Prestimulated PBMCs (2.5 × 105 cells per well) were pulsed with 1 µg/mL (HLA class I) or 2.5 µg/mL (HLA class II) peptide and incubated for 24 to 26 hours. Readout was performed according to the manufacturer’s instructions. Phytohemagglutinin was used as positive control. HLA-A*02 (KLFEKVKEV)- and B*07 (KPSEKIQVL)-restricted control peptides derived from benign tissues (HV-exclusive HLA ligands) served as negative control. Spots were counted using an ImmunoSpot S5 analyzer (CTL). T-cell responses were considered to be positive when >10 spots per well were counted and the mean spot count per well was at least threefold higher than the mean number of spots in the negative control wells (according to the cancer immunoguiding program guidelines51 ).
aAPC priming of peptide-specific T cells
For the generation of artificial antigen-presenting cells (aAPCs), 4 × 106 streptavidin-coated polystyrene particles (Bangs Laboratories) per milliliter were resuspended in PBE (phosphate-buffered saline/bovine serum albumin/EDTA; Gibco/Sigma Aldrich/Lonza) containing 200 pM biotinylated MHC-peptide monomer and 20 nM antihuman biotinylated CD28 antibody and incubated at room temperature for 30 minutes. After washing, the aAPCs were stored at 4°C prior to use.52 CD8+ T cells from MM patients and HV were enriched by positive selection using magnetic cell sorting (Miltenyi Biotec). Stimulations were initiated in 96-well plates with 1 × 106 T cells plus 2 × 105 aAPCs in 200 μL of T-cell medium complemented with 5 ng/mL human IL-12 (PromoKine). IL-2 (65 U/μL; R&D Systems) was added on day 5. aAPC stimulation was repeated on day 10, for a total of 3 cycles.
Tetramer staining
The frequency of peptide-specific CD8+ T cells was determined on a FACSCanto II cytometer (BD Bioscience) by staining with anti-CD8 (Biolegend) and HLA:peptide-tetramer-phycoerythrin as described previously.52 Staining with tetramers containing the cytomegalovirus (CMV) pp65 A*02 peptide NLVPMVATV served as positive control, tetramers containing irrelevant; nonprimed A*02-restricted control peptides served as negative controls. Successful priming was considered if frequency of peptide-specific CD8+ T cells was >0.1% of viable cells and at least threefold higher than the frequency of peptide-specific CD8+ T cells in the negative control.
Software and statistical analysis
Flow cytometric data analysis was performed using FlowJo 7.2 (TreeStar). In-house R and Python scripts were used for the generation of virtual ligandomes and definition of virtual TAAs in the analysis of TAA FDRs and for the TAA-plateau regression analysis. The standard R heatmap.2 script was used for the unsupervised cluster analysis of HLA ligand source proteins. GraphPad Prism 6.0 (GraphPad Software) was used for statistical analysis. Statistical analysis of HLA surface expression was based on unpaired t tests.
Results
HLA class I surface expression is elevated on myeloma cells
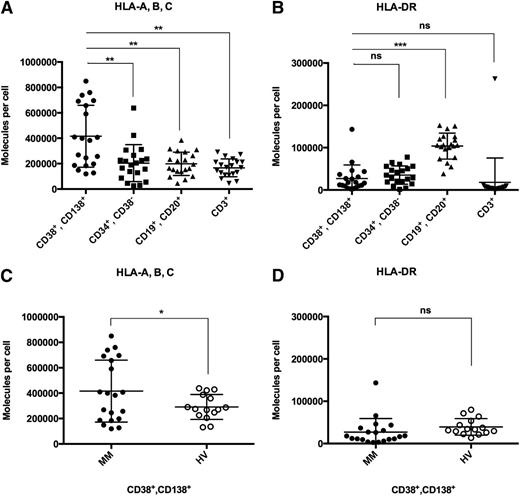
As loss or downregulation of HLA expression on target cells might severely hamper the effectiveness of T-cell–based immunotherapy, we quantified HLA class I and II surface molecule counts on primary myeloma cells compared with autologous hematopoietic cells and plasma cells derived from the bone marrow of HVs. In MM patients (n = 20), HLA class I expression was found to be heterogeneous with mean expression levels on CD38+CD138+ myeloma cells of 416 000 ± 54 500, which was found to be significantly higher as compared with autologous normal CD19+CD20+ B cells (198 5000 ± 20 500, P = .001), CD3+ T cells (167 500 ± 15 500, P = .0002), and CD34+CD38− HPCs (204 000 ± 32 500, P = .002, Figure 1A). In addition, HLA class I expression on primary MM cells was also found to be significantly higher than that on CD38+CD138+ plasma cells of HVs (n = 15, 291 500 ± 25 500, P < .05; Figure 1C). No significant differences in HLA class I expression were observed when comparing normal B cells, T cells, and HPCs of MM patients to the corresponding cell populations of HVs (supplemental Figure 1, see supplemental Data available on the Blood Web site). HLA-DR expression levels on myeloma cells were generally found to be much lower than HLA class I levels. Mean HLA-DR surface molecule counts on myeloma cells (27 000 ± 7000) showed no significant difference compared with autologous HPCs (35 000 ± 5000) and T cells (18 000 ± 13 000) or plasma cells of HVs (39 500 ± 5000) (Figure 1B,D). HLA-DR expression of MM patient CD19+CD20+ B cells (104 000 ± 7000) was significantly higher compared with myeloma cells (P < .0001). No correlation of HLA surface expression on myeloma cells with patient characteristics such as sex, age, disease stage, risk classification, or prior therapy was observed (supplemental Table 1).
HLA class I and II surface expression on myeloma patient and HV bone marrow cells. Quantification of HLA surface expression was performed using a bead-based flow cytometric assay. (A) HLA class I and (B) HLA-DR expression on CD38+CD138+ primary myeloma cells compared with autologous CD34+CD38− HPCs, CD19+CD20+ B cells, and CD3+ T cells. (C) HLA class I and (D) HLA-DR expression on primary MM cells compared with bone marrow–derived plasma cells of HVs. n.s., not significant; *P < .05; **P < .01; ***P < .001.
HLA class I and II surface expression on myeloma patient and HV bone marrow cells. Quantification of HLA surface expression was performed using a bead-based flow cytometric assay. (A) HLA class I and (B) HLA-DR expression on CD38+CD138+ primary myeloma cells compared with autologous CD34+CD38− HPCs, CD19+CD20+ B cells, and CD3+ T cells. (C) HLA class I and (D) HLA-DR expression on primary MM cells compared with bone marrow–derived plasma cells of HVs. n.s., not significant; *P < .05; **P < .01; ***P < .001.
MS identifies naturally presented HLA ligands of primary MM samples and MCLs
Mapping the HLA class I ligandomes of 10 myeloma patients and 5 MCLs (Table 1), we identified a total of 17 583 different peptides representing 7574 source proteins, attaining >80% of the maximum attainable coverage (Figure 2A). The mean number of unique peptide identifications (IDs) was 1059 IDs for primary myeloma samples and 2243 IDs for MCLs (supplemental Table 2). Overall, peptides restricted by 20 different HLA-A and -B allotypes were identified in this study, covering 99.3% of the German population (calculated according to Bui et al53 ). No mutated peptide identifications were obtained when processing the data set against a reference proteome containing recurrent mutations in hematologic malignancies.
Comparative HLA ligandome profiling and identification of myeloma-associated antigens. (A) Saturation analysis of HLA class I ligand source protein identifications in MM patients. Number of unique HLA ligand source protein identifications as a function of cumulative HLA ligand source protein identifications in 10 MM patients. Exponential regression allowed for the robust calculation (R2 = 0.99) of the maximum attainable number of different source protein identifications (dashed line). The dotted line depicts the source proteome coverage achieved in our MM patient cohort. (B) Overlap analysis of HLA class I ligand source proteins of primary MM samples (n = 10), MCLs (n = 5), and HV samples (total n = 45: PBMC [n = 30], BMNC [n = 10], granulocytes [n = 5]). (C) Comparative profiling of HLA ligand source proteins based on the frequency of HLA-restricted presentation in MM and HV ligandomes. Frequencies of MMs/HVs positive for HLA-restricted presentation of the respective source protein (x-axis) are indicated on the y-axis. The box on the left highlights the subset of myeloma-associated antigens showing MM-exclusive presentation in >25% of myeloma samples. (D) Statistical assessment of false-positive myeloma-antigen IDs at different threshold values. The numbers of original TAAs identified based on the analysis of the MM and HV cohorts were compared with random virtual TAAs. Virtual MM and HV samples were generated in silico based on random weighted sampling from the entirety of protein identifications in both original cohorts. These randomized virtual ligandomes of defined size (n = 957 proteins, which is the mean number of protein identifications in all analyzed samples) were used to define TAAs based on simulated cohorts of 15 MM vs 45 HV samples. The process of protein randomization, cohort assembly, and TAA identification was repeated 1000 times and the mean value of resultant virtual TAAs was calculated and plotted for the different threshold values. The corresponding FDRs for any chosen TAA threshold are listed below the x-axis. sum, summary.
Comparative HLA ligandome profiling and identification of myeloma-associated antigens. (A) Saturation analysis of HLA class I ligand source protein identifications in MM patients. Number of unique HLA ligand source protein identifications as a function of cumulative HLA ligand source protein identifications in 10 MM patients. Exponential regression allowed for the robust calculation (R2 = 0.99) of the maximum attainable number of different source protein identifications (dashed line). The dotted line depicts the source proteome coverage achieved in our MM patient cohort. (B) Overlap analysis of HLA class I ligand source proteins of primary MM samples (n = 10), MCLs (n = 5), and HV samples (total n = 45: PBMC [n = 30], BMNC [n = 10], granulocytes [n = 5]). (C) Comparative profiling of HLA ligand source proteins based on the frequency of HLA-restricted presentation in MM and HV ligandomes. Frequencies of MMs/HVs positive for HLA-restricted presentation of the respective source protein (x-axis) are indicated on the y-axis. The box on the left highlights the subset of myeloma-associated antigens showing MM-exclusive presentation in >25% of myeloma samples. (D) Statistical assessment of false-positive myeloma-antigen IDs at different threshold values. The numbers of original TAAs identified based on the analysis of the MM and HV cohorts were compared with random virtual TAAs. Virtual MM and HV samples were generated in silico based on random weighted sampling from the entirety of protein identifications in both original cohorts. These randomized virtual ligandomes of defined size (n = 957 proteins, which is the mean number of protein identifications in all analyzed samples) were used to define TAAs based on simulated cohorts of 15 MM vs 45 HV samples. The process of protein randomization, cohort assembly, and TAA identification was repeated 1000 times and the mean value of resultant virtual TAAs was calculated and plotted for the different threshold values. The corresponding FDRs for any chosen TAA threshold are listed below the x-axis. sum, summary.
As controls, we analyzed the HLA class I ligandomes of 45 HV-derived samples (30 PBMC, 10 BMNC, and 5 granulocyte specimens) identifying a total of 20 171 different peptides representing 7729 source proteins (supplemental Table 3). The HLA allotype distribution in the HV cohort covered >80% of HLA-A and -B alleles in the MM sample cohort.54 Analysis of HLA class II ligandomes was performed for 7 MM patients and 5 MCLs. A total of 6076 unique peptides representing 1743 source proteins were identified. The HLA class II HV cohort (13 PBMC, 5 BMNC, 5 granulocyte specimens) yielded 2899 different peptides representing 889 source proteins (supplemental Table 2).
HLA class I ligandome profiling identifies a novel panel of myeloma-associated antigens
To identify myeloma-associated antigens, we comparatively analyzed the HLA ligandomes of the MM sample and HV cohorts at the source protein level. Overlap analysis of HLA ligand source proteins identified 2412 proteins (corresponding to 31.8% of the mapped MM HLA source proteome) to be exclusively represented in the HLA ligandomes of MM samples. Of these MM-exclusive source proteins, 68.3% were solely identified on MCL samples, whereas 13.2% of proteins were found to be presented both on MCLs and primary MM samples. A fraction of 18.5% of myeloma-exclusive source proteins was found to be restricted to primary MM samples (Figure 2B). To identify broadly presented TAAs, myeloma-exclusive source proteins were ranked according to their frequencies of representation in the MM sample cohort (Figure 2C). To statistically assess and optimize the stringency of antigen identification, we simulated randomized virtual ligandomes in silico and calculated the resultant number of false-positive TAAs at different frequencies of representation (Figure 2D). We set the frequency threshold for HLA class I TAA definition to >25% of myeloma-exclusive antigen presentation, yielding 58 TAAs with an estimated FDR of 4.1% (supplemental Table 4). This novel panel of frequently presented myeloma-associated antigens was represented by 197 unique HLA class I ligands and constitutes 0.8% of the mapped myeloma HLA ligand source proteome. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis55 and functional annotation clustering of these antigens with respect to their biological function (GO Term BP FAT56 ) did not identify any statistically significant overrepresented pathways or functional clusters. Notably, the proto-oncogene multiple myeloma SET domain containing protein (MMSET) was identified as a TAA showing representation in 33% of MM patient ligandomes and was found to be represented by 3 different HLA ligands (ASNPSNPRPSK (HLA-A*30:01), KAMEAASSL (A*02:01), SLLEQGLVEA (A*02:01)). Moreover, MMSET was detected on both MM patients with the oncogenic translocation t(4;14), but only on 1 of 6 patients (17%) without this aberration.
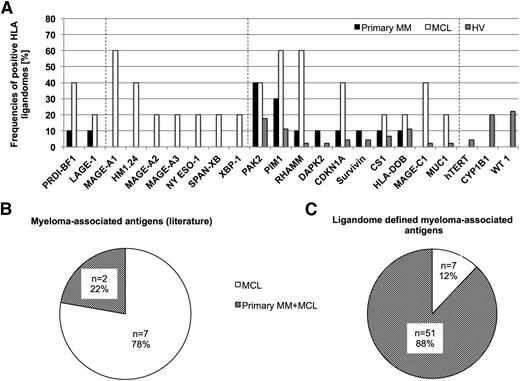
Representation of established myeloma-associated antigens in the HLA class I ligandome
Based on our extensive HLA ligandome data set, we investigated the presentation of established myeloma-associated antigens within the different sample cohorts. We identified 73 different HLA ligands representing 22 of 25 (88%) previously described myeloma antigens.42 We found 9 of the 22 detectable antigens (41%) to be exclusively presented on MM samples, 10 of 22 antigens (45.5%) to be represented both on MM and HV samples, and 3 of 22 (13.6%) exclusively presented on HV-derived samples (Figure 3A). Of note, 7 of 9 MM-exclusive antigens (77.8%) were only detectable on MCLs. Only 2 of 9 (22.2%) of these MM-exclusive antigens HLA ligands were detected on primary MM patient samples (Figure 3B). For reference, only 7 of 58 (12.1%) of the newly defined myeloma antigens showed presentation exclusively on MCLs, whereas the majority (51 of 58 [87.9%]) of antigens were also presented on primary MM patient samples as well, which underlines their potential as clinical target antigens (Figure 3C).
Representation of established myeloma-associated antigens in the HLA ligandomes of MM and HV. (A) Representation of previously described MM-associated antigens in HLA class I ligandomes. Bars indicate relative representation (%) of respective antigens by HLA class I ligands on primary MM samples, MCLs, and HV samples. Dashed lines divide the antigens into 4 groups according to their degree of MM association (MM and MCL-exclusive, MCL-exclusive, mixed presentation, HV-exclusive). (B-C) Distribution of myeloma-exclusive antigen presentation for (B) previously described antigens and (C) ligandome-defined TAAs on MCLs (white) and MM plus MCLs (shaded).
Representation of established myeloma-associated antigens in the HLA ligandomes of MM and HV. (A) Representation of previously described MM-associated antigens in HLA class I ligandomes. Bars indicate relative representation (%) of respective antigens by HLA class I ligands on primary MM samples, MCLs, and HV samples. Dashed lines divide the antigens into 4 groups according to their degree of MM association (MM and MCL-exclusive, MCL-exclusive, mixed presentation, HV-exclusive). (B-C) Distribution of myeloma-exclusive antigen presentation for (B) previously described antigens and (C) ligandome-defined TAAs on MCLs (white) and MM plus MCLs (shaded).
Moreover, unsupervised clustering of source protein presentation in the HLA ligandomes revealed the cluster of MCLs to be highly distinct from primary MM samples (supplemental Figure 2).
Analysis of HLA class II ligandomes identifies potentially synergistic vaccine candidates
As the direct involvement of CD4+ T cells in tumor control is established,57 we further aimed to identify HLA class II antigens. Overlap analysis of HLA class II ligand source proteins identified 1135 myeloma exclusive antigens (Figure 4A). Comparative profiling of HLA class II ligandomes identified a single antigen (TFRC) represented by 67 HLA class II ligands showing MM-exclusive presentation at FDR <5% (Figure 4B-C). Functional characterization of the most abundant TFRC peptide (NSVIIVDKNGRLV) by IFNγ ELISPOT revealed memory T-cell responses in 2 of 5 MM patients (supplemental Figure 3A).
Identification of synergistic HLA class II–restricted myeloma-associated antigens. (A) Overlap analysis of HLA class II ligand source proteins of primary MM samples (n = 7), MCLs (n = 5), and HV samples (total, n = 23: PBMC [n = 13], BMNC [n = 5], granulocytes [n = 5]). (B) Statistical analysis of false-positive myeloma-antigen identifications at different threshold values, as described in Figure 2. Randomized virtual ligandome sizes were set to 226 proteins and TAAs were defined based on simulated cohorts of 12 MM vs 23 HV samples. (C) Comparative profiling of HLA class II ligand source proteins based on the frequency of HLA-restricted presentation in MM and HV ligandomes. Frequencies of MMs/HVs positive for HLA-restricted presentation of the respective source protein (x-axis) are indicated on the y-axis. (D) Overlap analysis of HLA class I TAAs (n = 58) and HLA class II MM-exclusive antigens (n = 1135). (E) HLA class I TAAs, which also yield potentially synergistic HLA class II ligands. (F) Overlap analysis comprising the entire HLA class I and II ligand source proteomes of MM samples. sum, summary; rep., representation.
Identification of synergistic HLA class II–restricted myeloma-associated antigens. (A) Overlap analysis of HLA class II ligand source proteins of primary MM samples (n = 7), MCLs (n = 5), and HV samples (total, n = 23: PBMC [n = 13], BMNC [n = 5], granulocytes [n = 5]). (B) Statistical analysis of false-positive myeloma-antigen identifications at different threshold values, as described in Figure 2. Randomized virtual ligandome sizes were set to 226 proteins and TAAs were defined based on simulated cohorts of 12 MM vs 23 HV samples. (C) Comparative profiling of HLA class II ligand source proteins based on the frequency of HLA-restricted presentation in MM and HV ligandomes. Frequencies of MMs/HVs positive for HLA-restricted presentation of the respective source protein (x-axis) are indicated on the y-axis. (D) Overlap analysis of HLA class I TAAs (n = 58) and HLA class II MM-exclusive antigens (n = 1135). (E) HLA class I TAAs, which also yield potentially synergistic HLA class II ligands. (F) Overlap analysis comprising the entire HLA class I and II ligand source proteomes of MM samples. sum, summary; rep., representation.
As CD4+ T cells play pivotal roles in the induction and maintenance of antigen-specific CD8+ T-cell responses,58-60 we implemented a second approach to identify potentially synergistic HLA class II–restricted peptides derived from HLA class I TAAs. Overlap analysis of the 58 HLA class I antigens with the 1135 HLA class II presented MM-exclusive proteins identified a panel of 6 class-spanning antigens represented by 31 peptides (Figure 4D-E; supplemental Table 5). Functional characterization of synergistic HLA class II ligands revealed peptide-specific T-cell responses in myeloma patients for 3 of 5 tested peptides (Figure 4E, supplemental Figure 3B).
The overall comparison of the HLA class I and II ligandomes of MM samples revealed 80% (1622) of HLA class II–presented proteins to be also presented on HLA class I (Figure 4F). Functional annotation clustering (GO Term CC clustering using DAVID56 ) was performed on the top 500 most frequently presented proteins in each HLA class to identify the cellular compartments from which these proteins derive. Antigens presented on class I displayed highly enriched clusters for nuclear proteins as well as for ribosomal, cytoskeletal, and vesicle-derived proteins. Notably, this pattern was recapitulated in the clustering of proteins presented on both HLA classes, albeit with a higher ranking and an almost threefold higher enrichment for vesicle-derived proteins. HLA class II–presented antigens showed intermediate enrichment for plasma membrane, vesicle-derived, and lysosomal proteins (supplemental Table 6).
HLA class I TAAs are targeted by spontaneous T-cell responses in myeloma patients
Functional characterization of the novel myeloma antigens was performed in panels of 11 HLA-A*02– and 2 HLA-B*07–restricted peptides, including 2 HLA-A*02 ligands derived from MMSET (Figure 5A). Myeloma-associated peptides were evaluated in 12-day recall IFNγ ELISPOT assays using PBMC obtained from MM patients and HVs. We observed IFNγ secretion for 5 of 11 A*02 ligands and 1 of 2 B*07 ligands in myeloma patients, as shown exemplarily in Figure 5C. Both peptides (P1 and P2) derived from MMSET showed specific T-cell recognition in 2 of 16 (13%) and 1 of 8 (13%) MM patients, respectively. Importantly, no myeloma peptide-specific IFNγ secretion was observed in 10 HLA-matched healthy controls (Figure 5B). Notably, T-cell responses were only observed for myeloma-associated peptides identified on primary myeloma samples (10 of 13), and never for the 3 of 13 peptides identified on MCLs only. The frequencies of peptide-specific T-cell responses detected in MM patients by ELISPOT were generally in the same range as the frequencies of presentation of the respective peptide in allotype-matched ligandomes of MM patients (Figure 5A). Due to limitations in the numbers of cells available for analysis, further controls with target cells expressing the corresponding antigens could not be performed. We therefore cannot exclude that T-cell reactivity is directed against impurities contained in the synthetic peptide batch. Indeed, it is well known that synthetic peptides contain impurities, for example, peptides modified with a protecting group, and that these impurities are immunogenic. However, HLA-A*02– and -B*07–restricted control peptides derived from benign tissues (HV-exclusive HLA ligands) used in all ELISPOTs in the study at hand never resulted in significant IFNγ release (Figure 5C).
Functional characterization of myeloma-associated antigens. (A) Myeloma-associated T-cell epitopes with their corresponding HLA restrictions and frequencies of immune recognition by myeloma patient-derived T cells in IFNγ-ELISPOT assays. (B) Example of myeloma-associated T-cell epitopes evaluated in an IFNγ-ELISPOT using HV PBMCs. An EBV epitope mix containing the frequently recognized peptides BRLF109-117 YVLDHLIVV (A*02) and EBNA3247-255 RPPIFIRRL (B*07 served as positive control). Benign-tissue–derived peptides KLFEKVKEV (HLA-A*02) and KPSEKIQVL (B*07) served as negative control. (C) Examples of myeloma-associated T-cell epitopes evaluated in IFNγ-ELISPOTs using MM patient PBMCs (n = 3). Results are shown only for immunoreactive peptides. An EBV epitope mix containing 5 frequently recognized peptides (BRLF109-117 YVLDHLIVV [A*02], EBNA3471-479 RLRAEAQVK [A*03], EBNA3247-255 RPPIFIRRL [B*07], BZLF1190-197 RAKFKQLL [B*08], EBNA6162-171 AEGGVGWRHW [B*44]) was used as positive control. Benign-tissue–derived peptides KLFEKVKEV (HLA-A*02) and KPSEKIQVL (B*07) served as negative control. (D-E) Tetramer staining of CD8+ T cells after 3 cycles of aAPC-based in vitro primings using T cells derived from (D) a healthy individual and (E) a myeloma patient: Leftmost panels, P2-tetramer staining of CD8+ T cells primed with P2-aAPCs (SLLEQGLVEA, A*02); left middle panels, ex vivo P2-tetramer staining of CD8+ T cells; right middle panels, control staining with A*02-tetramer containing a nonrelevant A*02-restricted control peptide (KAMEAASSL, A*02) on CD8+ T cells derived from the same population as T cells depicted in the left panels. Rightmost panels, Positive control: tetramer staining of CD8+ T cells primed with CMV-aAPCs (NLVPMVATV, A*02). EBV, Epstein-Barr virus; neg., negative; pos., positive; UPN, uniform patient number.
Functional characterization of myeloma-associated antigens. (A) Myeloma-associated T-cell epitopes with their corresponding HLA restrictions and frequencies of immune recognition by myeloma patient-derived T cells in IFNγ-ELISPOT assays. (B) Example of myeloma-associated T-cell epitopes evaluated in an IFNγ-ELISPOT using HV PBMCs. An EBV epitope mix containing the frequently recognized peptides BRLF109-117 YVLDHLIVV (A*02) and EBNA3247-255 RPPIFIRRL (B*07 served as positive control). Benign-tissue–derived peptides KLFEKVKEV (HLA-A*02) and KPSEKIQVL (B*07) served as negative control. (C) Examples of myeloma-associated T-cell epitopes evaluated in IFNγ-ELISPOTs using MM patient PBMCs (n = 3). Results are shown only for immunoreactive peptides. An EBV epitope mix containing 5 frequently recognized peptides (BRLF109-117 YVLDHLIVV [A*02], EBNA3471-479 RLRAEAQVK [A*03], EBNA3247-255 RPPIFIRRL [B*07], BZLF1190-197 RAKFKQLL [B*08], EBNA6162-171 AEGGVGWRHW [B*44]) was used as positive control. Benign-tissue–derived peptides KLFEKVKEV (HLA-A*02) and KPSEKIQVL (B*07) served as negative control. (D-E) Tetramer staining of CD8+ T cells after 3 cycles of aAPC-based in vitro primings using T cells derived from (D) a healthy individual and (E) a myeloma patient: Leftmost panels, P2-tetramer staining of CD8+ T cells primed with P2-aAPCs (SLLEQGLVEA, A*02); left middle panels, ex vivo P2-tetramer staining of CD8+ T cells; right middle panels, control staining with A*02-tetramer containing a nonrelevant A*02-restricted control peptide (KAMEAASSL, A*02) on CD8+ T cells derived from the same population as T cells depicted in the left panels. Rightmost panels, Positive control: tetramer staining of CD8+ T cells primed with CMV-aAPCs (NLVPMVATV, A*02). EBV, Epstein-Barr virus; neg., negative; pos., positive; UPN, uniform patient number.
Antigen-specific T cells can be induced in vitro from naive T cells of MM patients or HVs
To assess whether myeloma antigen-specific T-cell responses can be induced from naive T cells in vitro, we isolated CD8+ T cells from 1 healthy individual and 1 MM patient. We performed aAPC primings using the MMSET-derived peptide SLLEQGLVEA (P2). Using HV-derived CD8+ T cells, a population of 0.403% P2-tetramer positive CD8+ T cells was detected after in vitro priming. No tetramer-positive T-cell populations >0.1% were detectable ex vivo. After priming of T cells from an MM patient without previous T-cell reactivity for P2 (as detected by 12-day-recall IFNγ-ELISPOT and ex vivo tetramer staining), we detected the induction of a small population of 0.236% P2-tetramer positive CD8+ T cells (Figure 5E). Importantly, control stainings performed with an A*02-tetramer containing a nonrelevant A*02 control peptide were performed in parallel on cells derived from the same wells as used for the relevant staining and did not yield any specific tetramer-positive T-cell populations (Figure 5D).
Discussion
With the advent of immune checkpoint modulation, T-cell–based immunotherapy has opened up new avenues for the treatment of a range of solid tumors61-68 and is undergoing clinical evaluation in hematologic malignancies.69,70 These novel therapeutic options may further be improved in combination with antigen-specific immunotherapy, which may help induce and guide specific anticancer immune responses. To this end, the exact knowledge of tumor-associated epitopes which can act as rejection antigens is indispensable.4,8 In myeloma, a multitude of studies have investigated MM-associated antigens, yielding a handful of promising targets.25-35 However, these studies were limited in scope of antigens and HLA allotypes.42 In our previous studies in acute myeloid leukemia and chronic lymphocytic leukemia, we demonstrated that the comprehensive and comparative analysis of the HLA-presented antigenome can identify extensive panels of broadly presented T-cell epitopes covering a multitude of HLA allotypes.12,13 Here, we translated this systematic approach to MM.
Quantification of HLA surface expression on different cell populations in the bone marrow of myeloma patients and healthy volunteers demonstrated that HLA loss or downregulation on malignant plasma cells is of no concern, even in patients who received prior therapy. Comparative analysis of the HLA ligandomes of these cell populations revealed distinct antigenic signatures and identified a panel of myeloma-associated antigens. These antigens did not cluster into any of the major tumor-associated pathways and did not show enrichment for highly prioritized targets characterized by tumor-associated overexpression. This indicates a more complex interplay of underlying mechanisms such as protein turnover71 and antigen processing as the source of altered antigen presentation on tumor cells and further underscores the isolated character of the HLA ligandome and the distorted correlation with its upstream sources.72,73
Importantly, a substantial proportion of established myeloma antigens was found to be only infrequently presented on primary myelomas or to show suboptimal degrees of myeloma specificity. Of note, the majority of these antigens was selectively detected on myeloma cell lines but not in primary samples, indicating that selection of pathophysiologically relevant antigens should be based on analysis of primary tumor samples.
A notable exception was the established myeloma-associated protein MMSET, which is currently being investigated as a target for the therapy of poor-prognosis t(4;14) myeloma patients.74-77 Although MMSET-derived peptides were frequently identified on t(4;14) myeloma samples, we also detected MMSET peptides in the HLA ligandomes of a t(4;14)-negative patient and 1 t(4;14)-negative MCL (U266). Strikingly, functional characterization by ELISPOT revealed memory T-cell responses targeting these MMSET-derived epitopes exclusively in myeloma patients and not in HV. This suggests myeloma-dependent priming of anti-MMSET T-cell responses in vivo in MM patients, which underscores the pathophysiological relevance of this antigen. In concordance with the HLA ligandomics data, we found these T-cell responses not to be restricted to t(4;14) myeloma patients. Results of in vitro primings suggest that MMSET-specific CD8+ T-cell responses can be induced from naive T cells, both in healthy individuals and, importantly, also in myeloma patients, albeit with limited magnitudes. With the current strategies focusing on inhibition of MMSET by small molecules or small interfering RNAs,78,79 our identification of myeloma-exclusive MMSET-derived T-cell epitopes provides new options for targeting MMSET by T-cell–based immunotherapy. Notably, this therapeutic strategy may not necessarily have to be restricted to t(4;14) myelomas, as we observed MMSET-presentation and immune recognition irrespective of the mutational status. This might be explained by the distorted correlation of gene expression and HLA-restricted antigen presentation as well as by the subclonal distribution of t(4;14) in myeloma cells and genomic plasticity occurring over the course of disease.73,80
Together, our findings illustrate how antigen identification guided by HLA ligandomics can pinpoint novel MM-associated T-cell epitopes and allow for direct assessment of antigen distribution patterns in patient cohorts. In parallel to our findings with MMSET, our study features an extensive panel of novel antigens previously not associated with myeloma or cancer in general. Analogously to MMSET, we detected preexisting T-cell responses against a substantial proportion of these targets in myeloma patients, indicating a high enrichment for relevant MM-associated antigens. In conclusion, our ligandome-centric study may guide the design of future antigen-specific T-cell immunotherapy in MM.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Claudia Falkenburger, Patricia Hrstic, Nicole Zuschke, Katharina Graf, and Beate Pömmerl for excellent technical support.
This work was supported by the German Cancer Consortium (DKTK), the Deutsche Forschungsgemeinschaft (DFG; SFB 685), and the European Union (EU; ERC AdG 339842 MUTAEDITING).
Authorship
Contribution: S.W., J.S.S., D.J.K., S.S., and H.-G.R. designed the study; S.W., J.S.S., D.J.K., H.-G.R., L.K., K.W., H.R.S., and S.S. drafted the manuscript; S.W., J.S.S., and H.S. performed flow cytometric analysis and HLA surface expression quantitation; S.W., D.J.K., J.S.S., and S.K. conducted MHC peptide immunoaffinity purification; S.W. and D.J.K. performed MS-based analyses and quantitation; S.W., A.N., T.S., J.S.S., and H.S. conducted in vitro T-cell experiments; J.S.S., L.K., K.W., M.H., and H.R.S. conducted patient data collection and medical evaluation; S.W., J.S.S., D.J.K., O.K., and L.B. performed statistical analyses; and S.S., H.-G.R., and J.S.S. supervised the study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Juliane S. Stickel, Department of Hematology and Oncology, University of Tübingen, Otfried-Müller Strasse 10, 72076 Tübingen, Germany; e-mail: juliane.stickel@med.uni-tuebingen.de.
References
Author notes
S.W. and J.S.S. contributed equally to this work.

![Figure 2. Comparative HLA ligandome profiling and identification of myeloma-associated antigens. (A) Saturation analysis of HLA class I ligand source protein identifications in MM patients. Number of unique HLA ligand source protein identifications as a function of cumulative HLA ligand source protein identifications in 10 MM patients. Exponential regression allowed for the robust calculation (R2 = 0.99) of the maximum attainable number of different source protein identifications (dashed line). The dotted line depicts the source proteome coverage achieved in our MM patient cohort. (B) Overlap analysis of HLA class I ligand source proteins of primary MM samples (n = 10), MCLs (n = 5), and HV samples (total n = 45: PBMC [n = 30], BMNC [n = 10], granulocytes [n = 5]). (C) Comparative profiling of HLA ligand source proteins based on the frequency of HLA-restricted presentation in MM and HV ligandomes. Frequencies of MMs/HVs positive for HLA-restricted presentation of the respective source protein (x-axis) are indicated on the y-axis. The box on the left highlights the subset of myeloma-associated antigens showing MM-exclusive presentation in >25% of myeloma samples. (D) Statistical assessment of false-positive myeloma-antigen IDs at different threshold values. The numbers of original TAAs identified based on the analysis of the MM and HV cohorts were compared with random virtual TAAs. Virtual MM and HV samples were generated in silico based on random weighted sampling from the entirety of protein identifications in both original cohorts. These randomized virtual ligandomes of defined size (n = 957 proteins, which is the mean number of protein identifications in all analyzed samples) were used to define TAAs based on simulated cohorts of 15 MM vs 45 HV samples. The process of protein randomization, cohort assembly, and TAA identification was repeated 1000 times and the mean value of resultant virtual TAAs was calculated and plotted for the different threshold values. The corresponding FDRs for any chosen TAA threshold are listed below the x-axis. sum, summary.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/126/10/10.1182_blood-2015-04-640532/4/m_1203f2.jpeg?Expires=1767796430&Signature=JDU8B4joQfCAvtqo792HVypiMYfv7Q0jdozFDTfX6E7wLLsPUkrZ7dfLqhMapYEw2m5r9pwfo0ALFJGjIPm3Uu18NEH0r366K8qCBISyLzYTORJYNRRutWQ9ab0kQY8YTB8oi-Xa~fpfwcba5dPwPiDHfXQYOcjrp7CN556BOJqYxulBRGJ3z1g5ETOasNUY0TqCOfR8xJgvrUQ1ZdzKFr-GrvFs7x6TjaKu3Pn0i6kU7Gg9bPgfVZHc9BdE5QvuHZkP4TWboUjVfyvGWSvJyVUQAc3VA4pRc6panTaLVbItL7hTxSs6uAiGvzZYU4w7OTGiE~eNtb2ShTVwX2xZLQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. Identification of synergistic HLA class II–restricted myeloma-associated antigens. (A) Overlap analysis of HLA class II ligand source proteins of primary MM samples (n = 7), MCLs (n = 5), and HV samples (total, n = 23: PBMC [n = 13], BMNC [n = 5], granulocytes [n = 5]). (B) Statistical analysis of false-positive myeloma-antigen identifications at different threshold values, as described in Figure 2. Randomized virtual ligandome sizes were set to 226 proteins and TAAs were defined based on simulated cohorts of 12 MM vs 23 HV samples. (C) Comparative profiling of HLA class II ligand source proteins based on the frequency of HLA-restricted presentation in MM and HV ligandomes. Frequencies of MMs/HVs positive for HLA-restricted presentation of the respective source protein (x-axis) are indicated on the y-axis. (D) Overlap analysis of HLA class I TAAs (n = 58) and HLA class II MM-exclusive antigens (n = 1135). (E) HLA class I TAAs, which also yield potentially synergistic HLA class II ligands. (F) Overlap analysis comprising the entire HLA class I and II ligand source proteomes of MM samples. sum, summary; rep., representation.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/126/10/10.1182_blood-2015-04-640532/4/m_1203f4.jpeg?Expires=1767796430&Signature=wDljeMTS1IMz3sYszHvWuqWmUF9DmEGbo7qsLijKkPPQzG6OAf6BmrgY~dQxpTyexvPJk7QYH6moKh39eiFqSXLNQMhuNwvHygwHzcLoUr~VVOdCHch7xvtyWaP1HrTv0eHb2Xbhx1M~mxVdCjSKDJWOoXL7mwE2kg5fjiC-XhYOoejj62v~Qocj1CFzeLyyXu4MJ69S-UnN1vhUTQAgpT--8Kim39YvDwS7eNoxhpSpsPnLYfJHLTGFh~Trh9z09XHrWno24hNYYhcE9y9ggCKbDZ9Qe4lUFuaMENZEAU-pOx-F4DGlrcDylB9mB9MD94hyuViRd8ix3358M2xqKA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. Functional characterization of myeloma-associated antigens. (A) Myeloma-associated T-cell epitopes with their corresponding HLA restrictions and frequencies of immune recognition by myeloma patient-derived T cells in IFNγ-ELISPOT assays. (B) Example of myeloma-associated T-cell epitopes evaluated in an IFNγ-ELISPOT using HV PBMCs. An EBV epitope mix containing the frequently recognized peptides BRLF109-117 YVLDHLIVV (A*02) and EBNA3247-255 RPPIFIRRL (B*07 served as positive control). Benign-tissue–derived peptides KLFEKVKEV (HLA-A*02) and KPSEKIQVL (B*07) served as negative control. (C) Examples of myeloma-associated T-cell epitopes evaluated in IFNγ-ELISPOTs using MM patient PBMCs (n = 3). Results are shown only for immunoreactive peptides. An EBV epitope mix containing 5 frequently recognized peptides (BRLF109-117 YVLDHLIVV [A*02], EBNA3471-479 RLRAEAQVK [A*03], EBNA3247-255 RPPIFIRRL [B*07], BZLF1190-197 RAKFKQLL [B*08], EBNA6162-171 AEGGVGWRHW [B*44]) was used as positive control. Benign-tissue–derived peptides KLFEKVKEV (HLA-A*02) and KPSEKIQVL (B*07) served as negative control. (D-E) Tetramer staining of CD8+ T cells after 3 cycles of aAPC-based in vitro primings using T cells derived from (D) a healthy individual and (E) a myeloma patient: Leftmost panels, P2-tetramer staining of CD8+ T cells primed with P2-aAPCs (SLLEQGLVEA, A*02); left middle panels, ex vivo P2-tetramer staining of CD8+ T cells; right middle panels, control staining with A*02-tetramer containing a nonrelevant A*02-restricted control peptide (KAMEAASSL, A*02) on CD8+ T cells derived from the same population as T cells depicted in the left panels. Rightmost panels, Positive control: tetramer staining of CD8+ T cells primed with CMV-aAPCs (NLVPMVATV, A*02). EBV, Epstein-Barr virus; neg., negative; pos., positive; UPN, uniform patient number.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/126/10/10.1182_blood-2015-04-640532/4/m_1203f5.jpeg?Expires=1767796430&Signature=V1zK0sq8KEdM4C50CS49cjcZeOwxNDuMaGxocN8Cx8YePBeSNACpBYhDBoxCQUvcPeMgT31VyvKFvS4wc6~pdcd94QkgMB9PQgMPcOk~5tTGnI0Xz4UI51V7aoY~BV7vIW6TwRpxxoHtfQbMbYy4Igx1Hr-H7sgFh8cYXgpd7VJ7gBKWGtsqOSLe4w5wDb-Kkk7fAPqsBzMuAAjDdA4wnpIzWk1BHeNT7bvscDiluTRC1hllxVq~FJ15t-9RzbCQ18FeUL621T6xC3i9b62eH-9o2dVsTiHd-6TtRKnn-GqSyn4YdcK0sdAkWemDT50EgbaVMmwgRsphbWjTgfVqZg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal