Immunotherapy is an exciting advance in tumor treatment and identifying relevant peptides presented by major histocompatibility complex (MHC) class I on tumors is critical for this process. In this issue of Blood, Walz et al describe a massive mass spectrometry experiment to identify relevant peptides presented by multiple myeloma (MM) cells and show that these represent just normal antigens. This expands the tumor-relevant peptidome beyond mutated antigens, implying that even tumors that are not highly mutated can be amenable to T-cell–based immunotherapies.1
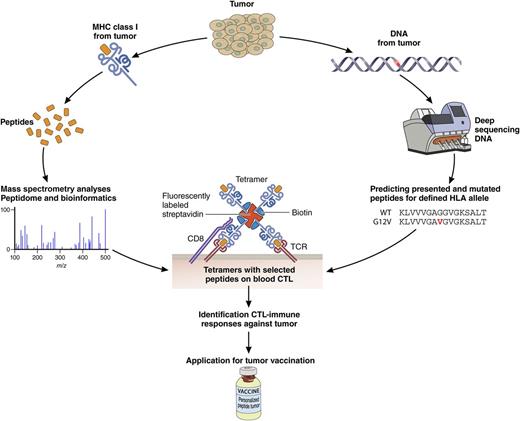
Two arms to embrace the oncopeptidome. The oncopeptidome can be determined by definition of mutations in the genome followed by peptide prediction and validation (right arm). A complementary arm (left) defines the oncopeptidome directly by analyzing MHC class I–associated peptides by mass spectrometry and then validation. These are mainly nonmutated peptides. Integration of the 2 arms will further expand the oncopeptidome associated with clinical responses and ultimately be applied in personalized tumor vaccination programs. Professional illustration by Patrick Lane, ScEYEnce Studios.
Two arms to embrace the oncopeptidome. The oncopeptidome can be determined by definition of mutations in the genome followed by peptide prediction and validation (right arm). A complementary arm (left) defines the oncopeptidome directly by analyzing MHC class I–associated peptides by mass spectrometry and then validation. These are mainly nonmutated peptides. Integration of the 2 arms will further expand the oncopeptidome associated with clinical responses and ultimately be applied in personalized tumor vaccination programs. Professional illustration by Patrick Lane, ScEYEnce Studios.
Tumor immunotherapy started almost a century ago with the first experiments with bacillus Calmette-Guérin in bladder cancer. It took another 70 years before the next step, the application of (humanized) antibodies against CD20, became clinical practice. Since the realization, some 25 years ago, that MHC class I presents intracellular antigens in the form of peptides, these have been used for vaccination against tumors. The recent development of checkpoint antibodies blocking PD-1/PD-1L and/or CTLA-4 proteins further illustrated the power of cytotoxic T lymphocytes (CTLs) in the control of tumors. These checkpoint antibodies allowed the activation of various CTLs present in tumor tissue where they were kept dormant, with stunning clinical effects in a significant percentage of cancer patients. The best responding tumors are melanoma and lung cancer, but effects in many other tumor types have been reported.2-4 Because melanoma and lung cancer are characterized by a high mutation rate, the theory predicting clinical effects of checkpoint antibodies became “the more mutations, the more cure.” This makes sense, as mutated antigens yield altered peptides that are considered by T cells as “nonself” followed by tumor elimination. Indeed, T cells responding to mutated antigens have been identified by first sequencing the genome of cancer cells. Subsequently, peptides presented by the patient’s MHC class I alleles are predicted from these mutated genes. Then, peptides are chemically synthesized and loaded onto corresponding MHC class I tetramers. These tetramers are subsequently used to detect corresponding T cells in cancer patients.5,6 This pipeline for detecting tumor immune responses by definition only detects mutated peptides, and these are frequently observed in the blood of immunotherapy-responding patients (see figure). The interpretation was simple: mutations make new peptides that are recognized by patient’s T cells responding to immune system activation by the checkpoint antibodies. These T cells then eliminate the corresponding tumor cells. And the result: successful immunotherapy of patients! At least, so goes the interpretation, when the tumor is specified by a high mutational load, for example, as a result of smoking (lung, head and neck, bladder, and other tumors) or sunlight (melanoma).
Although this concept fulfills the general dogma related to T-cell selection and antigen presentation developed over the last 20 years, it also implies that tumors with few if any mutations would fail immunotherapy. But is this correct? Walz et al report in this issue a massive mass spectrometry analysis of the MHC class I– and MHC class II–associated peptidome of MM cells of 29 patients by subtracting the normal B-cell peptidome.1 They identified over 58 MM-specific peptides that were all derived from normal unmutated proteins. They then performed experiments similar to those described earlier; they generated MHC class I tetramers with identified peptides to show that T cells against (some of) these original peptide-MHC class I combinations could be detected in the circulation of MM patients. The T cells are there; they only require a wake-up call.
This procedure does not imply that mutated antigens are entirely absent in the original peptidome of MM cells. The procedure to identify peptides may simply have ignored altered peptides when these could not be mapped on the reference human protein sequences. Incorporating the most common mutated peptides in cancer cells (including those identified through the genome sequencing approach) in these procedures may identify the mutated peptidome. Yet, this does not change a major conclusion from this elegant work: even normal antigens can yield tumor antigens! Every tumor may then in principle be a target of tumor immunotherapy,7 not only melanoma and other cancers specified by high mutation rates.
How then do normal peptides induce a tumor-specific immune response? This is only conceivable when such peptides have been missed in the negative selection steps in the thymus, thus allowing specific CTLs to progress into fully functional entities. CTLs with low-affinity T-cell receptors (TCRs) against self-antigens may escape negative selection. When tumors express these antigens in large quantities, a resulting high MHC-peptide concentration will allow low-affinity TCR to build up a sufficient signal for CTL activation, then following the laws of mass action.8 Also, CTLs can be made inactive by so-called peripheral tolerance and respond to tumor antigens, especially for antigens more or less selectively and highly expressed in tumor tissue, when activated by checkpoint antibodies. Of note, many patients undergo chemotherapy and/or radiotherapy as first-line treatment of their malignancies. These treatments have marked effects on the peptidome. Many drugs used in chemotherapy of cancer also affect the epigenome and thus the transcriptome, the proteome, and, ultimately, the MHC class I–associated peptidome.9 Radiotherapy activates the mammalian target of rapamycin pathway, which controls translation of a subset of transcripts and thus alters the proteome and the MHC class I peptidome as well.10 These and possibly many other options can affect the MHC class I peptidome in tumor cells. These nonmutated peptides can then be recognized by CTLs that have escaped negative selection and allow successful immunotherapy of tumors with low mutation rates, especially when checkpoint control is removed. Mass spectrometry analysis of the tumor-specific peptidome in combination with tetramer analyses identified these peptides for MM. But the study by Walz et al1 also implies that the full spectrum from tumors with many mutations to tumors with few or no mutations can in fact respond to immunotherapy with checkpoint antibodies and other immunotherapy approaches under development. Feeding the DNA sequencing activities into the programs for identifying the oncopeptidome by mass spectrometry may further expand the oncopeptidome for personal vaccination options of cancer patients. The recent developments in tumor immunotherapy may then have clinical consequences for more cancer types than currently appreciated. And this is good news for all cancer patients.
Conflict-of-interest disclosure: The author declares no competing financial interests.