Key Points
B-cell malignancies were efficiently recognized by T cells expressing high-affinity alloHLA-restricted TCRs specific for CD79b.
Aberrant expression of CD79b in non–B cells caused unwanted reactivity, rendering CD79b unsuitable for TCR-based immunotherapies.
Abstract
Immunotherapy of B-cell malignancies using CD19-targeted chimeric antigen receptor–transduced T cells or CD20-targeted therapeutic monoclonal antibodies has shown clinical efficacy. However, refractory disease and the emergence of antigen-loss tumor escape variants after treatment demonstrate the need to target additional antigens. Here we aimed to target the B-cell receptor–associated protein CD79b by a T-cell receptor (TCR)-based approach. Because thymic selection depletes high-avidity T cells recognizing CD79b-derived peptides presented in self-HLA molecules, we aimed to isolate T cells recognizing these peptides presented in allogeneic HLA. Peptide-HLA tetramers composed of CD79b peptides bound to either HLA-A2 or HLA-B7 were used to isolate T-cell clones from HLA-A*0201 and B*0702-negative individuals. For 3 distinct T-cell clones, CD79b specificity was confirmed through CD79b gene transduction and CD79b-specific shRNA knockdown. The CD79b-specific T-cell clones were highly reactive against CD79b-expressing primary B-cell malignancies, whereas no recognition of nonhematopoietic cells was observed. Although lacking CD79b-cell surface expression, intermediate reactivity toward monocytes, hematopoietic progenitor cells, and T-cells was observed. Quantitative reverse transcriptase polymerase chain reaction revealed low CD79b gene expression in these cell types. Therefore, aberrant gene expression must be taken into consideration when selecting common, apparently lineage-specific self-antigens as targets for TCR-based immunotherapies.
Introduction
Clinical studies using chimeric antigen receptor (CAR)-modified T-cells or targeted monoclonal antibodies have shown the efficacy of immunotherapy to treat different hematologic malignancies. The adoptive transfer of T cells modified to express CD19-targeted CARs have led to complete remission in patients with chronic lymphocytic leukemia (CLL) and acute lymphoblastic leukemia (ALL).1-4 Because of its high expression on malignant cells, CD19 can be efficiently targeted by CAR-transduced T cells. Similarly, CD20-targeting therapeutic monoclonal antibodies such as rituximab have caused complete remissions in patients treated for follicular lymphoma5,6 and diffuse large B-cell lymphoma7,8 in combination with chemotherapy. In addition to their expression on malignant B cells, CD19 and CD20 are also highly expressed on healthy B cells, causing depletion of the B-cell compartment in patients treated with CD19-targeted or CD20-targeted immunotherapies.1,3-5,9 However, side effects of long-term B-cell depletion are manageable, which indicates that B-cell aplasia can be tolerated.9-13
Although successful in several patients, malignancies refractory to CD20-targeted therapy and the emergence of CD19-negative tumor escape variants after treatment with CD19-targeting CAR-transduced T-cells4 demonstrate the need to identify additional targets for the treatment of B-lineage malignancies. We hypothesized that the B-cell receptor (BCR)-associated protein CD79b presents a suitable target for immunotherapy because CD79b membrane expression is restricted to the B-cell compartment and can be found on many B-cell malignancies.14-16 The BCR-associated heterodimer CD79a/b is critically involved in BCR transport17 and functionality by providing signaling capacity18-22 to regulate processes such as allelic exclusion, proliferation, differentiation, anergy, and apoptosis.23-25 CD79b is encoded by gene B2926 and through the event of alternative splicing can produce different transcripts.27 The truncated variant of CD79b, missing almost entirely its extracellular domain, has been implicated to render CLL unresponsive to BCR-induced apoptosis.28 Furthermore, a high percentage of malignant cells expressing cell-surface CD79b in combination with other phenotype markers has been associated with shortened overall survival in B-CLL patients.29,30
An alternative strategy for the treatment of cancer is the use of T-cell receptor (TCR) gene-transfer therapy.31,32 T cells engineered to express TCRs with defined antigen specificity have already successfully been used in the treatment of solid tumors.33 Furthermore, only very low quantities of agonistic peptide–HLA complex are required for TCR triggering and T-cell activation,34-36 whereas CAR and antibody-based approaches necessitate a greater abundance of the antigen.37 Therefore, T cells modified with CD79b-targeting TCRs can be a valuable addition to current immunotherapies.
T cells expressing high-affinity TCRs recognizing self-antigens such as CD79b in the context of self-HLA molecules are deleted from the naïve T-cell repertoire during thymic development by negative selection to prevent autoreactivity. In contrast, presentation of such self-antigens in the context of allogeneic HLA (alloHLA) can result in strong T-cell responses as was observed in the induction of PRAME-specific T cells after HLA-A*0201–mismatched hematopoietic stem cell transplantation.38 In an additional study we demonstrated that the high-affinity TCRs of these alloHLA-restricted T cells are single peptide–specific.39 Using the concept of self-antigen presented in alloHLA, Wilms tumor antigen (WT1)-reactive T cells have been generated by stimulating T cells from HLA-A2–negative donors with autologous B-lymphocytes coated with HLA-A2 monomers loaded with WT1-derived peptide.40 In addition, antigen-specific T cells were obtained by stimulating T cells from HLA-A2–negative donors with autologous dendritic cells (DCs) pulsed with RNA to express HLA-A2 in parallel with a variety of targeted antigens.41-43
In this study, from HLA-A*0201/B*0702–negative healthy individuals, we isolated 3 T-cell clones targeting 3 distinct CD79b peptides presented in HLA-A*0201 or HLA-B*0702. CD79b-positive primary CLL, ALL, and ALL cell lines were recognized by these T-cell clones, whereas no reactivity toward nonhematopoietic tissue was detected. However, low to intermediate recognition of CD14+ monocytes, T cells, and CD34+ hematopoietic precursor cells (HPCs) could be observed. Although no cell-surface CD79b was present on these cells, aberrant mRNA expression of CD79b could be detected in these cells.
Materials and methods
Culture conditions and cells
Peripheral blood mononuclear cells (PBMCs) were obtained from different individuals, after their informed consent, by isolation using Ficoll-gradient centrifugation and were then cryopreserved. T cells were cultured in T-cell medium consisting of Iscove’s modified Dulbecco’s medium (Lonza, Basel, Switzerland) supplemented with 100 IU/mL IL-2 (Proleukine; Novartis Pharma, Arnhem, The Netherlands), 5% fetal bovine serum (Gibco, Life Technologies, Carlsbad, CA), and 5% human serum. Isolation, generation, and culture of various hematopoietic and nonhematopoietic cell subsets and lines are described in supplemental Methods, available on the Blood Web site. The Leiden University Medical Center review board approved the use of human material, and research was conducted in accordance with the Declaration of Helsinki.
Generation of peptide-MHC complexes
All peptides were synthesized in-house using standard Fmoc chemistry. Recombinant HLA-A2 and HLA-B7 heavy-chain and human β2m light-chain were in-house produced in Escherichia coli. Major histocompatibility complex (MHC) class I refolding was performed as previously described, with minor modifications.44 MHC class I complexes were purified by gel filtration using high-performance liquid chromatography. pMHC tetramers were generated by labeling biotinylated pMHC monomers with streptavidin-coupled phycoerythrin (PE; Invitrogen, Carlsbad, CA) or Quantum dots (Qdots; Invitrogen) Q585, Q605, Q655, and Q705. Complexes were stored at 4°C and spun at 17 000g for 2 minutes before use.
Isolation of CD79b-specific T-cell clones using pMHC tetramers
T cells binding to pMHC tetramers containing CD79b peptides were isolated from cryopreserved PBMCs from healthy HLA-A*02:01 and HLA-B*07:02–negative individuals following a previously described protocol.45 PBMCs were incubated with PE-labeled pMHC tetramers for 1 hour at 4°C. Cells were washed twice and incubated with anti-PE magnetic microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany). PE-labeled cells were isolated by magnetic-activated cell sorting (MACS) on an LS column (Miltenyi Biotec) according to the manufacturer’s instruction. Positive fractions were cultured at 5000 cells per well in 96-well round-bottom culture plates containing 5 × 104 irradiated (35 Gy) autologous PBMCs and 11 000 anti-CD3/CD28 beads (Invitrogen) in 100 µL T-cell medium. After 2 weeks of expansion, CD79b-reactive T-cell clones were obtained from pMHC tetramer–enriched T-cell populations by sorting CD8+ T cells stained with PE-labeled pMHC tetramers containing CD79b peptides and an antibody against CD8 (Invitrogen/Caltag, Buckingham, United Kingdom) combined with antibodies against CD4, CD14, and CD19 (BD Pharmingen, San Jose, CA). Cells were first stained with PE-labeled pMHC tetramers for 1 hour at 4°C before antibodies were added for an additional 15 minutes at 4°C. Single pMHC tetramer+ CD8+ T cells were sorted into 96-well round-bottom culture plates containing 5 × 104 irradiated allogeneic PBMCs in 100 µL T-cell medium supplemented with 0.8 µg/mL phytohemagglutinin (PHA) (Remel, Lenexa, KS). Cell sorting was performed on a FACSAria III (BD Biosciences).
FACS analysis
Fluorescence-activated cell sorting (FACS) was performed on an LSRII (BD Biosciences) and analyzed using Diva Software (BD Biosciences) or on a FACS Calibur (BD Biosciences) and analyzed using FlowJo Software (TreeStar, Ashland, OR). T-cell lines generated by MACS enrichment using pMHC tetramers and T-cell clones were analyzed for pMHC tetramer+ CD8+ T cells by staining with pMHC tetramers CD79b173:A2 labeled with PE or Q705, CD79b20:A2 labeled with Q705 or Q655, CD79b9:B7 labeled with Q605 or Q585, and an Alexa700-conjugated antibody against CD8 (Invitrogen/Caltag) combined with fluorescein isothiocyanate–labeled antibodies against CD4, CD14, and CD19 (BD Pharmingen). Cells were first incubated with a mixture of pMHC tetramers (2 µg/mL per pMHC tetramer) for 15 minutes at 37°C before antibodies were added and incubated for an additional 15 minutes at 4°C. PBMCs, purified hematopoietic cell subsets, or activated cells were stained with antibodies against CD79b (clone 3A2-2E7; BD Pharmingen) or isotype control in combination with CD3, CD4, CD14, CD19, CD34 (BD Pharmingen), or CD271 (NGF-R; Sanbio, Uden, The Netherlands) at 4°C for 15 minutes.
Functional analysis
Stimulator cells were peptide-pulsed at various peptide concentrations for 30 minutes at 37°C. Responder T cells and peptide-pulsed or unloaded stimulator cells were coincubated at various stimulator to responder (S:R) ratios. After 18 hours of coincubation, supernatants were harvested and interferon (IFN)-γ production was measured by enzyme-linked immunosorbent assay (ELISA; Sanquin Reagents, Amsterdam, The Netherlands).
Cytotoxicity assay
Adapted from Jedema et al,46 20 000 PKH26GL-labeled (Sigma-Aldrich/Merck, St. Louis, MO) target cells were coincubated with T cells at various effector:target ratios in 100 µL T-cell medium for 17 hours. After coincubation, cells were stained with Sytox Blue dead-cell stain (Invitrogen/Caltag) in a final concentration of 1 µM for 5 minutes. 10 µL Flow-count fluorospheres (Beckman Coulter, Indianapolis, IN) were added and samples were analyzed using FACS. For each sample, 2500 Flow-count fluorospheres were acquired and the percent surviving cells was calculated as follows: (PKH26GL-labeled targets in the presence of effector cells)/(PKH26GL-labeled targets in absence of effector cells) × 100.
Transduction with CD79b retroviral vector and CD79b-specific shRNA
CD79b (accession number #NM_000626.2_variant1) was codon-optimized (GeneArt, Life Technologies) and expressed on a MP71 retroviral backbone in combination with NGF-R. CD79b-targeting short-hairpin RNAs (shRNA) TRC 057 648 and TRC 057 651 (Sigma-Aldrich) were a kind gift of Prof Rob Hoeben (Department of Molecular Cell Biology, Leiden University Medical Center, Leiden, The Netherlands). shRNAs were expressed on retroviral backbone pLKO.1 or pGreenPuro (Systembiosciences, Mountain View, CA), both containing a puromycin resistance gene (see supplemental Methods for production of retroviral supernatant).
For transduction, retroviral supernatant was loaded on 24-well nontissue culture–treated plates that had been coated with 30 µg/mL retronectine (Takara, Shiga, Japan) and blocked with 2% human serum albumin (Sanquin Reagents). Viral supernatant was spun down at 2000g for 20 minutes at 4°C. 1 × 105 K562-A2 or K562-B7 cells were added to CD79b retroviral supernatant and incubated for 18 hours. High-purity bulk CD79b-transduced K562-A2 and K562-B7 cell populations were obtained by sorting cells stained with an antibody against NGF-R. 1 × 105 cells of Epstein-Barr virus–transformed lymphoblastic cell lines (LCLs) or 3 × 105 CD40L-activated B cells were added to retroviral supernatant encoding for CD79b-targeting shRNA and incubated for 18 hours. Transduced cells were selected by supplementing medium with 2 to 10 µg/mL puromycin (Calbiochem, San Diego, CA) for 2 days. Optimal puromycin concentration for each cell type was determined by escalating dose experiments.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was isolated using the RNAqueous Micro-Kit and Small Scale Kit (Ambion, Life Technologies) for a maximum of 0.5 × 106 and 10 × 106 cells, respectively, following the manufacturer’s instructions. Total RNA was converted to cDNA using Moloney murine leukemia virus reverse transcriptase (Invitrogen). CD79b expression was measured on the Roche Lightcycler 480 (Roche) using Fast Start TaqDNA Polymerase (Roche) and EvaGreen (Biotium, Hayward, CA) with forward primer 5′-CCAGGCTGGCGTTGTCTCCTG-3′ and reverse primer 5′-GGTACCGGTCCTCCGATCTGGC-3′.
Results
Identification of naturally processed and presented CD79b peptides
To identify MHC-presented CD79b peptides, we adapted a previously described algorithm.45 From a peptide elution library that was generated by eluting and sequencing MHC-bound peptides from human HLA-A2/B7–positive B-LCLs,47 three peptides of CD79b were identified with mascot ion scores >35 and netMHC48,49 binding scores <250 nM (Table 1). Peptides CD79b9 and CD79b173 were newly identified. Peptide CD79b20 had previously been eluted.50,51 All 3 peptides are encoded by each isoform of CD79b. Matching mass spectrometry fragmentation patterns of eluted and newly synthesized CD79b peptides verified correct identification (data not shown). Stable pMHC tetramers of CD79b20 and CD79b173 bound to HLA-A2 (CD79b20:A2 and CD79b173:A2, respectively) and CD79b9 bound to HLA-B7 (CD79b9:B7) were generated by conjugating conventionally refolded pMHC monomers to streptavidin-labeled fluorophores.
Sequences and properties of CD79b-derived peptides
| . | Peptide sequence . | HLA restriction . | netMHC affinity (nM)* . | Source . |
|---|---|---|---|---|
| CD79b173 | IIVPIFLLL | A*0201 | 177 | Eluted47 |
| CD79b9 | VPSHWMVAL | B*0702 | 14 | Eluted47 |
| CD79b20 | LLSAEPVPA | A*0201 | 112 | Eluted,47 previously described50,51 |
Generation of pMHC tetramer+ CD8+ T-cell lines by tetramer-guided enrichment
T cells binding to pMHC tetramers containing CD79b peptides presented in HLA-A2 or HLA-B7 were isolated from PBMCs of 13 HLA-A2/B7–negative healthy individuals using PE-labeled pMHC tetramers and MACS isolation. From 6 × 107 to 3 × 108 total PBMCs between 10 000 and 160 000 total cells were recovered in the pMHC tetramer–labeled fraction. Among the isolated cells, 7% to 21% were CD8+ T cells, of which only a fraction stained positive for the pMHC tetramers (data not shown). Other cells included CD4+ T cells, monocytes, and B-cells. After 2 weeks of expansion, the generated T-cell lines were subjected to a second enrichment using the same pMHC tetramers. After 2 weeks of expansion, the presence of CD79b-reactive T cells were determined by pMHC-tetramer staining (supplemental Table 1 and supplemental Figure 1). In 11 of 13 individuals CD8+ T cells positive for pMHC tetramer CD79b173:A2 were detected with frequencies of 0.02% to 23.8% of CD8+ T cells. CD8+ T cells staining positive for CD79b20:A2 were present in 3 individuals with frequencies of 0.1% to 31.2%. CD8+ T cells staining positive for CD79b9:B7 were detected in 3 individuals with frequencies of 0.03% to 0.7% of the CD8+ T-cell compartment.
In summary, CD8+ T cells binding to pMHC tetramers presenting CD79b peptides in HLA-A2 or HLA-B7 could be isolated from the allo-repertoire of HLA-A2/B7–negative individuals.
Identification of 3 distinct T-cell clones recognizing CD79b peptides presented in alloHLA
To analyze the functional activity of the CD8+ T-cell populations binding to CD79b pMHC tetramers, pMHC tetramer+ CD8+ T cells were clonally expanded using single-cell sorting. We sorted CD8+ T cells binding to CD79b173:A2 from 4 individuals. CD8+ T-cell clones binding to CD79b20:A2 were obtained from 2 individuals. CD8+ T-cell clones binding to CD79b9:B7 were sorted from one. T-cell clones were analyzed for CD79b peptide-specificity and avidity by loading CD79b peptides on the CD79b-negative cell line K562 expressing HLA-A2 or HLA-B7 (K562-A2 or K562-B7, respectively). Recognition of endogenously processed and MHC-presented CD79b peptides was assessed on 3 HLA-A2/B7–positive B-LCLs, which naturally express CD79b.
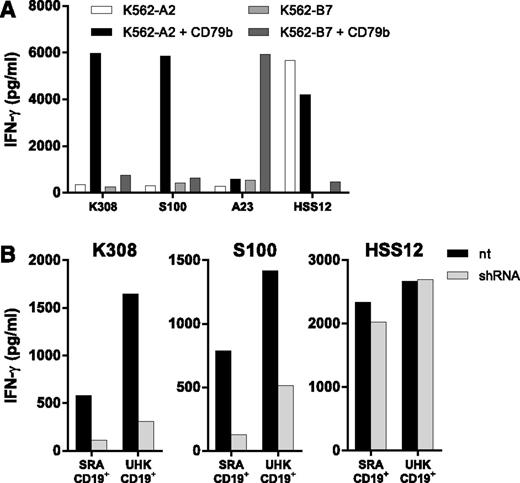
CD8+ T-cell clones binding to pMHC tetramer CD79b173:A2 demonstrated varying degrees of peptide specificity and levels of avidity. Figure 1A-C shows the tetramer staining and functional results of 5 representative clones. T-cell clone K110 showed recognition of all 3 HLA-A2+ B-LCLs, as well as unloaded K562-A2 cells, illustrating non-CD79b specificity of the T-cell clone. Clone K64 demonstrated no recognition of peptide-pulsed K562-A2 cells and no reactivity toward any CD79b-expressing B-LCL (Figure 1B-C, respectively). To exclude that absence of recognition could be attributed to failure to produce IFN-γ, granulocyte macrophage colony stimulating factor (GM-CSF) secretion was assessed, but no GM-CSF production was observed (data not shown), suggesting an insufficient sensitivity of clone K64 to recognize MHC-bound CD79b173 peptide. In contrast, T-cell clones K68, K116, and K308 showed CD79b173 peptide–dependent recognition of peptide-pulsed K562-A2 cells. Clone K308 demonstrated the highest peptide sensitivity (Figure 1B). Corresponding with this high peptide sensitivity, clone K308 demonstrated reactivity toward all 3 B-LCLs, whereas the recognition of 3 B-LCLs was low or absent for the low avidity clones K116 and K68 (Figure 1C). These results indicate that clone K308 exhibits sufficient avidity for CD79b173 peptide to be able to efficiently recognize endogenously processed and presented CD79b in the context of HLA-A2 (Figure 1C,G).
pMHC tetramer+ CD8+ T cells exhibit different degrees of CD79b peptide sensitivity and specificity. T cells were clonally expanded by single-cell sorting of CD8+ T cells binding to pMHC tetramers containing CD79b peptides. (A) Shown are pMHC-tetramer staining of 5 representative clones (K110, K64, K68, K116, and K308) isolated using pMHC tetramer CD79b173:A2. Control includes clone HSS12 specific for USP11-derived peptide FTWEGLYNV bound to HLA-A2. Shown are CD8+ cells. (B-C) The same T-cell clones as in (A) were coincubated with K562-A2 cells loaded with decreasing concentrations of CD79b173 peptide (B) or were cocultured with 3 HLA-A2/B7–positive B-LCLs endogenously expressing CD79b (C). Controls included CD79b-negative K562-A2 cells. (D-E) K562-A2 cells peptide-pulsed with decreasing concentration of peptide CD79b173 or CD79b20 as indicated were coincubated with clone K308 isolated using pMHC tetramer CD79b173:A2 (D) or clone S100 isolated using pMHC tetramer CD79b20:A2 (E). (F) K562 cells retrovirally transduced with HLA-B7 (K562-B7) were peptide-pulsed with decreasing concentration of peptide CD79b9 and coincubated with clone A23 isolated with pMHC tetramer CD79b9:B7. (G-I) Clones K308 (G), S100 (H), and A23 (I) were cocultured with HLA-A2 and HLA-B7–positive B-LCLs. CD79b-negative K562-A2 or K562-B7 cells were included as controls. IFN-γ secretion was measured using standard ELISA. Axes were adjusted to maximum cytokine release measured. (J-L) K308 was stained with pMHC tetramer CD79b173:A2 (J), S100 with CD79b20:A2 (K), and A23 with CD79b9:B7 (L). Staining with an irrelevant pMHC tetramer USP11:A2 was used as control. Dot plots are shown with biexponential axis.
pMHC tetramer+ CD8+ T cells exhibit different degrees of CD79b peptide sensitivity and specificity. T cells were clonally expanded by single-cell sorting of CD8+ T cells binding to pMHC tetramers containing CD79b peptides. (A) Shown are pMHC-tetramer staining of 5 representative clones (K110, K64, K68, K116, and K308) isolated using pMHC tetramer CD79b173:A2. Control includes clone HSS12 specific for USP11-derived peptide FTWEGLYNV bound to HLA-A2. Shown are CD8+ cells. (B-C) The same T-cell clones as in (A) were coincubated with K562-A2 cells loaded with decreasing concentrations of CD79b173 peptide (B) or were cocultured with 3 HLA-A2/B7–positive B-LCLs endogenously expressing CD79b (C). Controls included CD79b-negative K562-A2 cells. (D-E) K562-A2 cells peptide-pulsed with decreasing concentration of peptide CD79b173 or CD79b20 as indicated were coincubated with clone K308 isolated using pMHC tetramer CD79b173:A2 (D) or clone S100 isolated using pMHC tetramer CD79b20:A2 (E). (F) K562 cells retrovirally transduced with HLA-B7 (K562-B7) were peptide-pulsed with decreasing concentration of peptide CD79b9 and coincubated with clone A23 isolated with pMHC tetramer CD79b9:B7. (G-I) Clones K308 (G), S100 (H), and A23 (I) were cocultured with HLA-A2 and HLA-B7–positive B-LCLs. CD79b-negative K562-A2 or K562-B7 cells were included as controls. IFN-γ secretion was measured using standard ELISA. Axes were adjusted to maximum cytokine release measured. (J-L) K308 was stained with pMHC tetramer CD79b173:A2 (J), S100 with CD79b20:A2 (K), and A23 with CD79b9:B7 (L). Staining with an irrelevant pMHC tetramer USP11:A2 was used as control. Dot plots are shown with biexponential axis.
Among the T-cell clones isolated using pMHC tetramers CD79b20:A2 or CD79b9:B7, 2 T-cell clones exhibited peptide-specific recognition of peptide-pulsed K562 cells. T-cell clones S100 and A23 recognized peptide CD79b20 and CD79b9 when loaded on K562-A2 and K562-B7 cells, respectively (Figure 1E-F, respectively). Both T-cell clones efficiently recognized all 3 B-LCLs (Figure 1H-I).
Specificity for their cognate pMHC tetramers was confirmed for clones K308, S100, and A23 by FACS analysis (Figure 1J-L). None of the T-cell clones stained with the control pMHC tetramer composed of HLA-A2 presenting peptide FTWEGLYNV derived from USP11.
These data demonstrate that we have isolated high-avidity peptide-specific T-cell clones targeting CD79b peptides presented by alloHLA molecules. T-cell clones K308, S100, and A23 were identified as the most avid clones targeting peptides CD79b173, CD79b20, and CD79b9, respectively.
CD79b transduction and CD79b knockdown verified CD79b specificity of the T-cell clones
To verify CD79b-dependent reactivity by T-cell clones K308, S100, and A23, K562-A2 and K562-B7 cells were retrovirally transduced with CD79b. Clone K308 and S100 specific for HLA-A2–presented peptides CD79b173 and CD79b20, respectively, demonstrated efficient recognition of K562-A2 engineered to express CD79b, whereas reactivity toward nontransduced K562-A2 or K562 carrying the wrong HLA-restriction molecule was absent (Figure 2A). CD79b9-specific clone A23 efficiently recognized K562 simultaneously expressing HLA-B7 and CD79b, whereas no recognition was observed of nontransduced K562-B7. Recognition was also absent when HLA-A2 was expressed in CD79b-transduced K562 instead of HLA-B7.
CD79b expression and shRNA knockdown confirm CD79b specificity of isolated T-cell clones. (A) CD79b-negative K562 cells expressing either HLA-A2 (K562-A2) or HLA-B7 (K562-B7) were retrovirally transduced with codon-optimized CD79b (K562-A2 + CD79b or K562-B7 + CD79b) and sorted to high purity. CD79b-reactive T-cell clones K308 (CD79b173:A2), S100 (CD79b20:A2), and A23 (CD79b9-17:B7) were coincubated with transductants. Control T-cell clone HSS12 recognizes a peptide of the ubiquitously expressed gene USP11 in the context of HLA-A2. (B) CD40L-activated CD19+ B cells of 2 HLA-A2–positive healthy individuals (SRA CD19+ and UHK CD19+) were transduced with CD79b-specific shRNA (shRNA) and selected by puromycin treatment or were left untreated (nt) and coincubated with T-cell clones K308 or S100 or control clone HSS12. Clones K308 and S100 were cocultured at a S:R ratio of 2:1. Control clone HSS was cocultured at an S:R ration of 10:1. IFN-γ production was determined using standard ELISA. Antigen-presenting capacity of transduced and nontransduced target cells was controlled by clone HSS12.
CD79b expression and shRNA knockdown confirm CD79b specificity of isolated T-cell clones. (A) CD79b-negative K562 cells expressing either HLA-A2 (K562-A2) or HLA-B7 (K562-B7) were retrovirally transduced with codon-optimized CD79b (K562-A2 + CD79b or K562-B7 + CD79b) and sorted to high purity. CD79b-reactive T-cell clones K308 (CD79b173:A2), S100 (CD79b20:A2), and A23 (CD79b9-17:B7) were coincubated with transductants. Control T-cell clone HSS12 recognizes a peptide of the ubiquitously expressed gene USP11 in the context of HLA-A2. (B) CD40L-activated CD19+ B cells of 2 HLA-A2–positive healthy individuals (SRA CD19+ and UHK CD19+) were transduced with CD79b-specific shRNA (shRNA) and selected by puromycin treatment or were left untreated (nt) and coincubated with T-cell clones K308 or S100 or control clone HSS12. Clones K308 and S100 were cocultured at a S:R ratio of 2:1. Control clone HSS was cocultured at an S:R ration of 10:1. IFN-γ production was determined using standard ELISA. Antigen-presenting capacity of transduced and nontransduced target cells was controlled by clone HSS12.
Furthermore, downregulation of CD79b with specific shRNA abrogated recognition of CD40L-stimulated CD19+ B cells from HLA-A2+ healthy individuals by clones K308 and S100, whereas the reactivity of control clone HSS12 specific for a peptide derived from the ubiquitously expressed gene USP11 presented in HLA-A2 was unaffected (Figure 2B).39 Similar results were obtained for clone A23, where CD79b-specific knockdown abrogated recognition of LCL-JY (supplemental Figure 2). In summary, CD79b-dependent and specific recognition were confirmed for T-cell clones K308, S100, and A23.
CD79b-specific T-cell clones recognize B-cell malignancies
Next, CD79b-specific T-cell clones K308 and S100 were assessed for their potency to recognize a variety of primary B-cell malignancies and malignant B-cell lines. Clones K308 and S100 efficiently recognized HLA-A2–positive ALL cell lines (Figure 3A). Clone S100 also recognized 4 HLA-A2–positive primary CLL samples (Figure 3B). In addition, stimulation with primary HLA-A2–positive CLL and ALL samples resulted in strong IFN-γ production of clone K308 (Figure 3C-D). No recognition of the HLA-A2–negative CLL sample ERV 8300 was observed (Figure 3D). Coincubation of the ALL cell lines or primary HLA-A2–positive ALL and CLL samples with clone K308 resulted in efficient lysis of the malignant cells (Figure 3E-F), indicating a cytotoxic effect of clone K308. No lysis of the HLA-A2–negative CLL sample ERV 8300 (Figure 3F) after coincubation with clone K308 was detected. Killing of the malignant cells was not observed for a cytomegalovirus (CMV)-specific control clone recognizing the irrelevant peptide NLVPMVATV bound to HLA-A2 (Figure 3E-F).These results indicate efficient recognition of CD79b-expressing HLA-A2–positive B-cell malignancies by the CD79b-specific clones K308 and S100.
CD79b-specific T-cell clones recognize malignant B-cell lines and primary B-cell malignancies. (A) T-cell clones K308 and S100 recognizing CD79b173 and CD79b20 peptides in the context of HLA-A2, respectively, were stimulated with 3 HLA-A2–positive ALL cell lines ALL-BV, ALL-CM, and ALL-RL at an S:R ratio of 10:1. Controls included CD79b-negative K562-A2 cells and CD79b-positive LCL-JY. (B) T-cell clone S100 was stimulated with 4 primary HLA-A2–positive primary CLL samples at an S:R ratio 10:1. (C-D) Clone K308 was tested against primary CLL (C) and primary ALL (D) samples at the indicated S:R ratios. The HLA-A2 genotype of primary samples is indicated in parentheses. Controls included samples that were exogenously loaded with 100 nM CD79b173 peptide (3:1 + peptide). IFN-γ production was assessed using standard ELISA. (E-F) Cytotoxicity of K308 was determined for ALL cell lines and primary CLL and ALL samples. PKH26GL-labeled ALL cell lines (E) or primary CLL and ALL samples (F) were coincubated with K308 (K308) at different effector-to-target ratios for 17 hours. Living PKH26GL-labeled target cells were counted and analyzed by FACS, and the percent surviving cells was calculated (see Material and methods). Controls included a CMV-specific T-cell clone (CMV clone) recognizing irrelevant pp65-derived peptide NLVPMVATV presented in HLA-A2, and CD79b-negative K562-A2 cells. The experiment was carried out in triplicate showing mean ± standard deviation.
CD79b-specific T-cell clones recognize malignant B-cell lines and primary B-cell malignancies. (A) T-cell clones K308 and S100 recognizing CD79b173 and CD79b20 peptides in the context of HLA-A2, respectively, were stimulated with 3 HLA-A2–positive ALL cell lines ALL-BV, ALL-CM, and ALL-RL at an S:R ratio of 10:1. Controls included CD79b-negative K562-A2 cells and CD79b-positive LCL-JY. (B) T-cell clone S100 was stimulated with 4 primary HLA-A2–positive primary CLL samples at an S:R ratio 10:1. (C-D) Clone K308 was tested against primary CLL (C) and primary ALL (D) samples at the indicated S:R ratios. The HLA-A2 genotype of primary samples is indicated in parentheses. Controls included samples that were exogenously loaded with 100 nM CD79b173 peptide (3:1 + peptide). IFN-γ production was assessed using standard ELISA. (E-F) Cytotoxicity of K308 was determined for ALL cell lines and primary CLL and ALL samples. PKH26GL-labeled ALL cell lines (E) or primary CLL and ALL samples (F) were coincubated with K308 (K308) at different effector-to-target ratios for 17 hours. Living PKH26GL-labeled target cells were counted and analyzed by FACS, and the percent surviving cells was calculated (see Material and methods). Controls included a CMV-specific T-cell clone (CMV clone) recognizing irrelevant pp65-derived peptide NLVPMVATV presented in HLA-A2, and CD79b-negative K562-A2 cells. The experiment was carried out in triplicate showing mean ± standard deviation.
No reactivity against nonhematopoietic tissue but unexpected reactivity against different hematopoietic cell subsets
To further validate that the recognition pattern of the CD79b-specific T-cell clones is restricted to the B-cell compartment, their reactivity toward healthy nonhematopoietic and hematopoietic cells was assessed. No IFN-γ production was measured for K308 and S100 after overnight coculture with HLA-A2–positive fibroblasts or proximal tubular epithelial cells (PTECs) (Figure 4A). Recognition was also absent when fibroblast and PTECs were pretreated with IFN-γ for 4 days to simulate inflamed conditions. Control-clone HSS12 recognized fibroblasts and PTECs under all conditions, indicating the stimulatory capacity of the target cells. In addition, we assessed clone K308’s reactivity toward hematopoietic cell populations by isolating different cell subsets from the peripheral blood of HLA-A2–positive healthy individuals. As shown in Figure 4B, clone K308 did not recognize monocyte-derived immature and mature DCs. However, clone K308 exhibited intermediate reactivity against either activated or nonactivated CD4+ T cells, nonactivated CD8+ T cells, and CD34+ HPCs, as well as low reactivity against CD14+ monocytes (Figure 4B-D). Furthermore, coincubation of nonactivated and activated B cells and T cells, and CD34+ HPCs with clone K308, also led to their lysis, which was not observed when target cells were coincubated with a CMV-specific T-cell clone (Figure 4E-F). Similarly, clone S100 also recognized activated CD4+ T cells (Figure 4G).
CD79b-specific T-cell clones spare healthy nonhematopoietic tissue but demonstrate intermediate and low recognition of T cells, HPCs, and monocytes. CD79b-specific T-cell clones were assessed for their reactivity toward healthy nonhematopoietic tissue and hematopoietic cell subsets. (A) Recognition of HLA-A2–positive fibroblasts (FB) and proximal tubular epithelial cells (PTECs) untreated (–) or pretreated with IFN-γ (+) for 4 days was assessed after coincubation with CD79b-reactive T-cell clones K308 and S100 for 18 hours at an S:R ratio of 5:1. Controls included CD79b-negative K562-A2 cells, CD79b-positive LCL-JY, and T-cell clone HSS12 recognizing a peptide of the ubiquitously expressed USP11 presented in HLA-A2. (B-C) Reactivity toward HLA-A2–positive healthy hematopoietic cells was assessed by coculturing clone K308 with cultured (B) and nonactivated (C) cell subsets at the indicated S:R ratios for 18 hours. Controls included peptide-pulsing of stimulators at a concentration of 100 nM peptide (2:1 + peptide or 6:1 + peptide). CD19+ B cells were cocultured with irradiated CD40L-expressing mouse fibroblasts for 6 days before the experiment (CD19 [CD40L]). Immature dendritic cells (imDCs) and mature DCs (mDCs) were CD14+ monocyte–derived (see supplemental Data). Activated CD4+ cells were generated from CD4+ cells that were treated with 0.8 μg/mL PHA and cultured for 10 days. Nonactivated CD19+, CD14+, CD4+, and CD34+ cells were isolated from peripheral blood using MACS isolation. (D) Clone K308 was stimulated with primary CD4+ or CD8+ cells in an S:R ration of 6:1. (E-F) Cytotoxicity of clone K308 was determined for nonactivated CD4+ and CD8+ T cells and activated T cells (containing CD4+ and CD8+ T cells), activated and nonactivated CD19+ B cells (E), and CD34+ HPCs (F). PKH26GL-labeled target cells were coincubated with K308 (K308) at different effector to target ratios for 17 hours. Living PKH26GL-labeled target cells were counted and analyzed by FACS and the percent surviving cells was calculated (see Material and methods). Controls included a CMV-specific T-cell clone (CMV clone) recognizing irrelevant pp65-derived peptide NLVPMVATV presented in HLA-A2. The experiment was carried out in triplicate showing mean ± standard deviation. (G) T-cell clone S100 recognizing peptide CD79b20 in HLA-A2 was stimulated with activated CD4+ T cells or CD40L-activated CD19+ B cells from 2 HLA-A2–positive individuals in an S:R ratio of 10:1. IFN-γ secretion was assessed by standard ELISA.
CD79b-specific T-cell clones spare healthy nonhematopoietic tissue but demonstrate intermediate and low recognition of T cells, HPCs, and monocytes. CD79b-specific T-cell clones were assessed for their reactivity toward healthy nonhematopoietic tissue and hematopoietic cell subsets. (A) Recognition of HLA-A2–positive fibroblasts (FB) and proximal tubular epithelial cells (PTECs) untreated (–) or pretreated with IFN-γ (+) for 4 days was assessed after coincubation with CD79b-reactive T-cell clones K308 and S100 for 18 hours at an S:R ratio of 5:1. Controls included CD79b-negative K562-A2 cells, CD79b-positive LCL-JY, and T-cell clone HSS12 recognizing a peptide of the ubiquitously expressed USP11 presented in HLA-A2. (B-C) Reactivity toward HLA-A2–positive healthy hematopoietic cells was assessed by coculturing clone K308 with cultured (B) and nonactivated (C) cell subsets at the indicated S:R ratios for 18 hours. Controls included peptide-pulsing of stimulators at a concentration of 100 nM peptide (2:1 + peptide or 6:1 + peptide). CD19+ B cells were cocultured with irradiated CD40L-expressing mouse fibroblasts for 6 days before the experiment (CD19 [CD40L]). Immature dendritic cells (imDCs) and mature DCs (mDCs) were CD14+ monocyte–derived (see supplemental Data). Activated CD4+ cells were generated from CD4+ cells that were treated with 0.8 μg/mL PHA and cultured for 10 days. Nonactivated CD19+, CD14+, CD4+, and CD34+ cells were isolated from peripheral blood using MACS isolation. (D) Clone K308 was stimulated with primary CD4+ or CD8+ cells in an S:R ration of 6:1. (E-F) Cytotoxicity of clone K308 was determined for nonactivated CD4+ and CD8+ T cells and activated T cells (containing CD4+ and CD8+ T cells), activated and nonactivated CD19+ B cells (E), and CD34+ HPCs (F). PKH26GL-labeled target cells were coincubated with K308 (K308) at different effector to target ratios for 17 hours. Living PKH26GL-labeled target cells were counted and analyzed by FACS and the percent surviving cells was calculated (see Material and methods). Controls included a CMV-specific T-cell clone (CMV clone) recognizing irrelevant pp65-derived peptide NLVPMVATV presented in HLA-A2. The experiment was carried out in triplicate showing mean ± standard deviation. (G) T-cell clone S100 recognizing peptide CD79b20 in HLA-A2 was stimulated with activated CD4+ T cells or CD40L-activated CD19+ B cells from 2 HLA-A2–positive individuals in an S:R ratio of 10:1. IFN-γ secretion was assessed by standard ELISA.
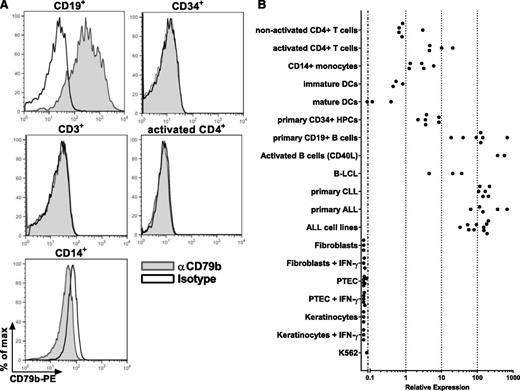
Aberrant CD79b expression in T cells, HPCs, and monocytes is responsible for reactivity of CD79b-specific T-cell clones
To resolve the unexpected recognition of T cells, HPCs, and monocytes, we examined whether these cell populations demonstrated unreported CD79b cell-surface expression. Staining of PBMCs with anti-CD79b antibody revealed no cell-surface expression of CD79b on CD3+ T cells and CD14+ monocytes (Figure 5A). In addition, activated CD4+ T cells and purified CD34+ HPCs did not show detectable levels of cell-surface CD79b, whereas CD79b was demonstrated on CD19+ B cells (Figure 5A). To investigate whether gene expression of CD79b in cells outside of the B-cell compartment occurred, we performed qRT-PCR. As expected, high expression of CD79b in CLL, ALL, B-LCL, and healthy CD19+ B-cell samples was demonstrated by qRT-PCR (Figure 5B), whereas no expression could be detected for nonhematopoietic cells such as fibroblasts, keratinocytes, and PTECs under normal and IFN-γ–treated conditions. In addition, mature and immature DCs showed no or very low gene expression of CD79b (100-1000–fold lower expression compared with CD19+ B cells). In activated CD4+ T cells and CD34+ HPCs, the expression of CD79b was approximately only tenfold lower than in CD19+ B cells correlating with intermediate recognition of these cell subsets. In weakly recognized CD14+ monocytes and nonactivated CD4+ T cells, the expression of CD79b was approximately ten- to 100-fold lower compared with CD19+ B cells. These results demonstrate a correlation between CD79b expression and the reactivity of the CD79b-specific clones, exhibiting strong reactivity against CD19+ B cells, no recognition of fibroblasts and PTECs, and immature and mature DCs but intermediate to low reactivity against T cells, HPCs, and monocytes.
Aberrant CD79b gene expression in T cells, HPCs, and monocytes in the absence of cell-surface expression. (A) PBMCs were stained with CD79b-specific antibody (αCD79b) or isotype control (Isotype) in combination with lineage-specific markers as indicated. Histograms show CD79b expression after gating on B cells (CD19+), T cells (CD3+), and monocytes (CD14+). CD34+ HPCs were isolated from granulocyte colony-stimulating factor–mobilized blood. Activated CD4+ T cells were stimulated with PHA and irradiated autologous feeders and cultured for 10 days. (B) CD79b mRNA expression was determined for indicated cell subsets by qRT-PCR. Expression was normalized to average CD79b expression in primary CD19+ B cells, which was set to 100. Dots indicate individual samples.
Aberrant CD79b gene expression in T cells, HPCs, and monocytes in the absence of cell-surface expression. (A) PBMCs were stained with CD79b-specific antibody (αCD79b) or isotype control (Isotype) in combination with lineage-specific markers as indicated. Histograms show CD79b expression after gating on B cells (CD19+), T cells (CD3+), and monocytes (CD14+). CD34+ HPCs were isolated from granulocyte colony-stimulating factor–mobilized blood. Activated CD4+ T cells were stimulated with PHA and irradiated autologous feeders and cultured for 10 days. (B) CD79b mRNA expression was determined for indicated cell subsets by qRT-PCR. Expression was normalized to average CD79b expression in primary CD19+ B cells, which was set to 100. Dots indicate individual samples.
In summary, aberrant gene expression of CD79b in CD4+ T cells, HPCs, and monocytes, although undetectable at the cell surface, caused unexpected reactivity of the CD79b-specific T-cell clones against these cell subsets.
Discussion
In this study, we aimed to isolate T cells expressing TCRs specific for BCR-associated protein CD79b that is expressed on normal and malignant B cells.14-16 MHC-presented peptides of CD79b were identified from peptide-elution studies.47,50,51 By exploiting the immunogenicity of alloHLA molecules, 3 distinct T-cell clones derived from different HLA-A*0201/B*0702– healthy individuals were isolated targeting distinct peptides of CD79b presented in HLA-A*0201 or HLA-B*0702. The CD79b-specific clones efficiently recognized primary CLL, ALL, and ALL cell lines, whereas no reactivity toward nonhematopoietic cells and professional antigen-presenting cells was detected. However, unexpected reactivity toward monocytes, HPCs, and T cells was observed. Aberrant intracellular CD79b expression in those cell subsets was detected in the absence of cell-surface CD79b.
Although all T-cell clones were isolated by binding to pMHC tetramers containing CD79b peptides, CD79b specificity was not observed in all cases. Various T-cell clones lacked specificity for CD79b peptides, indicated by recognition of CD79b-negative K562-A2 or K562-B7 cells. This reactivity is probably the result of crossreactivity with other alloHLA-bound peptides and resembles the reactivity previously observed for WT1-specific T cells derived from the allo-repertoire.52 Other isolated T-cell clones showed no reactivity toward peptide-loaded stimulator cells and CD79b-expressing B-LCLs, suggesting insufficient T-cell avidity. In contrast, we also isolated pMHC tetramer+ T-cell clones with variable sensitivity for the CD79b peptide-loaded stimulators. Among these T-cell clones, only the most avid clones were capable of recognizing endogenously-processed peptide. We have previously illustrated53 that not pMHC-tetramer binding but TCR-ligand koff-rate as measured by decay of pMHC:TCR interaction correlates with functional characteristics. Assessment of sensitivity and specificity for MHC-bound CD79b peptides on a clonal level was therefore imperative to select for the most avid and CD79b-specific clones.
For 3 high-avidity T-cell clones, we confirmed CD79b specificity by CD79b transduction into K562 cells and abrogation of recognition of B-LCLs and CD19+ B cells after shRNA-mediated CD79b-specific knockdown. Furthermore, absence of reactivity toward CD79b-negative cells such as nonhematopoietic cell lines pretreated with IFN-γ and professional antigen-presenting cells indicated that CD79b-independent activation of our CD79b-specific T-cell clones does not easily occur. However, recognition of T cells, HPCs, and monocytes was observed, which correlated with low aberrant intracellular CD79b expression in these cells. Although we cannot formally exclude the recognition of peptide lookalikes or mimitopes in these cells, the strong CD79b dependency of our CD79b-specific T-cell clones argues otherwise.
Recognition of primary B-cell malignancies by our T-cell clones suggested that the expressed CD79b-specific HLA-A*0201 or B*0702-restricted TCRs could be used for TCR gene transfer approaches to treat HLA-A*0201 or B*0702-positive patients with B-cell malignancies. However, we also observed recognition of monocytes, T cells, and HPCs. In contrast to B-cell depletion, long-term monocytopenia and T-cell and HPC ablation is not tolerable from a clinical perspective.
The high-avidity CD79b-specific T cells recognizing a self-antigen such as CD79b when presented in HLA-A2 or HLA-B7 were isolated from HLA-A2/B7–negative individuals. Negative selection during thymic development does not deplete these high-avidity T cells in these individuals because the HLA-A2 and HLA-B7 restriction molecules are not present. In HLA-A2 and HLA-B7–positive individuals, however, the elimination of such high-avidity CD79b-specific T cells is crucial to prevent loss of B cells, monocytes, and HPCs as the result of autoreactivity. Furthermore, the CD79b expression in HLA-A2/B7–positive T cells would also cause fratricide41 among T cells; T cells would simultaneously express the correct HLA restriction molecule as well as the CD79b antigen recognized by their TCR. In concordance, we have previously shown that self-HLA–restricted PRAME-specific T cells possess insufficient avidity to recognize endogenously expressed PRAME.38 Therefore, it is very unlikely that such high-avidity CD79b-specific T cells as demonstrated in this study could be isolated from the autologous repertoire of HLA-A2 and HLA-B7 individuals.
In conclusion, exploiting the immunogenicity of alloHLA molecules is an effective and attractive strategy to overcome self-tolerance toward self-antigens. TCRs raised from alloreactive T cells can meet the requirements for high affinity interaction with self-antigens while preserving peptide specificity and HLA-restriction. Their identification can be readily achieved by pMHC tetramer–based isolation but requires functional assessment on a clonal level. Aberrant expression of CD79b in monocytes, T cells, and HPCs eliminated CD79b as a suitable target for TCR-based immunotherapy to treat B-cell malignancies. However, CD79b may still be an attractive target for CAR or antibody-based immunotherapies because no cell-surface expression on monocytes, T cells, and HPCs was observed.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Prof Cees van Kooten (Department of Nephrology, Leiden University Medical Center, Leiden, The Netherlands) for providing PTECs. CD79b-specific shRNA constructs were a kind gift of Prof. Rob Hoeben (Department of Molecular Cell Biology, Leiden University Medical Center, Leiden, The Netherlands). Guido de Roo and Menno van der Hoorn provided technical assistance in flow cytometric cell sorting.
This research was supported by the financial assistance of the Dutch Cancer Society (2010-4832) and the Landsteiner Foundation for Blood Transfusion Research (LSBR0713).
Authorship
Contribution: L.J. designed, performed and analyzed research, and wrote the manuscript; P.H. and M.G.D.K. designed, performed, and analyzed research; C.H., D.M.v.d.S., and R.S.H. performed and analyzed research; P.A.v.V designed research; J.H.F.F. designed research and wrote the manuscript; M.H.M.H. designed and analyzed research, and wrote the manuscript; and all authors reviewed and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Lorenz Jahn, Department of Hematology, Leiden University Medical Center, PO Box 9600, 2300 RC Leiden, The Netherlands; e-mail: l.jahn@lumc.nl.



![Figure 4. CD79b-specific T-cell clones spare healthy nonhematopoietic tissue but demonstrate intermediate and low recognition of T cells, HPCs, and monocytes. CD79b-specific T-cell clones were assessed for their reactivity toward healthy nonhematopoietic tissue and hematopoietic cell subsets. (A) Recognition of HLA-A2–positive fibroblasts (FB) and proximal tubular epithelial cells (PTECs) untreated (–) or pretreated with IFN-γ (+) for 4 days was assessed after coincubation with CD79b-reactive T-cell clones K308 and S100 for 18 hours at an S:R ratio of 5:1. Controls included CD79b-negative K562-A2 cells, CD79b-positive LCL-JY, and T-cell clone HSS12 recognizing a peptide of the ubiquitously expressed USP11 presented in HLA-A2. (B-C) Reactivity toward HLA-A2–positive healthy hematopoietic cells was assessed by coculturing clone K308 with cultured (B) and nonactivated (C) cell subsets at the indicated S:R ratios for 18 hours. Controls included peptide-pulsing of stimulators at a concentration of 100 nM peptide (2:1 + peptide or 6:1 + peptide). CD19+ B cells were cocultured with irradiated CD40L-expressing mouse fibroblasts for 6 days before the experiment (CD19 [CD40L]). Immature dendritic cells (imDCs) and mature DCs (mDCs) were CD14+ monocyte–derived (see supplemental Data). Activated CD4+ cells were generated from CD4+ cells that were treated with 0.8 μg/mL PHA and cultured for 10 days. Nonactivated CD19+, CD14+, CD4+, and CD34+ cells were isolated from peripheral blood using MACS isolation. (D) Clone K308 was stimulated with primary CD4+ or CD8+ cells in an S:R ration of 6:1. (E-F) Cytotoxicity of clone K308 was determined for nonactivated CD4+ and CD8+ T cells and activated T cells (containing CD4+ and CD8+ T cells), activated and nonactivated CD19+ B cells (E), and CD34+ HPCs (F). PKH26GL-labeled target cells were coincubated with K308 (K308) at different effector to target ratios for 17 hours. Living PKH26GL-labeled target cells were counted and analyzed by FACS and the percent surviving cells was calculated (see Material and methods). Controls included a CMV-specific T-cell clone (CMV clone) recognizing irrelevant pp65-derived peptide NLVPMVATV presented in HLA-A2. The experiment was carried out in triplicate showing mean ± standard deviation. (G) T-cell clone S100 recognizing peptide CD79b20 in HLA-A2 was stimulated with activated CD4+ T cells or CD40L-activated CD19+ B cells from 2 HLA-A2–positive individuals in an S:R ratio of 10:1. IFN-γ secretion was assessed by standard ELISA.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/6/10.1182_blood-2014-07-587840/4/m_949f4.jpeg?Expires=1767758170&Signature=syuUgqmOHhj3JC3vj5py~bAib2pZYaCuhAFKqklxT0xpgWiU8ZmNFm~YgDpp1BNOFzjCgvszE83FykGYNlk8Iwtpt~dxjUsaKHI3yaVoM1HUfOsz~XHRitJLzGFKq2CJ9goVywvZNCxsxxC~6AaVqetskxBu4nw7KpDJbjYKEDtEVjP3tNQyLjuqe12Pp7PeG1uvCADahEwx4yS8UOGCpqnqwO18gpQBXLwBQjDsGYl19lquRtO3HErrxDpBwZ8xcBu~1XLH9I~J7gYfUdDb9h8FbklvR3FUcYirB2gl9GP9JLkK1ZtO21tSGAy6F1Se8b6Owc2mB9GsQ-pVtejmnA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal