Key Points
Ott1 regulates the alternative splicing of Mpl-TR, a truncated isoform of c-Mpl, which modulates Thpo-mediated signaling.
Mpl-TR expression impairs HSC engraftment.
Abstract
Thrombopoietin (Thpo) signaling through the c-Mpl receptor promotes either quiescence or proliferation of hematopoietic stem cells (HSCs) in a concentration-dependent manner; however, in vivo Thpo serum levels are responsive to platelet mass rather than HSC demands, suggesting additional regulation exists. Ott1 (Rbm15), a spliceosomal component originally identified as a fusion partner in t(1;22)-associated acute megakaryocytic leukemia, is also essential for maintaining HSC quiescence under stress. Ott1 controls the alternative splicing of a dominant negative isoform, Mpl-TR, capable of inhibiting HSC engraftment and attenuating Thpo signaling. Ott1, which associates with Hdac3 and the histone methyltransferase, Setd1b, binds to both c-Mpl RNA and chromatin and regulates H4 acetylation and H3K4me3 marks. Histone deacetylase or histone methyltransferase inhibition also increases Mpl-TR levels, suggesting that Ott1 uses an underlying epigenetic mechanism to control alternative splicing of c-Mpl. Manipulation of Ott1-dependent alternative splicing may therefore provide a novel pharmacologic avenue for regulating HSC quiescence and proliferation in response to Thpo.
Introduction
Hematopoietic stem cell (HSC) function depends on an incompletely understood combination of cytokine stimulation and niche interaction to simultaneously replenish differentiated hematopoietic cells, yet sustain a self-renewing population. The cytokine thrombopoietin (Thpo), the sole ligand for the receptor c-Mpl, has been identified as having an essential role in HSCs as well as megakaryocyte development. C-Mpl is a type 1 hematopoietic cytokine receptor composed of an extracellular domain capable of binding Thpo and an intracellular domain that functions as a “scaffold” to enable activation of Jak, Stat, Ras, and PI3K pathways upon Thpo-induced dimerization.1 Megakaryopoiesis requires Thpo for the early development of megakaryocyte precursors, and in many cases of congenital amegakaryocytic thrombocytopenia with c-MPL inactivating mutations, in addition to defects in megakaryocyte development, these patients are prone to early bone marrow failure caused by HSC exhaustion.2
c-Mpl−/− mice have reduced hematopoietic progenitors, although they maintain peripheral blood counts, with the exception of platelets.3,4 Even though c-Mpl−/− mice maintain hematopoiesis through adulthood, competitive transplantation yields only a third short-term engraftment and decreasing long-term engraftment over time. Conversely, wild-type (WT) engraftment is reduced in Thpo−/− recipients, and mice treated with a Thpo-blocking antibody lose quiescence and have enhanced cycling.5,6 At lower concentrations, Thpo enhances HSC quiescence and preserves or expands the self-renewing population, especially during periods of proliferative stress such as engraftment.5 In distinct contrast, high concentrations of Thpo, either in cell lines or in vivo, promotes HSC proliferation.6,7 It is unclear why this dual response is elicited in HSCs because Thpo levels are primarily regulated by platelet mass. Thpo is constitutively produced by the liver, kidneys, and bone marrow stroma and is cleared by platelet-associated c-Mpl binding and internalization.1 Therefore, Thpo levels appear primarily geared for the regulation of platelet mass rather than HSC function.
The physiologic role of Ott1 (Rbm15) in HSCs shares several similarities with c-Mpl. Ott1 was identified as the 5′ fusion partner in t(1;22)(p13;q13) acute megakaryocytic leukemia (AMKL), which generates a chimeric protein, OTT1-MAL (RBM15-MKL1).8,9 Hematopoiesis in Ott1-deleted mice is sustained under steady-state conditions; however, proliferative stress leads to loss of HSC quiescence and exhaustion similar to that observed with c-Mpl.10 Ott1 possesses a Spen paralog and ortholog C-terminal (SPOC) domain capable of transcriptional activation/repression in vitro, yet in vivo targets have not been previously identified.11,12 Binding partners to the SPOC domain include NCor/Smrt corepressors, the class I histone deacetylase Hdac3, and the histone methyltransferase (HMT) Setd1b.12,13 The N-terminus of Ott1 contains 3 RNA recognition motifs (RRMs). Ott1 was isolated in the spliceosome and has been identified in the binding and export of Kaposi sarcoma virus and Epstein-Barr virus viral RNAs; however, endogenous RNA targets have not been reported.11,14-16
In this report, we demonstrate that Ott1 regulates the production of Mpl-TR, an isoform that interferes with full-length Mpl (Mpl-FL) function and Thpo response. Exogenous expression of Mpl-TR in WT HSCs impairs short- and long-term engraftment. Ott1 binds to c-Mpl RNA and also to c-Mpl chromatin in regions flanking the alternatively spliced exons. C-Mpl histone methylation, H3K4me3, and H4 acetylation are regulated by Ott1, and chemical inhibition of Hdac and HMT activity reproduces an increase in the splicing ratio of Mpl-TR:Mpl-FL. Inhibition of Ott1-associated histone modification enzymes suggests this mechanism has an epigenetic basis and provides the potential for pharmacologic manipulation of the Thpo/c-Mpl axis.
Methods
Animals
Ott1flox/nullMx1-cre and Ott1flox/nullSox2-cre animals were generated as described previously.17,18 To generate Ott1 knockout (KO) bone marrow, 6-week-old Ott1flox/nullMx1-cre mice were injected intraperitoneally (IP) with 250 µg pIpC (GE Healthcare) every other day for 3 doses. Littermates possessing at least 1 WT Ott1 allele were used as controls. Experiments were performed at least 4 weeks post-pIpC, and excision was confirmed by polymerase chain reaction (PCR) of bone marrow at harvest, as described previously.17 For fetal liver harvest, timed matings of Ott1wt/nullSox2-cre males with Ott1flox/flox females were harvested at day E14.5 as described, with embryos with an Ott1 WT allele as controls.19 All experiments and animal handling were performed with UMass Medical School Institutional Animal Care and Use Committee approval.
Vector construction and virus production.
For MSCV-HA-Ott1-IRES-GFP, an HA tag was added by PCR amplifying murine Ott1 cDNA (Origene) using primers HA-Ott1-F/HA-Ott1-R (all primer sequences listed in Table 1). For MSCV-Mpl-FL-IRES-GFP and MSCV-Mpl-TR-IRES-GFP, murine c-Mpl cDNA (Origene) was PCR-amplified using primers Mpl-HA-F/Mpl-rev and Mpl-HA-R/Mpl-HA-R. Products were cloned into pENTR using a pCR8/GW/TOPO kit (Invitrogen) per the manufacturer’s instructions. The constructs were verified by sequencing then cloned into pMSCV-IRES-GFP using the GATEWAY recombination system (Invitrogen). 293T cells were transfected with the MSCV constructs and EcoPak plasmids using Fugene transfection reagent (Roche) per the manufacturer’s protocol. Viral supernatant was collected at 48 hours, and 0.45 µ was filtered postinfection and then stored at −80°C until use.
Primer sequences used
| Primer name . | Sequence 5′-3′ . |
|---|---|
| Mpl5-F | GGAAGCTGTCTCGTCTCAGG |
| Mpl5-R | CTAGTGCGGTCTTGTTGCTG |
| Mpl9-F | CCGAGCTCGCTACAGCTT |
| Mpl9-R | CTGTAGTGCGCAGGAAATTG |
| Mpl-TR-F | GAGGACTGGAAGGAGACTGAGGCA |
| Mpl-TR-R | AGGTTGCAGTCCTCTGTAGTCCAT |
| Gapdh-F | GGAGCCAAACGGGTCATCATCTC |
| Gapdh-R | GAGGGGCCATCCACAGTCTTCT |
| Mpl-chip-1F | TTGGTGAAATGGGGAGAAAG |
| Mpl-chip-1R | CTCAGATCCCTGAACCTGGA |
| Mpl-chip-2F | AGCCAGGGCTACACAGAGAA |
| Mpl-chip-2R | AAGCTGTTTGCTTGGGTTGT |
| Mpl-chip-3F | CAGGGCTGAAGCTGGTCTAC |
| Mpl-chip-3R | AGGAAACAAAAAGCATCTGACC |
| Mpl-chip-4F | CTGGACTCCACTCCACACCT |
| Mpl-chip-4R | ATGGCTCATGCCTGTAATCC |
| Mpl-chip-5F | ATGCTGGGGTAACAGGAGTG |
| Mpl-chip-5R | CTGACCACTCTGCCAAGACA |
| Mpl-chip-6F | GCACTGTGGGACAAGAGTCA |
| Mpl-chip-6R | TCCCAACTCAATCCCAAGTC |
| Mpl-chip-7F | ACAGGGCATGGCAACTTTAG |
| Mpl-chip-7R | GATCACTCCAGATGGCTGCT |
| Ott1-HA-F | GGGAATTCCATGTACCCATACGATGTTCCGGATTACGCTATGAGGTCTGCGGGGCGGGAG |
| Ott1-HA-R | TCATCCGCTGTTCACCAGTTTTG |
| Mpl-HA-F | CCGGAATTCGGGAATTCCATGTACCCATACGATGTTCCGGATTA GCTATGCCCTCTTGGGCCCTCTT ATGGTCACTCCTGCCTC |
| Mpl-HA-R | CGGATTCTTAGAAGGACTTAGGGCTGCAGTGTCTCTGAGGTACTGGCCTAGGACCCGGTGTAGGTCTGGAAGCGAGGGCCACAAAGCATGCCTCAGTCTCCTTCCAGTCCTCACGGCCT |
| Mpl-rev | TCAGGGCTGCTGCCAATAGCTTAG T |
| hMPL-TR-F | GAGACCGCCTGGATCTCCTTGGT |
| hMPL-TR-R | TCAGGGCTGCAGTGTCCCTAAG |
| hMPL-9-F | GCGATCTCGCTACCGTTTAC |
| hMPL-9-R | AGGAAACTGCCACCTCAGC |
| hGAPDH-F | AGGGCTGCTTTTAACTCTGGT |
| hGAPDH-R | CCCCACTTGATTTTGGAGGGA |
| Primer name . | Sequence 5′-3′ . |
|---|---|
| Mpl5-F | GGAAGCTGTCTCGTCTCAGG |
| Mpl5-R | CTAGTGCGGTCTTGTTGCTG |
| Mpl9-F | CCGAGCTCGCTACAGCTT |
| Mpl9-R | CTGTAGTGCGCAGGAAATTG |
| Mpl-TR-F | GAGGACTGGAAGGAGACTGAGGCA |
| Mpl-TR-R | AGGTTGCAGTCCTCTGTAGTCCAT |
| Gapdh-F | GGAGCCAAACGGGTCATCATCTC |
| Gapdh-R | GAGGGGCCATCCACAGTCTTCT |
| Mpl-chip-1F | TTGGTGAAATGGGGAGAAAG |
| Mpl-chip-1R | CTCAGATCCCTGAACCTGGA |
| Mpl-chip-2F | AGCCAGGGCTACACAGAGAA |
| Mpl-chip-2R | AAGCTGTTTGCTTGGGTTGT |
| Mpl-chip-3F | CAGGGCTGAAGCTGGTCTAC |
| Mpl-chip-3R | AGGAAACAAAAAGCATCTGACC |
| Mpl-chip-4F | CTGGACTCCACTCCACACCT |
| Mpl-chip-4R | ATGGCTCATGCCTGTAATCC |
| Mpl-chip-5F | ATGCTGGGGTAACAGGAGTG |
| Mpl-chip-5R | CTGACCACTCTGCCAAGACA |
| Mpl-chip-6F | GCACTGTGGGACAAGAGTCA |
| Mpl-chip-6R | TCCCAACTCAATCCCAAGTC |
| Mpl-chip-7F | ACAGGGCATGGCAACTTTAG |
| Mpl-chip-7R | GATCACTCCAGATGGCTGCT |
| Ott1-HA-F | GGGAATTCCATGTACCCATACGATGTTCCGGATTACGCTATGAGGTCTGCGGGGCGGGAG |
| Ott1-HA-R | TCATCCGCTGTTCACCAGTTTTG |
| Mpl-HA-F | CCGGAATTCGGGAATTCCATGTACCCATACGATGTTCCGGATTA GCTATGCCCTCTTGGGCCCTCTT ATGGTCACTCCTGCCTC |
| Mpl-HA-R | CGGATTCTTAGAAGGACTTAGGGCTGCAGTGTCTCTGAGGTACTGGCCTAGGACCCGGTGTAGGTCTGGAAGCGAGGGCCACAAAGCATGCCTCAGTCTCCTTCCAGTCCTCACGGCCT |
| Mpl-rev | TCAGGGCTGCTGCCAATAGCTTAG T |
| hMPL-TR-F | GAGACCGCCTGGATCTCCTTGGT |
| hMPL-TR-R | TCAGGGCTGCAGTGTCCCTAAG |
| hMPL-9-F | GCGATCTCGCTACCGTTTAC |
| hMPL-9-R | AGGAAACTGCCACCTCAGC |
| hGAPDH-F | AGGGCTGCTTTTAACTCTGGT |
| hGAPDH-R | CCCCACTTGATTTTGGAGGGA |
RNA quantitation
For murine Ott1 KO and control bone marrow populations, adult bone marrow cells were harvested, labeled, and flow-sorted as previously described.10,17 LT-HSC, ST-HSC, and megakaryocytic erythroid progenitor (MEP) populations were identified as Lin–c-Kit+Sca-1+Cd34–, Lin–c-Kit+Sca-1+Cd34+ and Lin–c-Kit+Sca-1– FcγRloCd34lo, respectively. Megakaryocytes were isolated from E14.5 fetal liver cultures grown in Dulbecco’s modified Eagle medium, 10% fetal calf serum, 1% Pen/Strep (Invitrogen), and 50 ng/mL mThpo (Peprotech) for 4 days followed by separation on a bovine serum albumin gradient as described.19 RNA was isolated using RNAeasy micro columns (Qiagen) and cDNA generated with either a Quantitect Reverse Transcriptase kit or Sensiscript RT kit (Qiagen). Quantitative PCR (qPCR) was performed using the indicated primers and the Quantifast SYBR green (Qiagen) on a Mastercycler Realplex (Eppendorf). Total c-Mpl RNA was determined using primers spanning the exons 5 and 6 junction (Mpl5-F/Mpl5-R) and the ratio of Mpl-FL using primers spanning the exon 9 and 10 junction (Mpl9-F/Mpl9-R). Mpl-TR amounts were confirmed using (Mpl-TR-F/Mpl-TR-R) overlapping the exon 8/11 junction. A Gapdh control (Gapdh-F/Gapdh-R) was used and expression was calculated using the ΔΔCT method.
For human cell populations, deidentified, pathologically confirmed normal human bone marrow samples were obtained through the UMass Medical School sample bank. Samples were RBC lysed and positively selected using mouse anti-human CD34 (BD Pharmingen) bound to anti-mouse IgG Magnabeads (Thermo Scientific) per the manufacturer’s protocol. The CD34-depleted supernatant was then positively selected using mouse anti-CD41(eBiosciences) bound to anti-IgG Magnabeads. Total RNA was isolated using the RNeasy micro plus kit (Qiagen) and cDNA generated using Sensiscript RT (Qiagen) according to the manufacturer’s instructions. MPL isoforms were quantified as described before using primers hMPL-TR-F/hMPL-TR-R, hMPL-9-F/hMPL-9-R, and hGAPDH-F/hGAPDH-R to amplify MPL-TR, MPL-FL, and GAPDH, respectively.
Western blotting.
Ott1 KO and control E14.5 fetal liver cells were harvested and cultured in 50 ng/mL mThpo as described before. 293T cells were transfected with the indicated construct using Fugene (Roche) according to the manufacturer’s instructions. Cells were collected and lysed in RIPA buffer (Santa Cruz Biotech) with Complete Protease inhibitor cocktail (Roche). Protein concentrations were determined using a BCA kit (Pierce), and equivalent concentrations were loaded onto a 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis gel followed by transfer to polyvinylidene fluoride membrane. Western blotting was performed using anti-Mpl, anti-Gapdh (Sigma), or anti-HA (Thermo Scientific), and then with either anti-Mouse-HRP(Sigma) or anti-Rabbit-HRP (Thermo Scientific).
Flow cytometry.
Lineage, c-Kit, Sca-1, and Cd34 was performed as previously described and run on a BD Biosciences LSR II flow cytometer then analyzed using FlowJo software (Treestar).10 For phosphoflow analysis, cells were surface-stained, treated with growth factor, and then fixed and stained using Phosflow Lyse/Fix and Phosflow Perm buffer (BD Bioscience) according to the manufacturer’s protocol with anti-Stat5-pY694-PerCP-Cy5.5 or control antibody (BD Biosciences).
Bone marrow/fetal liver transduction.
Retroviral transduction of WT bone marrow with MSCV-Mpl-TR IRES-GFP, MSCV-Mpl-FL IRES-GFP, and MSCV IRES-GFP and transplantation into CD45.1+ recipients was performed as described.20 Facial vein blood samples were analyzed by flow cytometry for GFP+ populations and normalized to GFP+ donor input. Ott1 KO and control E14.5 fetal livers were harvested, spin-fected as described20 with either MSCV-Mpl-FL IRES-GFP or MSCV IRES-GFP, and then grown for 48 hours in RPMI, 10% fetal calf serum, and 6 ng/mL IL3, 20 ng/mL IL6, and 10 ng/mL stem cell factor (Peprotech) before analysis.
RNA and chromatin immunoprecipitation.
For RNA immunoprecipitation, 1.5 × 107 NIH3T3 cells infected with MSCV HA-Ott1 IRES-GFP or GFP only control were used for the RNA ChIP-IT kit (Active Motif) and precipitated with anti-HA (Abcam) according to the manufacturer’s instructions. Output and input were used to generate cDNA and amplified using primers Mpl5-F/Mpl5-R as described before. For ChIP experiments, the aforementioned HA-Ott1–expressing NIH3T3 cells and controls were used with the ChIP-IT kit (Active Motif) per protocol, precipitated with anti-HA (Abcam) and output/input used for qPCR with primers Mpl-chip-(1-7)F/Mpl-chip(1-7)R. For ChIP using anti-H3K4me3 (Abcam) and anti–H4-pan-Acetyl (Active Motif), lineage-depleted adult bone marrow from Ott1 KO or control mice were used followed by qPCR of input/output with primers Mpl-chip-7F/Mpl-chip-7R.
Statistical analysis
Statistical analysis was performed using a 2-sided Student t test. Error bars equal ± 1 standard deviation.
Results
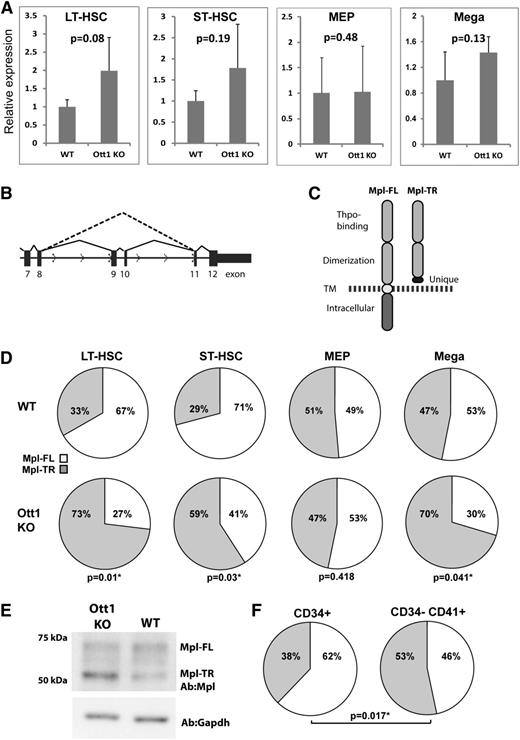
Ott1 regulates the ratio between full-length and truncated c-Mpl isoforms
Given the shared requirement for Ott1 and c-Mpl in HSCs during stress hematopoiesis, we used qPCR to measure mRNA expression of c-Mpl in Lineage–Sca-1+c-Kit+Cd34– (LT-HSC–enriched) and Lineage–Sca-1+c-Kit+Cd34+ (ST-HSC–enriched) sorted populations from Ott1-deleted (Ott1 KO) adult mice compared with WT controls to determine whether Ott1 regulated c-Mpl expression (Figure 1A). qPCR primers were directed to a sequence spanning exons 5 and 6. A small but nonsignificant increase in c-Mpl expression was observed in the Ott1 KO, which is in agreement with our previously generated Affymetrix microarray data.10 Sorted MEPs from adult Ott1 KO mice also showed no significant difference from WT controls. Purified primary cultured Ott1 KO fetal liver megakaryocytes generated using Ott1flox/null Sox2-Cre mice also had a slightly higher expression than WT controls but did not rise to the level of significance.
Ott1 regulates the ratio between full-length and truncated c-Mpl isoforms. (A) Relative c-Mpl expression determined through qPCR of total c-Mpl RNA between wild-type (WT) and Ott1 KO cells. Total RNA isolated from flow-sorted LT-HSC (Lin–c-Kit+Sca1+Cd34–), ST-HSC (Lin–c-Kit+Sca1+Cd34+) (WT, n = 4; Ott1 KO, n = 3), and Lin–c-Kit+Sca1–CD34–FcγR– (MEP, n = 4) from pIpC-treated Ott1 WT or Ott1flox/nullMx1-cre (Ott1 KO) adult bone marrow and isolated megakaryocytes (n = 3) from Ott1 WT or Ott1flox/nullSox2-cre E14.5 fetal liver cultures. Error bars represent 1 standard deviation. (B) Schematic of c-Mpl pre-mRNA splicing for Mpl-FL (solid line) and Mpl-TR (dotted line). (C) Diagram of predicted proteins generated from Mpl-FL and Mpl-TR isoforms. TM, transmembrane domain; dotted line, plasma membrane. (D) Ratio of Mpl-TR to Mpl-FL isoforms from qPCR of total RNA isolated in (A). (E) Western blot of lysates from E14.5 fetal liver megakaryocyte cultures of Ott1 WT and Ott1 KO (Ott1flox/nullSox2-cre) probed with anti-c-Mpl and anti-GAPDH. Performed in triplicate. (F) Ratio of MPL-TR (gray) to MPL-FL (white) in HSC-enriched (CD34+) and megakaryocyte-enriched (CD34–CD41+) human bone marrow populations (n = 5).
Ott1 regulates the ratio between full-length and truncated c-Mpl isoforms. (A) Relative c-Mpl expression determined through qPCR of total c-Mpl RNA between wild-type (WT) and Ott1 KO cells. Total RNA isolated from flow-sorted LT-HSC (Lin–c-Kit+Sca1+Cd34–), ST-HSC (Lin–c-Kit+Sca1+Cd34+) (WT, n = 4; Ott1 KO, n = 3), and Lin–c-Kit+Sca1–CD34–FcγR– (MEP, n = 4) from pIpC-treated Ott1 WT or Ott1flox/nullMx1-cre (Ott1 KO) adult bone marrow and isolated megakaryocytes (n = 3) from Ott1 WT or Ott1flox/nullSox2-cre E14.5 fetal liver cultures. Error bars represent 1 standard deviation. (B) Schematic of c-Mpl pre-mRNA splicing for Mpl-FL (solid line) and Mpl-TR (dotted line). (C) Diagram of predicted proteins generated from Mpl-FL and Mpl-TR isoforms. TM, transmembrane domain; dotted line, plasma membrane. (D) Ratio of Mpl-TR to Mpl-FL isoforms from qPCR of total RNA isolated in (A). (E) Western blot of lysates from E14.5 fetal liver megakaryocyte cultures of Ott1 WT and Ott1 KO (Ott1flox/nullSox2-cre) probed with anti-c-Mpl and anti-GAPDH. Performed in triplicate. (F) Ratio of MPL-TR (gray) to MPL-FL (white) in HSC-enriched (CD34+) and megakaryocyte-enriched (CD34–CD41+) human bone marrow populations (n = 5).
In addition to Mpl-FL, c-Mpl has 2 alternatively spliced isoforms in the mouse: Mpl-II, which affects exon 4, and Mpl-TR (or MPL-S), which excludes exons 9 and 10 and is the only shared isoform with humans (Figure 1B).21,22 Mpl-TR codes for a truncated protein possessing the Thpo-binding extracellular domain and a unique 30-amino-acid sequence resulting from an out-of-frame junction with exon 11. Mpl-TR lacks the transmembrane and C-terminal intracellular signaling domains but is not secreted (Figure 1C).22 Exogenous expression of Mpl-TR in cell lines shows a dominant-negative effect on c-Mpl/Thpo signaling.22 Because of the incorporation of Ott1 in the spliceosome complex, we hypothesized Ott1 might affect c-Mpl signaling through regulation of c-Mpl alternative splicing.
Using qPCR primer sets to measure the ratio of Mpl-TR to Mpl-FL isoforms, Ott1 KO LT-HSC and ST-HSC cells showed a significant increase in the proportion of Mpl-TR compared with controls (Figure 1D). A similar increase was observed in Ott1 KO fetal liver megakaryocytes but not in MEPs, suggesting the effect of Ott1 loss on c-Mpl alternative splicing is cell context–dependent. No significant amount of Mpl-II was detected (data not shown). Western blot analysis of control and Ott1 KO fetal liver megakaryocyte cultures using an antibody directed toward the N-terminus of c-Mpl detected both Mpl-FL and a band of the predicted molecular weight of 51 kDa for Mpl-TR, which was increased in the Ott1 KO (Figure 1E).
To examine the ratio of MPL-TR and MPL-FL in humans, CD34+ and CD34–CD41+ cells representing HSCs and megakaryocyte/megakaryocyte progenitor populations were isolated from normal bone marrow using antibody-bound magnetic beads (Figure 1F). qPCR using specific primers to the MPL-TR and MPL-FL isoforms demonstrated a significantly higher ratio of MPL-TR:MPL-FL in megakaryocyte populations compared with HSCs. The ratios of MPL-TR:MPL-FL in human HSCs and megakaryocytes are similar to corresponding murine cell populations.
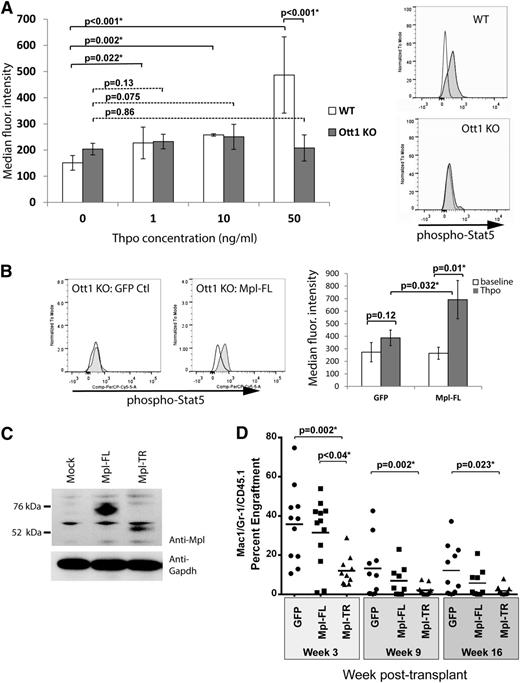
Thpo response in HSCs is Ott1-dependent
Expression of Mpl-TR was observed to interfere with Thpo response when coexpressed with Mpl-FL in cell lines.22 To determine whether loss of Ott1, which increases Mpl-TR expression, affects Thpo signaling, freshly harvested E14.5 Ott1flox/nullSox2-cre (Ott1 KO) and control with a WT Ott1 allele were surface-stained for LT-HSC markers (LSK Cd34−), serum-starved for 1 hour, and then incubated for 5 minutes with different concentrations of Thpo followed by fixation and intracellular staining for phospho-Stat5 (Figure 2A). Normally, Stat5 is rapidly phosphorylated after Thpo binding to c-Mpl through Jak2 activation. HSCs lacking Ott1 have impaired Stat5 phosphorylation over baseline at both low and high Thpo concentrations compared with WT, consistent with loss of c-Mpl signaling. Although baseline Stat5 phosphorylation appears to be slightly higher in the Ott1 KO, the reduction in Thpo response is most prominent at high concentrations. Similar results were obtained using pIpC-treated Ott1flox/nullMx1-cre adult bone marrow (data not shown). To exclude the possibility that Ott1 affects the integrity of Stat5 signaling, we examined whether Mpl-FL overexpression rescued Stat5 response to Thpo. Ott1 KO fetal liver cells were transduced with retrovirus expressing Mpl-FL-IRES-GFP (Figure 2C) or an IRES-GFP control virus. Cells were serum-starved and 50 ng/mL Thpo was added as described and stained for phospho-Stat5/LSKCd34–. Ott1 KO cells transduced with Mpl-FL regained phospho-Stat5 response to Thpo in contrast to GFP controls (Figure 2B). Therefore, downstream signaling pathways from c-Mpl to Stat5 are intact in Ott1-deleted cells, and loss of Thpo response likely stems from interference with Mpl function itself.
C-Mpl response is reduced by Ott1 loss, and Mpl-TR expression impairs HSC engraftment. (A) Phospho-Stat5 levels in HSCs post-Thpo stimulation. E14.5 fetal liver from Ott1flox/null Sox2-cre (Ott1 KO) or WT Ott1 controls at baseline (white) or 5 minutes poststimulation with 0 to 50 ng of Thpo (gray) and analyzed through flow cytometry using anti-phospho-Stat5 and surface markers for LT-HSC (LSK CD34–) (0, 50 ng/mL n = 6 Ott1 KO; n = 7 WT; 1, 10 ng/mL n = 3; error bars ± SD). Representative histograms of flow data at right showing baseline (white) and post 50 ng/mL Thpo stimulation (gray). (B) Phospho-Stat5 in LSKCD34– GFP+ HSCs pre- and post-Thpo stimulation. Median fluorescence intensity of phospho-Stat5 staining in Ott1 KO (Ott1flox/nullSox2-cre) fetal liver cells expressing either MSCV-IRES-GFP (GFP Ctl) or MSCV-Mpl-FL-IRES-GFP (Mpl-FL) retroviruses at baseline (white) or stimulated with 50 ng/mL Thpo for 5 minutes (gray). Representative histograms (left) and graph of median fluorescence intensity for experiments (n = 3, right). (C) Expression of Mpl constructs. Western blots of 293T cell lysates transfected with no vector, MSCV-HA-Mpl-FL-IRES-GFP, and MSCV-Mpl-TR-IRES-GFP using anti-Mpl antibody or anti-Gapdh antibody. (D) Engraftment of bone marrow overexpressing c-Mpl isoforms. Wild-type bone marrow infected with control GFP (n = 11), Mpl-FL-IRES- GFP (n = 12), and Mpl-TR-IRES-GFP (n = 8) expressing retroviruses was transplanted into irradiated recipients. Serial peripheral blood sampling underwent flow cytometric evaluation for GFP+/Mac1+/Gr1+ percentage of graft normalized to initial GFP+ input (bar = mean).
C-Mpl response is reduced by Ott1 loss, and Mpl-TR expression impairs HSC engraftment. (A) Phospho-Stat5 levels in HSCs post-Thpo stimulation. E14.5 fetal liver from Ott1flox/null Sox2-cre (Ott1 KO) or WT Ott1 controls at baseline (white) or 5 minutes poststimulation with 0 to 50 ng of Thpo (gray) and analyzed through flow cytometry using anti-phospho-Stat5 and surface markers for LT-HSC (LSK CD34–) (0, 50 ng/mL n = 6 Ott1 KO; n = 7 WT; 1, 10 ng/mL n = 3; error bars ± SD). Representative histograms of flow data at right showing baseline (white) and post 50 ng/mL Thpo stimulation (gray). (B) Phospho-Stat5 in LSKCD34– GFP+ HSCs pre- and post-Thpo stimulation. Median fluorescence intensity of phospho-Stat5 staining in Ott1 KO (Ott1flox/nullSox2-cre) fetal liver cells expressing either MSCV-IRES-GFP (GFP Ctl) or MSCV-Mpl-FL-IRES-GFP (Mpl-FL) retroviruses at baseline (white) or stimulated with 50 ng/mL Thpo for 5 minutes (gray). Representative histograms (left) and graph of median fluorescence intensity for experiments (n = 3, right). (C) Expression of Mpl constructs. Western blots of 293T cell lysates transfected with no vector, MSCV-HA-Mpl-FL-IRES-GFP, and MSCV-Mpl-TR-IRES-GFP using anti-Mpl antibody or anti-Gapdh antibody. (D) Engraftment of bone marrow overexpressing c-Mpl isoforms. Wild-type bone marrow infected with control GFP (n = 11), Mpl-FL-IRES- GFP (n = 12), and Mpl-TR-IRES-GFP (n = 8) expressing retroviruses was transplanted into irradiated recipients. Serial peripheral blood sampling underwent flow cytometric evaluation for GFP+/Mac1+/Gr1+ percentage of graft normalized to initial GFP+ input (bar = mean).
Mpl-TR expression impairs HSC engraftment
To test whether Mpl-TR affects HSC function, WT bone marrow was transduced with retroviruses expressing Mpl-TR, Mpl-FL, or an empty control vector (Figure 2C-D), along with an IRES-GFP cassette, and used to repopulate lethally irradiated recipients. Exogenous Mpl-FL has been previously shown to decrease long-term engraftment but does not impair short-term engraftment.23 Mice were monitored through peripheral blood sampling through 16 weeks to measure the percent GFP positive cells in donor granulocytes (Figure 2D). Mpl-TR–expressing cells had both markedly decreased short- and long-term engraftment compared with Mpl-FL–expressing cells and GFP controls. These data show that increased expression of Mpl-TR impairs HSC function.
To determine whether loss of c-Mpl signaling in Ott1-deleted HSCs was the primary defect, a rescue experiment was performed. Bone marrow from Ott1-deficient mice was transduced with either Mpl-FL-IRES-GFP or IRES-GFP control retrovirus, resulting in ∼5% GFP+ donor marrow in each arm. We were unable to use 5-FU-pretreatment to increase transduction efficiency because of the established toxicity on Ott1 KO bone marrow.10 106 total cells/mouse were injected into lethally irradiated recipients along with 2 × 105 CD45.1+/CD45.2+ WT helper marrow (n = 4). Less than 0.1% GFP+ engraftment was observed in each arm at 4 weeks through analysis of Mac1+Gr1+ peripheral blood samples, with no significant difference between the Mpl-FL and GFP controls. Similar experiments transducing Ott1-IRES-GFP into Ott1 KO cells are able to rescue engraftment (our unpublished results). Therefore, despite the ability of Mpl-FL to rescue Thpo signaling in Ott1 KO HSCs, it is insufficient to rescue the engraftment defect by itself.
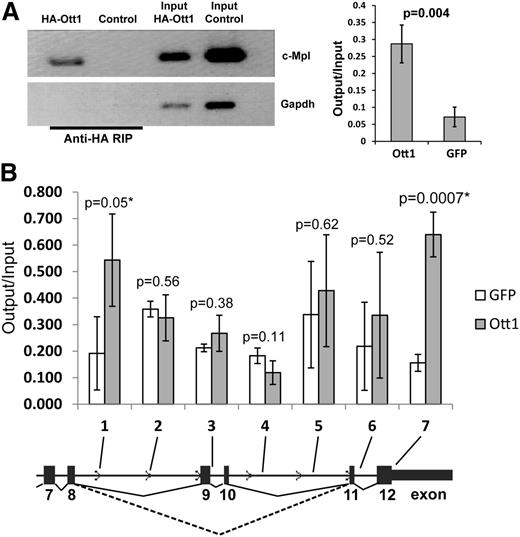
Ott1 interacts with c-Mpl RNA and chromatin
In light of Ott1 containing RRMs and its inclusion in the spliceosome, we tested whether Ott1 might bind either directly or in complex to c-Mpl pre-mRNA. NIH3T3 cells, which express low levels of endogenous c-Mpl (data not shown), were infected with a retrovirus expressing an HA-tagged Ott1 protein or a control GFP vector. RNA-immunoprecipitation (RIP) was performed from these cells using anti-HA antibody (Figure 3A). Conventional and qPCR analysis using primers spanning c-Mpl exon 5/6 detected significant enrichment with HA-Ott1 compared with control, indicating that Ott1 can complex with c-Mpl RNA.
Ott1 binds c-Mpl RNA and chromatin. (A) RNA immunoprecipitation (RIP) from HA-Ott1 or GFP control–infected NIH3T3 cells using anti-HA antibody. RNA immunoprecipitates and input lysate RNA were reverse transcription-PCR (RT-PCR)–amplified for exon 5/6 of c-Mpl and Gapdh controls. Agarose gel of RT-PCR products from RIP and input (left panel). Ratio of output:input of qPCR of RIP and input lysate cDNA (n = 3; error bars ± SD). (B) Chromatin immunoprecipitation (ChIP) from HA-Ott1 (gray columns) or GFP control (white)-infected NIH3T3 cells using anti-HA antibody. Ratio of output:input from HA-Ott1–expressing cells compared with GFP-expressing controls using qPCR, with primers directed toward the indicated regions (n = 3; error bars, SD; dotted line, Mpl-TR splice; solid line, Mpl-FL splice).
Ott1 binds c-Mpl RNA and chromatin. (A) RNA immunoprecipitation (RIP) from HA-Ott1 or GFP control–infected NIH3T3 cells using anti-HA antibody. RNA immunoprecipitates and input lysate RNA were reverse transcription-PCR (RT-PCR)–amplified for exon 5/6 of c-Mpl and Gapdh controls. Agarose gel of RT-PCR products from RIP and input (left panel). Ratio of output:input of qPCR of RIP and input lysate cDNA (n = 3; error bars ± SD). (B) Chromatin immunoprecipitation (ChIP) from HA-Ott1 (gray columns) or GFP control (white)-infected NIH3T3 cells using anti-HA antibody. Ratio of output:input from HA-Ott1–expressing cells compared with GFP-expressing controls using qPCR, with primers directed toward the indicated regions (n = 3; error bars, SD; dotted line, Mpl-TR splice; solid line, Mpl-FL splice).
Alternative splicing is postulated to occur primarily through a cotranscriptional mechanism, in which case, Ott1 would also be predicted to bind to c-Mpl chromatin as well as RNA.24 Using HA-Ott1–expressing NIH3T3 cells, chromatin immunoprecipitation (ChIP) was performed to analyze for enrichment of HA-Ott1 binding to the alternatively spliced region of c-Mpl (Figure 3B). Sequential qPCR primer sets were used and identified regions approximately flanking the region of exon skipping in the Mpl-TR isoform. Ott1 binding to both c-Mpl RNA and chromatin supports a potential cotranscriptional model of Ott1 regulating c-Mpl alternative splicing.
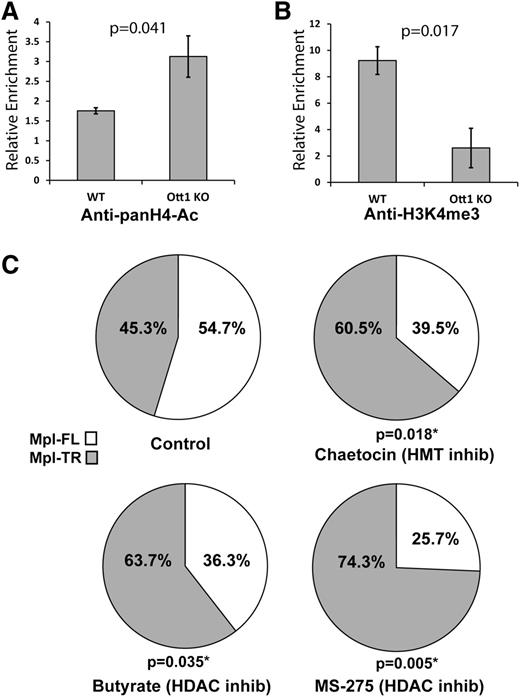
c-Mpl undergoes Ott1-dependent epigenetic modifications
Mechanisms for the regulation of cotranscriptional alternative splicing include histone modifications including H4-acetyl and H3K4me3 through control of transcriptional speed and spliceosome recruitment, respectively.24 The Ott1 SPOC domain binds epigenetic modifiers Hdac3 and Setd1b; therefore, the region of c-Mpl binding Ott1 was examined to determine whether histone modifications at the site were Ott1-dependent. Lin– Ott1-deleted and control bone marrow were used in a ChIP assay with anti–pan-acetyl H4 and tested for enrichment using c-Mpl qPCR primer sets (Figure 4A). In the absence of Ott1, H4 acetylation increased, consistent with the loss of Ott1-associated Hdac activity. ChIP using anti-H3K4me3 in Lin–Ott1-deleted bone marrow demonstrated a marked decrease in H3K4me3 marks on the region of c-Mpl binding to Ott1, correlating with the loss of Ott1-associated Setd1b HMT function (Figure 4B).
Histone modifications on c-Mpl are Ott1-dependent and inhibitors manipulate alternative splicing. (A) H4-Acetylation of WT and Ott1 KO bone marrow. ChIP using anti-pan-acetyl-H4 on pIpC-induced Ott1 WT or Ott1flox/nullMx1-cre (Ott1 KO) adult lineage–depleted bone marrow and then quantified using qPCR to determine relative enrichment. (B) H3K4me3 of WT and Ott1 KO bone marrow. ChIP using anti-pan-H3K4me3 on pIpC-induced Ott1 WT or Ott1flox/nullMx1-cre (Ott1 KO) adult lineage–depleted bone marrow and then quantified using qPCR to determine relative enrichment (n = 3; error bars, ± SD). (C) Ratio of Mpl-TR:Mpl-FL after Hdac and/or HMT inhibitor treatment. Thpo-cultured WT fetal livers were incubated for 24 hours with control (n = 6), 100 nm chaetocin (n = 6), 5 mM sodium butyrate (n = 7), or 5 µM MS-275 (n = 4). qPCR was performed on total RNA from isolated megakaryocytes to determine Mpl-TR:Mpl-FL isoform ratio.
Histone modifications on c-Mpl are Ott1-dependent and inhibitors manipulate alternative splicing. (A) H4-Acetylation of WT and Ott1 KO bone marrow. ChIP using anti-pan-acetyl-H4 on pIpC-induced Ott1 WT or Ott1flox/nullMx1-cre (Ott1 KO) adult lineage–depleted bone marrow and then quantified using qPCR to determine relative enrichment. (B) H3K4me3 of WT and Ott1 KO bone marrow. ChIP using anti-pan-H3K4me3 on pIpC-induced Ott1 WT or Ott1flox/nullMx1-cre (Ott1 KO) adult lineage–depleted bone marrow and then quantified using qPCR to determine relative enrichment (n = 3; error bars, ± SD). (C) Ratio of Mpl-TR:Mpl-FL after Hdac and/or HMT inhibitor treatment. Thpo-cultured WT fetal livers were incubated for 24 hours with control (n = 6), 100 nm chaetocin (n = 6), 5 mM sodium butyrate (n = 7), or 5 µM MS-275 (n = 4). qPCR was performed on total RNA from isolated megakaryocytes to determine Mpl-TR:Mpl-FL isoform ratio.
If the Ott1-dependent epigenetic modifications on the c-Mpl locus are physiologically important to alternative splice choice, pharmacologic inhibition of associated chromatin modifiers, Hdac3 and Setd1b, would be predicted to have a similar effect on the ratio of Mpl-TR:Mpl-FL splicing ratio as Ott1 loss. WT fetal liver megakaryocytes were incubated for 24 hours with 5 mM of sodium butyrate, a primarily Class I Hdac inhibitor, and then harvested for total RNA extraction and qPCR analysis of the Mpl-TR:Mpl-FL isoform ratio (Figure 4C).25 The ratio of Mpl-TR significantly increased after sodium butyrate treatment. Treatment with 5 µM MS-275 (Selleck), an Hdac inhibitor specific for Hdac1 and Hdac3, resulted in an even greater increase in the Mpl-TR ratio. Similarly, treatment with 100 nM of chaetocin, an inhibitor with activity against Setd1b class HMTs, also resulted in a significant increase in the ratio of Mpl-TR:Mpl-FL isoforms.26
Discussion
Thpo/c-Mpl signaling plays critical roles in HSC and megakaryocyte function; however, little is known about how different c-Mpl isoforms contribute to regulation. Four major isoforms have been reported in humans; MPL-P (MPL-FL), the full length transcript; MPL-K; MPL-D; and MPL-TR (or MPL-S).1 MPL-K results from termination within an unspliced intron 10 and codes for a shortened intracellular domain but has not been demonstrated to affect Thpo signaling.7 MPL-D has a 72-bp inframe deletion from use of an alternate splice acceptor within exon 9, which deletes a portion of the extracellular domain and has reduced or absent Thpo response.27,28 The MPL-TR isoform, generated by exclusion of exons 9 and 10, is the sole isoform shared by humans and mice, and interspecies conservation of isoforms often predicts physiologic importance.22 Mpl-FL is insufficient to rescue Mpl−/− HSC engraftment when expressed by a transgene using the c-Mpl promoter, thus supporting a necessary physiologic role for alternatively spliced Mpl isoforms.29 In addition, we observed that human HSC and megakaryocyte populations have similar ratios of MPL-TR:MPL-FL as murine populations, supporting an evolutionarily conserved process, although the presence of additional MPL isoforms in humans may signify further complexity.
Thpo-mediated Stat5 activation is Ott1-dependent at low and high concentrations of Thpo. In vitro coexpression of Mpl-TR with Mpl-FL was previously shown to increase degradation of Mpl-FL.22 We did not observe reduced Mpl-FL protein levels with higher ratios of Mpl-TR, yet Mpl signaling was still decreased, suggesting degradation may not be the primary dominant negative pathway in vivo. Modulation of growth-factor signaling through alternative splicing has been demonstrated as a physiologic mechanism in multiple cytokine receptors such as granulocyte macrophage colony stimulating factor (GM-CSF) recetor, IL6R, and vascular endothelial growth factor receptor (VEGFR).30-32 In the case of the IL6R and VEGFR, alternative splicing yields a soluble receptor capable of sequestering growth factor extracellularly; however, Mpl-TR has been shown not to be secreted, and our Thpo-stimulation experiments were not performed in conditioned media.22 Potentially, Mpl-TR:Mpl-FL interaction could occur intracellularly. For example, an alternatively spliced isoform of the GM-CSF–receptor β chain (CSF2RB), βGMRIT, which has a truncation of the intracellular domain, reduces proliferative response to GM-CSF stimulation.30
Consistent with a dominant negative effect on c-Mpl function, overexpression of Mpl-FL is able to rescue Stat5 activation in response to Thpo in Ott1 KO HSCs. Mpl-TR has a significant in vivo effect on HSC function because its expression impairs both long- and short-term HSC engraftment. This effect is similar to the c-Mpl−/− phenotype, where HSC quiescence and proliferation are altered under stress conditions.5 Although Mpl-FL overexpression has been shown to reduce long-term engraftment in WT cells, short-term engraftment is not reduced, as was observed with Mpl-TR.23 These data provide evidence that reduced Ott1-dependent Thpo/c-Mpl signaling caused by increased Mpl-TR expression contributes to the engraftment defect in Ott1-deficient HSCs; however, the failure of Mpl-FL to rescue engraftment suggests Mpl-independent functions of Ott1 may be required. Given that Ott1 is essential for multiple non–Mpl-expressing tissues, it would not be unexpected for additional HSC-essential genes to be affected.17 Nevertheless, an increase in Mpl-TR expression would be sufficient to confer defects in long- and short-term HSC engraftment to Ott1 KO-deleted cells.
Splicing has been identified as a cotranscriptional process, in which the spliceosome complex functions in conjunction with RNA Pol II transcription.33 In the kinetic model, RNA Pol II processivity determines exon inclusion, whereby slower transcriptional speeds or pausing allows spliceosome assembly on both strong and weak splice acceptors in contrast to rapid transcription, which favors strong splice acceptors.24 Acetylated histones keep chromatin in an open conformation and are associated with faster transcription. Inversely, deacetylated histones yield a slower transcription rate. H4 acetylation on c-Mpl is elevated in the absence of Ott1, potentially because of loss of associated Hdac3 activity, which predicts faster transcription and exon skipping. Consistent with this observation, Hdac inhibition is also sufficient to promote exon skipping of c-Mpl exons 9 and 10. Taken together, these data support histone acetylation as a possible contributory mechanism for Ott1-dependent alternative splicing.
An adaptor model of splicing uses histone lysine methylation marks to recruit the spliceosome to exons destined for inclusion. Although histone marks may be associated with increased transcription in promoter regions, they can affect splicing when located intragenically. Increased H3K36me3, H2K27me2, and H3K4me3 have all been associated with exon inclusion.24 Setd1b,which preferentially catalyzes H3K4me3 marks, binds to the SPOC domain of Ott1.13 H3K4me3 enrichment on c-Mpl is highly dependent on Ott1, potentially affecting spliceosomal recruitment. Finally, HMT inhibition with chaetocin reproduced the increased ratio of Mpl-TR. Because both Hdac inhibitors and chaetocin affected splicing of the Mpl-TR isoform, an integrated model dependent on both processes may be possible.24 The regulation of alternative splicing by Ott1 is a novel function for a Spen-family member. Although other RRM-containing proteins such as Rbm5, Rbm6, Rbm10, and Rbm20 have been associated with alternative splicing, only Ott1 possesses a SPOC domain and the associated epigenetic modifiers, Hdac3 and Setd1b.34,35 Family members Spen (or Mint/SHARP) and Ott3 (or Rbm15b), which share RRM and SPOC homology, may potentially use a similar mechanism.
Ott1-dependent splicing of c-Mpl isoforms adds an additional layer of regulation to Thpo response in HSCs and may explain how HSCs could have different responses to similar concentrations of Thpo in vivo, thus determining quiescence or proliferation. Alternative explanations for the function of Ott1 on the Mpl-TR:Mpl-FL ratio independent of a direct involvement in splicing include effects on mRNA stability or export as well as indirect regulation. In contrast to HSCs, Ott1-deficient MEPs do not have an alteration in the Mpl-TR:Mpl-FL ratio. Megakaryocyte proliferation is Thpo-dependent only during early development and may explain why Ott1 KO CFU-Meg numbers are normal.36 Ott1-deleted megakaryocytes, however, have reduced ploidy, which may reflect changes in Thpo-dependent terminal megakaryocyte development.37
An understanding of the upstream regulation of Ott1 would identify additional contributors to Thpo response and HSC function. Ott1 binds Rbpjκ, which is a key effector of the Notch pathway.11 Ott1 is an estrogen-responsive gene, and estrogen has been found to increase HSC self-renewal.38,39 Manipulating the Thpo/c-Mpl axis has shown therapeutic promise in HSC function as well as for thrombocytopenia through the use of THPO or c-MPL agonists to expand HSC populations ex vivo and to treat aplastic anemia.40,41 The ability to enhance or attenuate THPO-mediated effects to tailor HSC or megakaryocyte function would be a valuable adjunct. Clinical use of HDAC inhibitors is confounded by severe thrombocytopenia and leukopenia.42 Although thrombocytopenia may be partially explained by inhibitory effects on megakaryocyte ρ-family proteins, HDAC-induced leukopenia has an unknown etiology, and shifts in c-MPL isoforms may provide an explanation.43 In summary, production of the dominant-negative isoform Mpl-TR is dependent on Ott1, which negatively regulates HSC Thpo response and engraftment, potentially through alteration of histone acetylation and methylation within the c-Mpl locus.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Melissa Moore for valuable advice and reviewing the manuscript, Amit Sinha for advice on data analysis, Jing Chen and Wei Tong for supplying reagents, and Xinyang Zhao for sharing unpublished data and helpful discussions.
This work was supported in part by a grant from the National Institutes of Health, National Heart, Lung and Blood Institute (R01 HL116392).
Authorship
Contribution: N.X., S.L., and G.D.R. designed and performed experiments, analyzed data, and wrote the manuscript; S.P.D. and K.M. performed experiments and analyzed data; and J.L.J. analyzed data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Glen D. Raffel, Division of Hematology-Oncology, University of Massachusetts Medical School, 364 Plantation St LRB 319, Worcester, MA 01605; e-mail: glen.raffel@umassmed.edu.
References
Author notes
N.X. and S.L. contributed equally to this study.