Key Points
A single α(2,3) sialyltransferase, ST3Gal-4, controls sLeX biosynthesis on N- and O-glycans in cells of human myeloid lineage.
Blocking this enzyme activity prevents human neutrophil adhesion to E-, P-, and L-selectin.
Abstract
The precise glycosyltransferase enzymes that mediate selectin-ligand biosynthesis in human leukocytes are unknown. This knowledge is important because selectin-mediated cell tethering and rolling is a critical component of both normal immune response and various vascular disorders. We evaluated the role of 3 α(2,3)sialyltransferases, ST3Gal-3, -4, and -6, which act on the type II N-Acetyllactosamine structure (Galβ1,4GlcNAc) to create sialyl Lewis-X (sLeX) and related sialofucosylated glycans on human leukocytes of myeloid lineage. These genes were either silenced using lentiviral short hairpin RNA (shRNA) or functionally ablated using the clustered regularly interspaced short palindromic repeat/Cas9 technology. The results show that ST3Gal-4, but not ST3Gal-3 or -6, is the major sialyltransferase regulating the biosynthesis of E-, P-, and L-selectin ligands in humans. Reduction in ST3Gal-4 activity lowered cell-surface HECA-452 epitope expression by 75% to 95%. Glycomics profiling of knockouts demonstrate an almost complete loss of the sLeX epitope on both leukocyte N- and O-glycans. In cell-adhesion studies, ST3Gal-4 knockdown/knockout cells displayed 90% to 100% reduction in tethering and rolling density on all selectins. ST3Gal-4 silencing in neutrophils derived from human CD34+ hematopoietic stem cells also resulted in 80% to 90% reduction in cell adhesion to all selectins. Overall, a single sialyltransferase regulates selectin-ligand biosynthesis in human leukocytes, unlike mice where multiple enzymes contribute to this function.
Introduction
P- (CD62P), E- (CD62E), and L-selectin (CD62L) are C-type lectins that specialize in the capture of leukocytes from flowing blood onto the inflamed vascular endothelium.1 The ligands of this family of adhesion molecules, expressed on the leukocyte cell surface, are carbohydrates posttranslationally synthesized by the sequential action of various enzymes of the glycosyltransferase (glycoT) family. The binding of selectins to ligands under shear is characterized by high on and off rates.2 This results both in the frequent capture of leukocytes from flowing blood and their rolling interactions. Cell adhesion via selectins also results in signaling that may contribute to integrin activation and the transition of rolling cells to firm arrest.3,4 The sialofucosylated glycans that bind all 3 selectins include the tetrasaccharide sialyl Lewis-X (sLeX) and related structures.5,6 Although many cell-surface glycoconjugates express such epitopes, functional selectin ligands are only expressed on specific glycoproteins containing O-/N-linked glycans or glycosphingolipids (GSLs).
Whereas much of our current knowledge of the glycoTs that contribute to selectin-ligand biosynthesis comes from knockout mice, accumulating evidence suggests that the function of these enzymes and also the scaffolds bearing the selectin ligands are different between humans and mice.4,7 This is particularly relevant for the E-selectin ligands because P-selectin glycoprotein ligand-1 (PSGL-1, CD162) is the major ligand for L- and P-selectin in both humans and mice, and the glycoTs constructing functional selectin ligand(s) on this glycoprotein are similar in both species.7,8 With regard to E-selectin, however, human but not mouse granulocyte rolling on this selectin is insensitive to pronase treatment.9,10 Thus, protease-insensitive gangliosides may be physiological E-selectin ligands that are unique to humans.11 Among the α1,3fucosyltransferases, the enzyme FUT9 reportedly plays a more significant role during human leukocyte adhesion, compared with FUT7 and FUT4 which are the dominant players in mice.8 Among additional differences, L-selectin in human but not mouse neutrophils are considered to act as an E-selectin ligand,12 though its relative roles in direct E-selectin binding vs secondary neutrophil-neutrophil adhesion remains unresolved.13 ESL-1 is a functional E-selectin ligand on murine but not human myeloid cells.14 A glycoform of CD44 called “hematopoietic cell E- and L-selectin ligand (HCELL),” containing an N-linked sialofucosylated carbohydrate is an E-/L-selectin ligand in human but not murine hematopoietic stem and progenitor cells.15 CD44 expressed on murine neutrophils and some lymphocytes, but not mature human leukocytes, acts as an E-selectin ligand.16 Overall, although there is a greater consensus on the players in mice, the precise E-selectin ligands in human cells are as-yet unknown.16,17
We sought to identify the human α2,3 sialyltransferases (sialylTs) that regulate myeloid cell rolling on selectins. In this regard, among the 6 mammalian α2,3sialyTs (ST3Gal-1-6), we choose to focus on ST3Gal-3, -4, and -6 because these enzymes transfer sialic acid (NeuAc in humans) to the 3-position of galactose on type II n-Acetyllactosamine/LacNAc (Galβ1,4GlcNAc) structures to generate sLeX and related glycans18-22 (Figure 1). Among these, ST3Gal-3 also acts on type I lactosamine substrates (Galβ1,3GlcNAc) to create sialyl Lewis-a (sLea)–type epitopes.20 ST3Gal-1 and 2 are less important because they sialylate type III lactosamine (Galβ1,3GalNAc).23,24 Moreover, ST3Gal-5 uniquely sialylates the GM3 ganglioside (Neu5Acα2,3Galβ1,4Glcβ1Cer).25 To quantify the contributions of ST3Gal-3, -4, and -6, these enzymes were specifically disrupted in myeloid cells using either lentivirus-based short hairpin RNA (shRNA) or clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 nuclease-RNA guided genome editing.8,26 Although a number of studies were performed with HL-60 promyeloid leukemic cells because they resemble primary neutrophils in terms of glycosyltransferase expression and selectin binding phenotype,27 critical validation was performed using neutrophils derived from CD34+ human hematopoietic stem cells (hHSCs). The study demonstrates a dominant role for ST3Gal-4 in human leukocyte adhesion to all 3 selectins. Whereas studies with murine models show that ST3Gal-4 collaborates with ST3Gal-6 to control mouse leukocyte recruitment and rolling on E- and P-selectin,28,29 1 primary enzyme regulates this function in human cells. Thus, in the long run, the disruption of ST3Gal-4 function in human myeloid cells may provide a means for anti-cell adhesion and anti-inflammatory therapy.
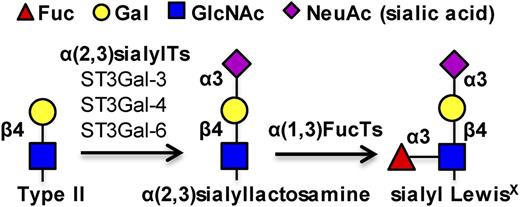
sLeX biosynthesis. Three α(2,3) sialylTs, ST3Gal-3, -4, and -6, act on the type II N-acetyllactosamine substrate to add the terminal sialic acid. α(1,3)fucosyltransferases act on the resulting sialylated lactosamine to form the sLeX antigen.
sLeX biosynthesis. Three α(2,3) sialylTs, ST3Gal-3, -4, and -6, act on the type II N-acetyllactosamine substrate to add the terminal sialic acid. α(1,3)fucosyltransferases act on the resulting sialylated lactosamine to form the sLeX antigen.
Methods
Supplemental methods
Cell culture, molecular biology, lentivirus preparation, ST3Gal shRNA silencing efficiency quantification, quantitative reverse transcription–polymerase chain reaction (RT-PCR), flow cytometry, and mouse neutrophil isolation were similar to previous studies (Buffone et al8 and Marathe et al27 ; supplemental Methods, available on the Blood Web site).
HL-60 carrying 1 or more ST3Gal shRNA
CHO-S cells stably expressing either ST3Gal-3, -4, or -6 enhanced green fluorescent protein (EGFP) fusion proteins were created using lentivirus vector 1 (Figure 2A, supplemental Methods). shRNA that can reduce EGFP fluorescence in these cells by >80% were identified using vector 2 (Figure 2A, supplemental Table 2). These efficacious shRNA were coexpressed with a DsRed reporter using vector 3. Corresponding lentivirus were used to transduce wild-type (WT) HL-60s to generate single knockdown HL-60 cells lacking ST3Gal-3 (ST3Gal-3−HL-60), ST3Gal-4 (ST3Gal-4−HL-60), or ST3Gal-6 (ST3Gal-6−HL-60). The negative control “DsRed HL-60” cells were created using vector 3, only the shRNA was replaced by the stuffer sequence from the TRC cloning vector (supplemental Methods). Dual knockdowns lacking ST3Gal-4 along with either ST3Gal-3 (ST3Gal-3−4−HL-60) or ST3Gal-6 (ST3Gal-4−6−HL-60) were created by applying shRNA against ST3Gal-3 or ST3Gal-6 to stable ST3Gal-4−HL-60s using vector 2. Whereas stable silencing in the single knockdowns was monitored using DsRed fluorescence, the dual knockdowns were additionally selected and maintained in growth media containing 0.5 μg/mL puromycin. Gene silencing was verified using quantitative real-time PCR and flow cytometry 4 to 20 days after transduction.
Effect of α(2,3) sialylT silencing on cell-surface glycan expression. (A) Lentiviral vectors used for shRNA screening and gene silencing. Stable CHO-S cells expressing the ST3Gal-EGFP fusion protein were made using the pCS-CG construct (vector 1). pLKO.1 vectors 2 and 3 carry the shRNA along with either puromycin drug selection (vector 2) or DsRed fluorescent reporter (vector 3). (B) Solid empty histograms in all panels present CHO-S cells stably expressing 1 of the ST3Gal-EGFP fusion proteins. Transduction of these cells with lentivirus carrying shRNA against corresponding ST3Gal variants (gray filled histograms), but not control shRNA (black filled histogram), resulted in a reduction in EGFP signal as measured using flow cytometry. Dashed empty histograms WT CHO-S cells without EGFP fusion protein. Efficient shRNA that reduced EGFP fluorescence by >80% were selected for functional studies. (C) Gel electrophoresis analysis of RT-PCR products demonstrate the absence of target mRNA (white arrow) in single and dual knockdown HL-60 cells. Similar results were obtained using quantitative RT-PCR. (D-G) Flow cytometry measured cell-surface expression of the CLA using mAb HECA-452 (D), sLeX/CD15s epitope using mAb CSLEX-1 (E), Lewis-X/CD15 using mAb HI98 (F), and Galβ1,4GlcNAc/N-acetyllactosamine using ECL lectin (G). All data were normalized with respect to WT HL-60s (dashed line). *P < .05 with respect to all other treatments, †P < .05 with respect to all other treatments except daggers (†) are not different from each other.
Effect of α(2,3) sialylT silencing on cell-surface glycan expression. (A) Lentiviral vectors used for shRNA screening and gene silencing. Stable CHO-S cells expressing the ST3Gal-EGFP fusion protein were made using the pCS-CG construct (vector 1). pLKO.1 vectors 2 and 3 carry the shRNA along with either puromycin drug selection (vector 2) or DsRed fluorescent reporter (vector 3). (B) Solid empty histograms in all panels present CHO-S cells stably expressing 1 of the ST3Gal-EGFP fusion proteins. Transduction of these cells with lentivirus carrying shRNA against corresponding ST3Gal variants (gray filled histograms), but not control shRNA (black filled histogram), resulted in a reduction in EGFP signal as measured using flow cytometry. Dashed empty histograms WT CHO-S cells without EGFP fusion protein. Efficient shRNA that reduced EGFP fluorescence by >80% were selected for functional studies. (C) Gel electrophoresis analysis of RT-PCR products demonstrate the absence of target mRNA (white arrow) in single and dual knockdown HL-60 cells. Similar results were obtained using quantitative RT-PCR. (D-G) Flow cytometry measured cell-surface expression of the CLA using mAb HECA-452 (D), sLeX/CD15s epitope using mAb CSLEX-1 (E), Lewis-X/CD15 using mAb HI98 (F), and Galβ1,4GlcNAc/N-acetyllactosamine using ECL lectin (G). All data were normalized with respect to WT HL-60s (dashed line). *P < .05 with respect to all other treatments, †P < .05 with respect to all other treatments except daggers (†) are not different from each other.
Genome editing using CRISPR/Cas9
The CRISPR Design tool26 was used to define the ST3Gal-4 target in the first exon by minimizing exonic off-target sites. The target sequence (5′-AGAAATAATCCTCAAGCCGC) was cloned into the pX330-U6-Chimeric_BB-CBh-hSpCas9 vector (Addgene, plasmid 42230) after BbsI digestion. HL-60 cells were electroporated with this vector using the Neon Transfection System (Invitrogen) and cultured in RPMI 1640 without antibiotics. Genomic DNA was isolated from the transfected HL-60s after 2 weeks, the region surrounding the CRISPR target site was PCR amplified (supplemental Table 3), and the product was screened using the SURVEYOR nuclease assay (Transgenomic).26 Subsequently, cells staining negative with monoclonal antibody (mAb) HECA-452 were single-cell sorted using a FACSAria (BD Biosciences) into HL-60 cell-culture conditioned RPMI 1640. Isogenic clones derived in this manner were validated using gene sequencing. Indels were detected on the target (ST3GAL4) but not potential off-target genes.
Gene silencing in hHSCs
All human subject protocols were approved by the State University of New York-Buffalo Institutional Review Board. Human cord and placental blood was kindly obtained from donors by Dr Dennis Weppner at the Millard Fillmore Suburban Hospital (Amherst, NY). CD34+ hHSCs were purified using the EasySep human cord blood CD34+ selection kit (Stem Cell Technologies). Isolated hHSCs were then cultured in complete Iscove's Modified Dulbecco's Medium supplemented with 50 ng/mL recombinant human stem cell factor (SCF) and interleukin-3 (IL-3) for 2 days.30,31 The concentration of SCF and IL-3 was reduced twofold every 48 hours thereafter until day 7 when an additional 25 ng/mL recombinant human granulocyte colony-stimulating factor was added. On day 5, a portion of the differentiating cells were transduced with lentivirus containing either ST3Gal-4 shRNA or the TRC cloning vector, both expressing the DsRed reporter. CD11b and CD34 levels were measured every 2 days to monitor differentiation toward neutrophils. Mature neutrophils obtained on days 12 to 14 were analyzed using cytospin. Thirty percent to 40% of these cells had segmented nuclei with some of these cells undergoing diapedesis across stimulated human umbilical vein endothelial cell (HUVEC) monolayers.
Microfluidic cell adhesion assay
HL-60 variants, mouse neutrophils, and transduced hHSCs at 2 × 106 cells per mL were perfused at a wall shear stress of 1 to 2 dyne/cm2 over substrates bearing either physisorbed recombinant L-/P-selectin–immunoglobulin G (IgG), P-selectin–bearing CHO-P cells, E-selectin–bearing L/E cells, or IL-1β–stimulated HUVECs (Buffone et al8 ; supplemental Methods). Here, the E-selectin density on HUVECs was ∼102 sites per μm2, the CHO-P cell P-selectin density was ∼167 sites per μm2, and all recombinant human selectins were adsorbed at ∼180 sites per μm2. During analysis, the interacting cells were defined to travel at velocities less than the theoretical free stream velocity of noninteracting 10-µm diameter spheres at the flow chamber wall.8 Among these, cells moving <1 cell diameter in a 10-second period were defined to be “adherent,” whereas the remaining were classified to be “rolling.” Rolling velocity was calculated by tracking individual cell trajectories. In the case of hHSCs, the number of DsRed-positive cells varied from 25% to 45% depending on the individual viral transduction. In this case, the rolling and adhesion density data were normalized according to: rolling/adherent cell density = measured cell density × 100/% DsRed-positive cells.
Mass spectrometry
N-linked, O-linked, and GSL-derived glycans were extracted from ∼70 to 100 × 106 WT and variant HL-60 cells as described previously (Jang-Lee et al32 ; supplemental Methods). Glycans were permethylated prior to matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry (MS) and MALDI-TOF-TOF MS/MS analysis. Data were annotated using the glycobioinformatics tool, GlycoWorkBench.33 The proposed assignments for the selected peaks were based on 12C isotopic composition together with knowledge of the biosynthetic pathways. The proposed structures were confirmed using MS/MS. In some cases, relative changes in glycan composition upon genomic editing were quantified based on an N-glycan library generated using the software Glycosylation Network Analysis Toolbox (supplemental Methods).34
Statistics
Error bars represent standard error mean for ≥3 independent experiments. The Student t test was performed for dual comparison. One-way analysis of variance followed by the Tukey posttest was performed for multiple comparisons. P < .05 was considered to be statistically significant.
Results
ST3Gal-4 regulates HECA-452 expression
shRNA were applied to WT HL-60s to create stable single (ST3Gal-3−, −4−, and 6−) and dual (ST3Gal-3−4− and ST3Gal-4−6−) knockdown HL-60 cells (Figure 2C). The transduction conditions and plasmid vectors were optimized so that ∼100% of the HL-60s were stably DsRed-positive for at least 30 to 45 days during which time functional studies were completed.8 The high transduction efficiency enabled experimentation with the original HL-60s, without the need to sort or enrich a particular population. More than 90% gene silencing was confirmed using quantitative RT-PCR.
Flow cytometry measured changes in sLeX and related antigens (Figure 2D-G). Most prominently, silencing ST3Gal-4 resulted in ∼80% reduction in cutaneous lymphocyte antigen (CLA) expression as measured using mAb HECA-452. This epitope, which resembles sLeX, was further reduced to ∼95% in the dual knockdowns (Figure 2D). ST3Gal-3−HL-60s displayed ∼35% reduction in CLA expression, consistent with the notion that this enzyme shows higher activity toward type I (Galβ1,3GlcNAc) rather than type II (Galβ1,4GlcNAc) LacNAc.20 Silencing ST3Gal-6 augmented CLA expression by >twofold, though the precise reason for this is unknown. Similar observations, as with HECA-452, were made when mAb CSLEX-1 measured cell-surface sLeX expression (Figure 2E). Here, ST3Gal-4−HL-60s and the dual knockdowns displayed ∼80% reduction in CSLEX-1 binding and the ST3Gal-3−HL-60s exhibited a 20% decrease.
The changes in cell-surface LeX levels were inversely related to the changes observed in CLA/sLeX expression, with a 2.5- to 5-fold increase in LeX expression being noted in the ST3Gal-4− single and dual knockdowns (Figure 2F). Erythrina cristagalli lectin (ECL) recognition (binds Galβ1,4GlcNAc) was upregulated ∼threefold in the ST3Gal-4− cells (Figure 2G). Lectin-binding specificity was confirmed by blocking with 200 mM lactose. In additional controls, Peanut agglutinin lectin binding (binds unsialylated Galβ1,3GalNAc) and PSGL-1 levels were unaffected in the ST3Gal knockdowns (supplemental Figure 1). Overall, ST3Gal-4 regulates HECA-452 and CSLEX-1 epitope biosynthesis.
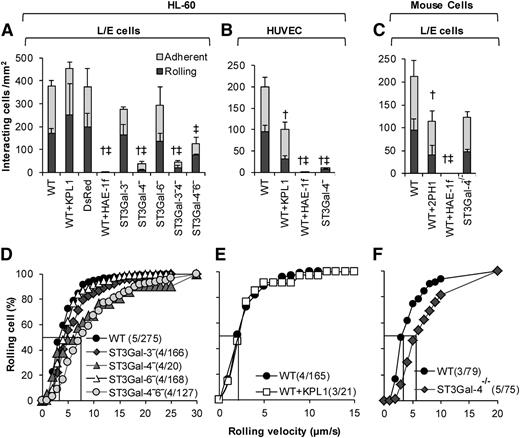
Silencing ST3Gal-4 abrogates HL-60 rolling on E-selectin
The rolling phenotype of the ST3Gal knockdown HL-60s was compared with that of neutrophils from ST3Gal-4−/− mice under hydrodynamic shear (Figure 3). Both substrates composed of E-selectin–bearing L/E cells (Figure 3A,C,D,F) and IL-1β–stimulated HUVECs (Figure 3B,E) were studied. Here, most notably, the ST3Gal-4−HL-60s displayed > 90% reduction in cell rolling and adhesion compared with the WT HL-60s (Figure 3A-B, supplemental Video A). The residual ST3Gal-4− cells interacting with the L/E substrate exhibited a ∼twofold increase in median rolling velocity (Figure 3D). Knocking down ST3Gal-3 and ST3Gal-6 did not have an additive effect in the dual knockdowns. Additionally, the rolling phenotype of single ST3Gal-3− and ST3Gal-6−HL-60s was no different from the WT cells. The binding in these experiments was abrogated by an anti-E-selectin mAb HAE-1f (Figure 3A-C). The anti-human (KPL-1, Figure 3A-B) and murine PSGL-1 mAbs (2PH1, Figure 3C) had a minor effect,35,36 possibly due to the inhibition of secondary leukocyte-leukocyte capture in addition to primary leukocyte recruitment onto the substrate.
E-selectin–mediated leukocyte rolling. A total of 2 × 106/mL WT and variant HL-60s were perfused over monolayers containing either E-selectin–expressing L/E cells (A,D) or IL-1β–stimulated HUVECs (B,E) at 1 dyne/cm2. WT and ST3Gal-4−/− mouse neutrophils were also perfused over L/E cells (C,F). Top panels present rolling and adherent cell density data 2 minutes after start of perfusion. Bottom panels present rolling velocity in the form of cumulative histogram plots. Median rolling velocity is indicated by straight line at 50% in the lower panels. Number in parentheses present “(number of independent experiments/total number of cells analyzed).” Rolling velocity was not measured when <5 cells rolled in a given run. † and ‡ denote statistically significance differences (P < .05) for rolling and adherent cell density, respectively. Here, † and ‡ are different with respect to all other treatments, except that treatments marked by † and ‡ in a given panel are not different from each other. HAE-1f, KPL-1, and 2PH1 are blocking mAbs against human E-selectin, human PSGL-1, and murine PSGL-1, respectively. Elimination of ST3Gal-4 activity has a more drastic effect on human, rather than murine, leukocyte rolling on E-selectin.
E-selectin–mediated leukocyte rolling. A total of 2 × 106/mL WT and variant HL-60s were perfused over monolayers containing either E-selectin–expressing L/E cells (A,D) or IL-1β–stimulated HUVECs (B,E) at 1 dyne/cm2. WT and ST3Gal-4−/− mouse neutrophils were also perfused over L/E cells (C,F). Top panels present rolling and adherent cell density data 2 minutes after start of perfusion. Bottom panels present rolling velocity in the form of cumulative histogram plots. Median rolling velocity is indicated by straight line at 50% in the lower panels. Number in parentheses present “(number of independent experiments/total number of cells analyzed).” Rolling velocity was not measured when <5 cells rolled in a given run. † and ‡ denote statistically significance differences (P < .05) for rolling and adherent cell density, respectively. Here, † and ‡ are different with respect to all other treatments, except that treatments marked by † and ‡ in a given panel are not different from each other. HAE-1f, KPL-1, and 2PH1 are blocking mAbs against human E-selectin, human PSGL-1, and murine PSGL-1, respectively. Elimination of ST3Gal-4 activity has a more drastic effect on human, rather than murine, leukocyte rolling on E-selectin.
The above observations made with the human HL-60s are in marked contrast to results obtained using murine neutrophils under identical conditions (Figure 3C,F). Mouse ST3Gal-4−/− neutrophil rolling and adhesion was only reduced by 40% compared with WT cells, compared with >90% in the case of human cells. Similar observations were made using substrates bearing recombinant mouse selectins (supplemental Table 4).
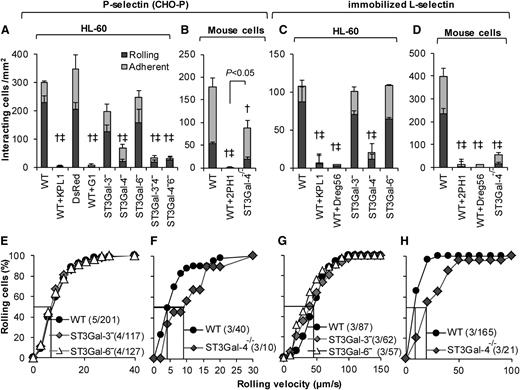
ST3Gal-4−HL-60s display negligible interaction with P-selectin and L-selectin
ST3Gal knockdown interactions were evaluated on P-selectin–bearing CHO-P substrates (Figure 4, left half) and also surfaces bearing immobilized recombinant L-selectin–IgG (Figure 4, right half). Here, the ST3Gal-4−HL-60s displayed ∼80% to 90% reduction in the cell rolling and adhesion density on both substrates compared with WT HL-60s (Figure 4A,C). ST3Gal-3− and ST3Gal-6−HL-60s behaved similar to WT HL-60s. No additive effect was observed in the dual knockdowns on P-selectin (Figure 4A). The number of ST3Gal-4−HL-60s interacting with P-/L-selectin was too few to perform rolling velocity analysis (Figure 4E,G).
P-/L-selectin mediated rolling. Cell-adhesion studies were performed identically to Figure 3 only the substrates were composed of either P-selectin–expressing CHO-P cells (A-B,E-F), or recombinant L-selectin (C-D,G-H). Top panels present rolling and adherent cell density data, whereas bottom panels present cumulative rolling velocity distribution. A, E and C, G present data for HL-60 and its derivatives, whereas the remaining panels present murine neutrophil data. All statistical symbols are identical to Figure 3, except that WT+2PH1 is significantly different from ST3Gal-4−/− in panel B. Silencing ST3Gal-4 reduced HL-60 rolling on both P- and L-selectin by >90%. ST3Gal-4−/− mouse neutrophils rolled well on P-selectin unlike the ST3Gal-4− HL-60s. mAbs G1 and Dreg56 are anti-P- and L-selectin blocking mAbs.
P-/L-selectin mediated rolling. Cell-adhesion studies were performed identically to Figure 3 only the substrates were composed of either P-selectin–expressing CHO-P cells (A-B,E-F), or recombinant L-selectin (C-D,G-H). Top panels present rolling and adherent cell density data, whereas bottom panels present cumulative rolling velocity distribution. A, E and C, G present data for HL-60 and its derivatives, whereas the remaining panels present murine neutrophil data. All statistical symbols are identical to Figure 3, except that WT+2PH1 is significantly different from ST3Gal-4−/− in panel B. Silencing ST3Gal-4 reduced HL-60 rolling on both P- and L-selectin by >90%. ST3Gal-4−/− mouse neutrophils rolled well on P-selectin unlike the ST3Gal-4− HL-60s. mAbs G1 and Dreg56 are anti-P- and L-selectin blocking mAbs.
In contrast to the human cells, similar to earlier work,28 ST3Gal-4−/− mouse neutrophils only displayed a 40% to 50% reduction in P-selectin–mediated cell adhesion compared with WT cells (Figure 4B, supplemental Table 4). Mouse ST3Gal-4 deletion reduced cell binding on L-selectin by ∼85% (Figure 4D; Sperandio et al37 ). ST3Gal-4−/− mouse neutrophil rolling velocity was augmented ∼twofold on both substrates (Figure 4F,H). Control studies performed with anti-P-selectin, L-selectin and anti-PSGL-1 function blocking mAbs demonstrate that cell adhesion to P-/L-selectin is primarily PSGL-1 dependent in both humans and mice. Overall, ST3Gal-4 is a key regulator of human leukocyte adhesion to all 3 selectins.
ST3Gal-4 HL-60 knockouts exhibit loss of selectin binding
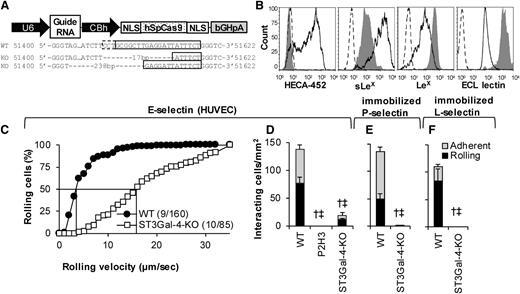
Although gene silencing is a powerful methodology, data interpretation is complicated due to the 10% to 20% residual enzyme activity. To overcome this, ST3Gal-4 was knocked out in HL-60s using CRISPR/Cas9-based editing (Figure 5). Figure 5A presents the vector used.26 When electroporated into HL-60s, CLA/HECA-452 expression decreased in 56% of the cells within 10 days (supplemental Figure 2). Isogenic ST3Gal-4-KO clones were then isolated based on low HECA-452 expression using flow sorting. These clones displayed indels in ST3GAL4. The particular clone used in this study contained a 17-bp and 238-bp deletion in the 2 ST3GAL4 alleles (Figure 5A).
Genomic deletion of STGal-4. (A) Schematic of vector used to create knockout cells. ST3Gal-4 genomic DNA sequencing results for WT HL-60s (top row) and the 2 alleles in the ST3Gal-4-KO cells (middle and bottom row). Here, the solid box highlights the guide RNA sequence and the dashed box denotes the protospacer adjacent motif (PAM/NGG). Seventeen- and 238-bp chromosomal deletion are noted upon genomic editing. (B) Flow cytometry histograms measuring CLA/mAb HECA-452, sLeX/CSLEX-1, LeX/HI98, and ECL lectin binding. Solid empty histograms correspond to WT cells; dashed histograms correspond to isotype control except for last panel where it is secondary Ab alone; and gray shaded histograms are for ST3Gal-4-KO HL-60s. (C-F) WT and ST3Gal-4-KO cell rolling data on IL-1β–stimulated HUVECs (C-D), recombinant P-selectin-IgG (E), and L-selectin-IgG (F) at 1 dyne/cm2. Statistics symbols are identical to Figure 3. P2H3 is an anti-E-selectin blocking mAb. ST3Gal-4-KOs did not roll on L- and P-selectin. Residual cells that rolled on E-selectin displayed fivefold higher rolling velocity compared with WT HL-60s.
Genomic deletion of STGal-4. (A) Schematic of vector used to create knockout cells. ST3Gal-4 genomic DNA sequencing results for WT HL-60s (top row) and the 2 alleles in the ST3Gal-4-KO cells (middle and bottom row). Here, the solid box highlights the guide RNA sequence and the dashed box denotes the protospacer adjacent motif (PAM/NGG). Seventeen- and 238-bp chromosomal deletion are noted upon genomic editing. (B) Flow cytometry histograms measuring CLA/mAb HECA-452, sLeX/CSLEX-1, LeX/HI98, and ECL lectin binding. Solid empty histograms correspond to WT cells; dashed histograms correspond to isotype control except for last panel where it is secondary Ab alone; and gray shaded histograms are for ST3Gal-4-KO HL-60s. (C-F) WT and ST3Gal-4-KO cell rolling data on IL-1β–stimulated HUVECs (C-D), recombinant P-selectin-IgG (E), and L-selectin-IgG (F) at 1 dyne/cm2. Statistics symbols are identical to Figure 3. P2H3 is an anti-E-selectin blocking mAb. ST3Gal-4-KOs did not roll on L- and P-selectin. Residual cells that rolled on E-selectin displayed fivefold higher rolling velocity compared with WT HL-60s.
Consistent with the shRNA studies (Figure 2), CLA and sLeX epitopes on this knockout were reduced by 97% and 91%, respectively (Figure 5B). Further, LeX and ECL expression were augmented 5- and 12-fold. Thus, ST3Gal-4 primarily controls cell-surface sLeX expression. ST3Gal-4-KOs exhibited dramatic reduction in selectin binding under fluid shear with ∼90% reduction in leukocyte rolling and adhesion density on stimulated HUVECs (Figure 5D) and complete ablation on substrates bearing immobilized recombinant P-selectin (Figure 5E) and L-selectin (Figure 5F). The residual rolling cells on HUVECs displayed fivefold increased median rolling velocity compared with the WT cells (Figure 5C). Although data presented here are for a single clone, identical observations were made using a mixed population of HL-60s sorted based on low CLA expression (data not shown). Overall, ST3Gal-4 is the dominant α(2,3)sialylT regulating HL-60 rolling on all selectins.
ST3Gal-4 controls sLeX expression on O- and N-glycans
MS experiments compared the glycomic profiles of WT, ST3Gal-4−, and ST3Gal-4-KO HL-60s. Such studies analyzed the N- and O-glycans released from glycoproteins and also carbohydrates from the GSLs. Here, the N-glycans of WT HL-60s included both high mannose (data not shown) and complex N-glycans (supplemental Figure 3). The latter consisted mainly of core fucosylated and nonbisected N-glycans, corresponding to bi-, tri-, and tetra-antennary structures occasionally extended with LacNAc repeats. Most of these glycans were decorated with fucose and/or NeuAc to form LeX, sialylated-LacNAc, or sLeX epitopes. The O-glycans of WT HL-60s contained core 1 and core 2 structures including the core 2 sLeX at m/z = 1879 (supplemental Figure 4).
Upon ST3Gal-4 disruption, the sLeX epitope was drastically reduced on both N- and O-glycans. For example, at m/z = 3140-3147, WT HL-60s contain 2 overlapping isotope clusters corresponding to a biantennary structure carrying a sLeX epitope and a nonsialylated tetra-antennary N-glycan (monoisotopic m/z = 3140 and 3142, respectively; Figure 6Ai). MALDI-TOF-TOF MS/MS analysis of this molecular cluster showed that the sLeX N-glycan (m/z 3140) was more abundant compared with the tetra-antennary glycan (m/z 3142), based on the relative abundance of fragment ions at m/z = 2765 vs 2679 (Figure 6Aii). Using isotope distribution analysis of MS data (Figure 6Ai), we estimate a biantennary (m/z 3140) to tetra-antennary glycan (m/z 3142) relative abundance ratio of ∼4 in WT HL-60s. This abundance dropped to 0.4 (ie, ∼90% decrease) in the ST3Gal-4−HL-60s (Figure 6Bi). Similarly, the intensity of the m/z = 2679 fragment ion was greater than that at m/z = 2765 (Figure 6Bii), and this indicates an increase in the relative abundance of the tetra-antennary N-glycan in the knockdown. Both isotope analysis and MS/MS analysis demonstrate the complete absence of the sLeX N-glycan (m/z 3140) in the CRISPR/Cas9 knockout HL-60s (Figure 6C). Similar disruption of the sLeX glycan was also noted for a triantennary N-glycan carrying a single sLeX (supplemental Figure 5). With respect to the O-glycans, the ratio of the core 2 glycan carrying a single sLeX epitope (m/z 1879) to the corresponding non-sLeX glycan (m/z 1344) was 0.51 in WT HL-60s (Figure 6Di). This ratio dropped to 0.12 in the ST3Gal-4− HL-60s (∼76% decrease, Figure 6D, middle panel). Core-2 sLeX was absent in the ST3Gal-4-KO (Figure 6Dii). Overall, the absence of the sLeX on N- and O-glycans in the CRISPR/Cas9 KOs demonstrates that ST3Gal-4 is the primary enzyme responsible for this epitope.
MS analysis of HL-60 N- and O-glycans. MALDI-TOF MS and TOF-TOF MS/MS spectra of permethylated N-glycans of the zoomed scan m/z 3140 to 3147 molecular ion cluster of (A) WT, (B) ST3Gal-4− HL-60s (knockdowns), (C) CRISPR/Cas9 ST3Gal-4-KO HL-60 cells. Panels Ai, Bi, Ci depict the zoomed scan area; panels Aii, Bii, Cii depict the MALDI TOF-TOF MS/MS of the corresponding molecular ion cluster. (D) Partial MALDI-TOF MS spectra of permethylated O-glycans derived from the WT (i), ST3Gal-4− knockdown (ii), and ST3Gal-4-KO (iii) HL-60 cells. All spectra of N- and O-glycans were derived from the 50% MeCN fraction. All molecular ions are [M+Na]+. For the MS/MS panels, arrows indicate the losses indicated from the molecular ion. Putative structures are based on composition, tandem MS, and biosynthetic knowledge. Full MALDI-TOF MS spectra of the N- and O-glycans can be found in supplemental Figures 3 and 4.
MS analysis of HL-60 N- and O-glycans. MALDI-TOF MS and TOF-TOF MS/MS spectra of permethylated N-glycans of the zoomed scan m/z 3140 to 3147 molecular ion cluster of (A) WT, (B) ST3Gal-4− HL-60s (knockdowns), (C) CRISPR/Cas9 ST3Gal-4-KO HL-60 cells. Panels Ai, Bi, Ci depict the zoomed scan area; panels Aii, Bii, Cii depict the MALDI TOF-TOF MS/MS of the corresponding molecular ion cluster. (D) Partial MALDI-TOF MS spectra of permethylated O-glycans derived from the WT (i), ST3Gal-4− knockdown (ii), and ST3Gal-4-KO (iii) HL-60 cells. All spectra of N- and O-glycans were derived from the 50% MeCN fraction. All molecular ions are [M+Na]+. For the MS/MS panels, arrows indicate the losses indicated from the molecular ion. Putative structures are based on composition, tandem MS, and biosynthetic knowledge. Full MALDI-TOF MS spectra of the N- and O-glycans can be found in supplemental Figures 3 and 4.
In addition to sLeX, the cumulative relative abundance of other epitopes was quantified in the partial MALDI-TOF MS spectrum (m/z = 2645-3604, supplemental Figure 6). Here, knocking out ST3Gal-4 increased the LeX epitope by ∼28%, augmented LacNAc by ∼35%, and decreased sialylated-LacNAc by ∼24%. These data are qualitatively consistent with flow cytometry results.
The glycans from the polar GSLs of HL-60s consisted of linear LacNAc repeating units decorated with fucose and/or NeuAc (supplemental Figures 7-8). The monosialogangliosides detected in the high mass region (above m/z = 3000) consisted of sialylated lactosylceramides with 4 to 6 LacNAc repeats and multiple internal fucoses as expected on putative selectin ligands11,38 (supplemental Figure 8A,C; m/z = 3449, 3898). A minor abundance of sLeX epitope was also detected (supplemental Figure 8A-B). No glycans were detected in the nonpolar GSL fraction (data not shown). Even though some of these glycans at m/z = 3623 and 3898 were reduced in the ST3Gal-4− (supplemental Figure 7B) and ST3Gal-4-KOs (supplemental Figure 7C), a majority of the gangliosides remained with some of them containing putative selectin ligands and the sLeX epitope (supplemental Figure 8I-J). Thus, although blocking ST3Gal-4 activity dramatically reduces myeloid cell rolling on all selectins, additional studies are necessary to determine the degree to which this is due to altered ganglioside biosynthesis.
ST3Gal-4 controls primary human neutrophil rolling
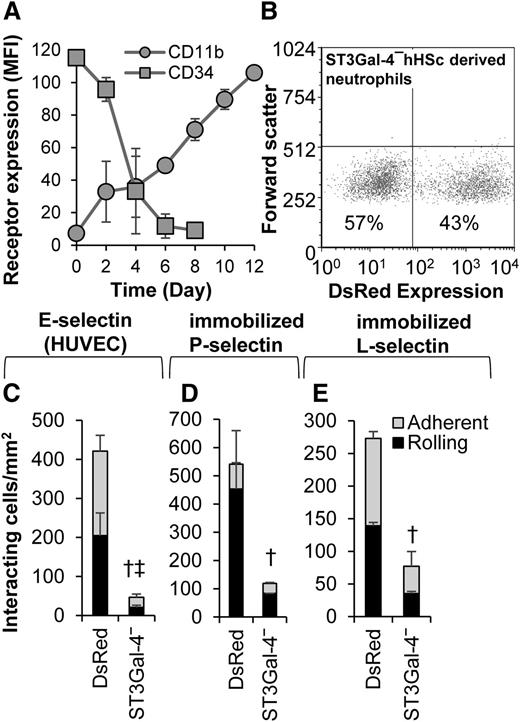
We tested the hypothesis that ST3Gal-4 controls human neutrophil rolling in primary cells. Thus, CD34+ hHSCs were purified and transduced with lentivirus carrying either ST3Gal-4 shRNA or TRC/DsRed control during their differentiation toward neutrophils (Figure 7). Cell differentiation was accompanied by a decrease in cell-surface CD34 and increase in CD11b expression (Figure 7A). Approximately 25% to 45% of the differentiated cells displayed red fluorescence due to the DsRed reporter present in the viral construct (Figure 7B). In functional studies that characterized the rolling of the DsRed-positive cells, knocking down ST3Gal-4 resulted in an 88%, 78%, and 72% decrease in rolling and total interacting cell density on E-selectin-bearing stimulated HUVECs (Figure 7C), recombinant P-selectin (Figure 7D), and L-selectin (Figure 7E), respectively. These data, using donor-derived primary cells, confirm a primary role for ST3Gal-4 in regulating selectin-dependent human neutrophil tethering and rolling.
Neutrophils derived from hHSCs. hHSCs were differentiated to mature neutrophils over 12 days. On the fifth day of the differentiation process, the cells were transduced with lentivirus containing either ST3Gal-4 shRNA (“ST3Gal-4−”) or TRC control virus (“DsRed”). In both cases, DsRed serves as a reporter of transduction. (A) CD34 and CD11b expression changes with time confirming differentiation to granulocytes. (B) Transduction efficiency quantified based on the percentage of cells that are DsRed positive. (C-E) Rolling and adherent cell density data for neutrophils derived from hHSCs on IL-1β–stimulated HUVECs (C), recombinant P-selectin (D), and recombinant L-selectin (E). Fluorescence microscopy was applied in panels C-E, so that only DsRed-positive cells were analyzed in the flow chamber assay. Knocking down ST3Gal-4 reduced cell adhesion to all 3 selectins. Data are presented for hHSCs derived from 4 different human donors. Statistics symbols are the same as Figure 3.
Neutrophils derived from hHSCs. hHSCs were differentiated to mature neutrophils over 12 days. On the fifth day of the differentiation process, the cells were transduced with lentivirus containing either ST3Gal-4 shRNA (“ST3Gal-4−”) or TRC control virus (“DsRed”). In both cases, DsRed serves as a reporter of transduction. (A) CD34 and CD11b expression changes with time confirming differentiation to granulocytes. (B) Transduction efficiency quantified based on the percentage of cells that are DsRed positive. (C-E) Rolling and adherent cell density data for neutrophils derived from hHSCs on IL-1β–stimulated HUVECs (C), recombinant P-selectin (D), and recombinant L-selectin (E). Fluorescence microscopy was applied in panels C-E, so that only DsRed-positive cells were analyzed in the flow chamber assay. Knocking down ST3Gal-4 reduced cell adhesion to all 3 selectins. Data are presented for hHSCs derived from 4 different human donors. Statistics symbols are the same as Figure 3.
Discussion
This study evaluated the relative contributions of 3 α(2,3)sialylTs, ST3Gal-3, -4, and -6, during human myeloid cell adhesion under hydrodynamic shear. It demonstrates that ST3Gal-4 is the primary enzyme regulating selectin-ligand biosynthesis in humans, with ST3Gal-3 and ST3Gal-6 playing negligible roles. Our observations upon silencing ST3Gal-3 are consistent with previous studies with ST3Gal-3−/− mice.28 In contrast to murine knockouts that suggest a role for both ST3Gal-4 and ST3Gal-6 in regulating leukocyte rolling on E- and P-selectin29 ; however, the current study demonstrates that ST3Gal-4 is the exclusive enzyme responsible for this function in humans.
Application of CRISPR/Cas9 for studies of leukocyte function
Two complementary experimental modalities were applied to alter sialylT activity, lentiviral shRNA knockdown and genomic editing using CRISPR/Cas9. Similar results were obtained using both methods and thus the overall findings are unlikely to be due to off-target effects. With respect to the genomic editing, several key protocols were developed for the rapid creation of isogenic human leukocyte cell populations. These and related methods may find wide applications in future studies of leukocyte cell adhesion function. A key advantage of the CRISPR/Cas9 method is that the product is a complete knockout, unlike shRNA which only results in 80% to 90% reduction in messenger RNA (mRNA) expression. Thus, unlike the RNA interference approach, functional studies performed with the knockouts are less ambiguous, and they allow the quantitative evaluation of the relative roles of specific glycosyltransferases. A limitation of the CRISPR/Cas9 approach is that it cannot be currently applied to CD34+ hHSCs.
Regulation of human myeloid cell binding to P- and L-selectin
A core 2–based O-linked glycan bearing the sLeX epitope, located at the N terminus of PSGL-1, at Thr-57 in humans,35,39 is the primary ligand for L- and P-selectin. Studies with knockout mice show that the biosynthesis of this O-glycan is initiated by the formation of the core 1 structure (Galβ1,3GalNAcα1-Thr) catalyzed by the core-1 β1,3galactosyltransferase/T-synthase.40 Following this, the extended core-2 tetrasaccharide (Galβ1,4GlcNAcβ1,6(Galβ1,3)GalNAcα1–Thr) is formed by the sequential action of the β1,6 N-Acetyl glucosaminyltransferase C2GnT-141 and the galactosyltransferase β1,4GalT-I.42 Additionally, the current study shows that silencing ST3Gal-4 in ST3Gal-4−HL-60s and neutrophils derived from hHSCs reduced cell adhesion to L- and P-selectin by 80% to 90%. Knocking out this enzyme using CRISPR/Cas9 abrogated leukocyte recruitment on both substrates. In contrast, consistent with previous studies,28,29 ST3Gal-4−/−mouse neutrophils only exhibit 40% to 50% reduction in cell rolling and adhesion on P-selectin. Neutrophils from ST3Gal-4−/− mice also displayed reduced binding to L-selectin, as reported previously in models of inflammation.37 Overall, in humans, the enzyme ST3Gal-4 may exclusively sialylate the type II LacNAc (Galβ1,4GlcNAc) structure at the N terminus of PSGL-1. In mice, however, ST3Gal-6 likely acts in synergy with ST3Gal-4 to sialylate this epitope. Following sialylation, the PSGL-1 N terminus is primarily α(1,3)fucosylated by the enzyme FUT7, in both humans and mice, with the enzyme FUT4 having a partial role.8,43,44
Consistent with the concept that ST3Gal-4 almost exclusively controls sLeX biosynthesis in human leukocytes, flow cytometry studies with knockdown and knockout cells exhibited a 90% to 97% reduction in the HECA-452 epitope and 70% to 85% reduction in mAb CSLEX-1 binding. These data are corroborated by MS-based O-glycan profiling which demonstrates the absence of core 2 sLeX on HL-60s lacking ST3Gal-4. Additionally, ST3Gal-4 almost exclusively controls sLeX biosynthesis on complex N-glycans in humans.
E-selectin–mediated human leukocyte adhesion
Unlike L- and P-selectin which primarily bind sLeX on O-glycans, many different sialofucosylated carbohydrates bind human E-selectin. These are potentially expressed on CD43, CD44, L-selectin, CD18-integrin Mac-1, and gangliosides.4,7 For example, in the case of GSLs, E-selectin binds gangliosides bearing non-sLeX epitopes.45 Regardless of the scaffold protein/lipid, our data suggest that a single enzyme ST3Gal-4 may α(2,3)sialylate a majority of these physiologically relevant human E-selectin ligands. Thus, in the knockdown and knockout studies, ST3Gal-4 reduction decreased cell rolling and firm adhesion density by ∼90% on stimulated HUVECs under flow. Similar observations were made with neutrophils derived from hHSCs. In contrast, ST3Gal-4−/− mouse neutrophils displayed only 40% to 50% reduction in cell rolling and adhesion.28 Overall, the contribution of ST3Gal-4 activity to leukocyte-endothelial cell adhesion is markedly greater in humans.
In addition to acting on glycoproteins, ST3Gal-4 disruption may also affect the gangliosides that facilitate myeloid cell recruitment. In support of this, human ST3Gal-4 exhibits broad substrate specificity and has been shown to act on both glycoprotein and glycolipid substrates.19,21,22 Further, our glycomics profiling data show that the disruption of ST3Gal-4 activity reduces some of the high-molecular-weight sialofucosylated gangliosides called “myeloglycans.”11 These GSLs, which contain 4 or more LacNAc repeats with multiple internal α(1,3)fucose residues, are considered to be physiological E-selectin ligands.38
Besides altering sLeX, the reduction in ST3Gal-4 activity lead to a 3.5- to 5-fold increase in LeX expression and 3- to 12-fold increase in ECL lectin binding, as measured using flow cytometry. Glycomics profiling of N-glycans showed a similar pattern of alteration though the changes were less pronounced. The difference in the 2 assays could be due to the specificity of the lectins/mAbs, whose recognition may depend on additional factors like the protein/lipid scaffolds bearing the glycan epitope and their preference to bind cell-surface exposed rather than cryptic sites. Concerning the opposing effects on LeX and sialylated epitopes, our results corroborate recent findings, where the selective inhibition of sialylation (in our case because of the ST3Gal-4 knockout) allows for greater fucosylation due to the competition between fucosyltransferases and sialylTs for the common LacNac substrate.46
Targeting ST3Gal-4 to reduce cell adhesion
The identification of critical rate limiting steps that regulate cell function is significant in the context of recent efforts to metabolically alter glycosylation pathways in order to fine tune selectin-binding function.46-48 In this regard, the current study identifies ST3Gal-4 as a key regulator of human myeloid cell adhesion. Thus, small molecules that can perturb this enzyme activity may be effective anti-inflammatory or antiadhesive drugs. That said, it remains to be determined whether ST3Gal-4 is also a critical regulator of selectin-mediated adhesion for other human nonmyeloid cell types. Also, the disruption of ST3Gal-4 activity in humans may perturb additional pathways that have been shown to be modified in ST3Gal-4−/− mice, like CXCR2-dependent activation,49 and von Willebrand factor and platelet circulation half-life.50 Finally, the remarkable species-specific function of the glycosyltransferases suggests the possibility that evolutionary divergence may have conditioned human immunity.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the National Institutes of Health, National Heart, Lung, and Blood Institute grants HL103411 and HL63014 (S.N.), a Program of Excellence in Glycosciences grant HL107146 (J.T.Y.L.), and the Biotechnology and Biological Sciences Research Council grant BB/K016164/1 (A.D. and S.M.H. for Core Support for Collaborative Research).
Authorship
Contribution: N.M., A.B., G.S., and A.A. designed and performed experiments and wrote the manuscript; J.T.Y.L. contributed vital reagents; and S.M.H., A.D., and S.N. designed research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sriram Neelamegham, State University of New York, 906 Furnas Hall, Buffalo, NY 14260; e-mail: neel@buffalo.edu.
References
Author notes
N.M. and A.B. contributed equally to this study.


![Figure 6. MS analysis of HL-60 N- and O-glycans. MALDI-TOF MS and TOF-TOF MS/MS spectra of permethylated N-glycans of the zoomed scan m/z 3140 to 3147 molecular ion cluster of (A) WT, (B) ST3Gal-4− HL-60s (knockdowns), (C) CRISPR/Cas9 ST3Gal-4-KO HL-60 cells. Panels Ai, Bi, Ci depict the zoomed scan area; panels Aii, Bii, Cii depict the MALDI TOF-TOF MS/MS of the corresponding molecular ion cluster. (D) Partial MALDI-TOF MS spectra of permethylated O-glycans derived from the WT (i), ST3Gal-4− knockdown (ii), and ST3Gal-4-KO (iii) HL-60 cells. All spectra of N- and O-glycans were derived from the 50% MeCN fraction. All molecular ions are [M+Na]+. For the MS/MS panels, arrows indicate the losses indicated from the molecular ion. Putative structures are based on composition, tandem MS, and biosynthetic knowledge. Full MALDI-TOF MS spectra of the N- and O-glycans can be found in supplemental Figures 3 and 4.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/4/10.1182_blood-2014-07-588590/4/m_687f6.jpeg?Expires=1765904361&Signature=kJaD~MJXp8nuKQ5beVY~RFrVoiDSWdOMF4ktJ2mtkVvfHTBH7FRqUANjbs4n8REFXPfvRBieZ8vcgk4KURc-e0NRHifzijckEda-oIBqJi4QfLnKfsMotkRo4Is0ZmqYJugN6fburY8HPX9a9Tm00SKu~fq990j9LTgUffUx1RROVKZ3Bg-L~EYNXF06b6Cs0Gn-hA4baD0TiF0q8LdgdBmSwj3V~E29~BX47laGdC9C2n8M5381Zlouadq~hauAODK0uq9e-J5G4Cz6cRGRzx6IeZ-Di6fQkHJLBdONk1P8~XVtlhdGGr75pY18HuCOkp-HS-kfU3meykdoVg-~dg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal