Key Points
Deubiquitinases Usp9x and Usp24 regulate Mcl-1 and myeloma cell survival.
Small-molecule–mediated Usp9x/Usp24 inhibition induces apoptosis and blocks myeloma tumor growth in vivo.
Abstract
Usp9x was recently shown to be highly expressed in myeloma patients with short progression-free survival and is proposed to enhance stability of the survival protein Mcl-1. In this study, we found that the partially selective Usp9x deubiquitinase inhibitor WP1130 induced apoptosis and reduced Mcl-1 protein levels. However, short hairpin RNA–mediated knockdown (KD) of Usp9x in myeloma cells resulted in transient induction of apoptosis, followed by a sustained reduction in cell growth. A compensatory upregulation of Usp24, a deubiquitinase closely related to Usp9x, in Usp9x KD cells was noted. Direct Usp24 KD resulted in marked induction of myeloma cell death that was associated with a reduction of Mcl-1. Usp24 was found to sustain myeloma cell survival and Mcl-1 regulation in the absence of Usp9x. Both Usp9x and Usp24 were expressed and activated in primary myeloma cells whereas Usp24 protein overexpression was noted in some patients with drug-refractory myeloma and other B-cell malignancies. Furthermore, we improved the drug-like properties of WP1130 and demonstrated that the novel compound EOAI3402143 dose-dependently inhibited Usp9x and Usp24 activity, increased tumor cell apoptosis, and fully blocked or regressed myeloma tumors in mice. We conclude that small-molecule Usp9x/Usp24 inhibitors may have therapeutic activity in myeloma.
Introduction
The success of bortezomib and lenalinomide in treating some B-cell tumors highlights the potential for additional agents that target components of the ubiquitin/proteasome cycle to have therapeutic value.1 Recent studies provide evidence for the overexpression of specific deubiquitinases (DUBs) in cancer, including some specific to multiple myeloma (MM) and possibly other B-cell malignancies.2 Usp7 was recently shown to be overexpressed in MM patients, where it regulates ubiquitination and turnover of p53 and other proteins. One recently described Usp7 inhibitor has demonstrated anti-MM activity in preclinical animal models.3 Other inhibitors targeting two DUBs that are components of the 19S proteasome have been shown to have activity against several tumors, including myeloma.4 Usp9x has received considerable attention as a potential therapeutic target for several cancers after it was reported to regulate the ubiquitination and half-life of Mcl-1, a key antiapoptotic protein detected in many tumors.5 Usp9x was also recently shown to suppress chemotherapy, radiotherapy, and targeted therapy in several tumor types through its regulation of Mcl-1 and other proteins.6,7 Mcl-1 is essential for tumor cell survival, and high levels are noted in patients with drug-resistant MM.8 By using proteomics, ubiquitinated Mcl-1 was recently shown to be bound and deubiquitinated by Usp9x, thus preventing its destruction by the proteasome.5 Usp9x expression was shown to be highly elevated in MM patients, and it correlated with poor patient prognosis.5 In other B-cell malignancies, Usp9x levels were associated with elevated Mcl-1 protein content.5 Therefore, inhibitors of Usp9x activity may function as a form of targeted therapy for patients with B-cell tumors.
DUB inhibitor WP1130, previously known as Degrasyn,9 is a partially selective DUB inhibitor shown to inhibit deubiquitinating activity of Usp9x, Usp5, Usp14, and UCH37.10,11 Usp9x inhibition by WP1130 has been shown to promote apoptosis by reducing Mcl-1 levels and increasing tumor cell sensitivity to chemotherapy.10,11 WP1130-mediated Usp9x inhibition also blocks the growth of ERG-positive prostate tumors in vitro and in mouse xenograft models of prostate cancer.12
To understand the role of Usp9x overexpression in MM and other B-cell tumors, we interrogated the impact of its silencing and small-molecule–mediated inhibition. We found that knockdown (KD) of Usp9x in MM cells led to an early onset of apoptosis, followed by sustained growth suppression by blocking G2→M cell cycle transition. Unexpectedly, we also noted that Usp9x KD led to a compensatory activation and upregulation of the closely related DUB Usp24,13 which shows a great degree of biochemical and sequence similarity to Usp9x. We found that Usp24 also plays a role in myeloma cell survival through its regulation of Mcl-1 levels, particularly in the absence of Usp9x. We finally assessed the antimyeloma activity of a small-molecule DUB inhibitor derived from WP1130 with more specific activity against Usp9x and Usp24. The results suggest that DUB inhibition presents an additional therapeutic approach to treating myeloma and other B-cell tumors.
Methods
Compounds WP113014 and EOAI3402143 (G9)15 were synthesized by methods outlined in patent applications.15 Bortezomib was provided by Millennium Pharmaceuticals, and MG132 was purchased from Cayman Chemical Company. All reagents were made up and stored frozen as 10-mM stock solutions. DUB activity in cell lines and patient samples was measured as previously described.10,11 Lysate preparation and western blotting were performed as previously described.10,11 Other methods used in this study can be found in the supplemental Data available at the Blood Web site.
Results
WP1130 inhibits Usp9x activity and promotes Mcl-1 destruction in B-cell tumors
Previously, we described small-molecule DUB inhibitor WP1130, which has activity against Usp9x.10,11 To determine whether WP1130-mediated Usp9x inhibition induced a corresponding change in Mcl-1 protein levels in myeloma and lymphoma, 4 established myeloma cell lines (MM1.R, H929, MM1.S, and RPMI-8226) and 2 mantle cell lymphoma (MCL) cell lines (Z138 and Mino) were treated with WP1130 before assessing Usp9x activity and Mcl-1 protein levels. As shown in Figure 1A-B, WP1130 induced time-dependent inhibition of Usp9x activity, effective reduction in Mcl-1 protein levels, and induction of apoptosis detected by poly (ADP-ribose) polymerase (PARP) cleavage. Both WP1130 and proteasome inhibitors (MG132 and bortezomib) increased Ub protein content (Figure 1C); however, WP1130-mediated Mcl-1 reduction was blocked by pretreatment with proteasome inhibitors (MG132 and bortezomib), confirming a role for ubiquitin-regulated/proteasomal-dependent destruction of Mcl-1 in WP1130-treated cells.5
WP1130 inhibits DUB activity and reduces Mcl-1 protein expression in myeloma cells. (A) Myeloma cells (as indicated) were treated with 5 μM WP1130 for 0, 2, or 4 hours before DUB activity was assessed by hemagglutinin-ubiquitin vinylsulfone (HA-UbVS) labeling. The labeled DUBs were detected by immunoblotting with anti-HA antibody. HA-labeled Usp9x is denoted. The protein lysates were also immunoblotted for total Ub, Usp9x, Mcl-1, and PARP. β-actin was used as a protein loading control. (B) Z138 and Mino cells were treated as noted before cell lysates were screened for Usp9x activity (HA-labeled Usp9x), Usp9x, Mcl-1, PARP, and actin protein level by blotting. (C) Myeloma cells were treated with WP1130 alone or were pretreated for 30 minutes with proteasome inhibitors MG-132 (5 µM) or bortezomib (Bz) (50 nM) prior to WP1130 treatment. Lysates were prepared after 4 hours and immunoblotted for Ub and Mcl-1. Equal protein load was confirmed by immunoblotting with β-actin.
WP1130 inhibits DUB activity and reduces Mcl-1 protein expression in myeloma cells. (A) Myeloma cells (as indicated) were treated with 5 μM WP1130 for 0, 2, or 4 hours before DUB activity was assessed by hemagglutinin-ubiquitin vinylsulfone (HA-UbVS) labeling. The labeled DUBs were detected by immunoblotting with anti-HA antibody. HA-labeled Usp9x is denoted. The protein lysates were also immunoblotted for total Ub, Usp9x, Mcl-1, and PARP. β-actin was used as a protein loading control. (B) Z138 and Mino cells were treated as noted before cell lysates were screened for Usp9x activity (HA-labeled Usp9x), Usp9x, Mcl-1, PARP, and actin protein level by blotting. (C) Myeloma cells were treated with WP1130 alone or were pretreated for 30 minutes with proteasome inhibitors MG-132 (5 µM) or bortezomib (Bz) (50 nM) prior to WP1130 treatment. Lysates were prepared after 4 hours and immunoblotted for Ub and Mcl-1. Equal protein load was confirmed by immunoblotting with β-actin.
To determine whether Usp9x activity and Mcl-1 levels were measureable and sensitive to inhibition by WP1130 in primary tumor cells, freshly isolated CD138+ cells from newly diagnosed or drug-refractory myeloma patients (Figure 2A-B) (characteristics provided in supplemental Table 1) or highly purified chronic lymphocytic leukemia (CLL) cells from patients (Figure 2C)16,17 were treated with WP1130 for 4 hours before assessing total DUB activity. WP1130 displayed partially selective Usp9x inhibitory activity and was associated with reduced expression of Mcl-1 protein (Figure 2A). WP1130 dose-dependent DUB inhibition was also assessed in patient samples (MM5500 and CLL60) if there was sufficient sample, and a close association between Usp9x inhibition and Mcl-1 destruction was confirmed (Figure 2B-C). DUB inhibition by WP1130 was also associated with a reduction in myeloma and plasmacytoma cell survival as assessed by the percentage of CD138+/annexin V–negative cell population (Figure 2A-B).
WP1130 inhibits DUB activity in primary B-cell tumors. (A) Top: primary myeloma cells were treated with 5 μM WP1130 for 4 hours before assessing DUB activity as described in the legend for Figure 1. The HA-labeled Usp9x protein is denoted. Middle: total protein levels for Usp9x, Mcl-1, and β-actin were also assessed by immunoblotting. Bottom: after 24 hours, aliquots of the same cells were subjected to staining with annexin V-fluorescein isothiocyanate (FITC)/propidium iodide and analyzed by flow cytometry. Viability of untreated control cells was set at 100%, and relative reduction in survival in WP1130-treated cells is noted below each blot. (B) Plasmacytoma cells from a newly diagnosed patient were treated with 0, 1.25, 2.5, and 5.0 µM WP1130 for 4 hours before measuring DUB activity by HA-UbVS labeling, and the labeled DUBs were visualized by anti-HA immunoblotting. Mcl-1 and actin protein levels were also measured by blotting. Viability of CD138+ cells was determined as described in (A). (C) Primary CLL cells were left untreated or were treated with WP1130 as described above. Patient CLL60 cells were treated with the WP1130 concentration indicated before assessing DUB activity and Usp9x, Mcl-1, and β-actin expression by immunoblotting.
WP1130 inhibits DUB activity in primary B-cell tumors. (A) Top: primary myeloma cells were treated with 5 μM WP1130 for 4 hours before assessing DUB activity as described in the legend for Figure 1. The HA-labeled Usp9x protein is denoted. Middle: total protein levels for Usp9x, Mcl-1, and β-actin were also assessed by immunoblotting. Bottom: after 24 hours, aliquots of the same cells were subjected to staining with annexin V-fluorescein isothiocyanate (FITC)/propidium iodide and analyzed by flow cytometry. Viability of untreated control cells was set at 100%, and relative reduction in survival in WP1130-treated cells is noted below each blot. (B) Plasmacytoma cells from a newly diagnosed patient were treated with 0, 1.25, 2.5, and 5.0 µM WP1130 for 4 hours before measuring DUB activity by HA-UbVS labeling, and the labeled DUBs were visualized by anti-HA immunoblotting. Mcl-1 and actin protein levels were also measured by blotting. Viability of CD138+ cells was determined as described in (A). (C) Primary CLL cells were left untreated or were treated with WP1130 as described above. Patient CLL60 cells were treated with the WP1130 concentration indicated before assessing DUB activity and Usp9x, Mcl-1, and β-actin expression by immunoblotting.
Effect of Usp9x KD on myeloma cell survival
To assess the role of Usp9x in myeloma, short hairpin RNA (shRNA) retroviral vectors targeting control (LMP) or Usp9x sequences (HS1, HS2) were expressed in MM1.S cells, and effects on Usp9x levels and cell survival were examined. Both HS1 and HS2 were effective in reducing Usp9x levels and MM1.S cell survival when compared with LMP-expressing cells (supplemental Figure 1A). To examine whether the Usp9x KD triggered a reduction in cell survival mediated through Mcl-1, subsequent studies compared cell survival in control, Usp9x KD, and Mcl-1 KD MM1.S and H929 cells. As shown in Figure 3A, Usp9x KD H929 cells show a slight reduction in Mcl-1 basal levels, but no effect was observed in Usp9x KD MM1.S cells. Usp9x KD in both H929 and MM1.S cells was not as effective as direct Mcl-1 KD in increasing apoptosis as detected by annexin positivity (Figure 3A, right). We also noted that viable cells emerged from the Usp9x KD that had reduced proliferative activity (Figure 3B, top), primarily because of a block in exit from G2→M (Figure 3B, bottom). After 4 to 5 division cycles, Usp9x KD cells with low-level Usp9x protein expression remained 90% to 95% viable but grew at a reduced rate when compared with control KD cells. We assessed basal and WP1130-sensitive DUB activity in control KD and chronic Usp9x KD H929 cells and determined that, although Usp9x protein levels were reduced by >90%, DUB activity associated with Usp9x (Figure 3C, top band; compare lanes 1 and 3) could not be distinguished between control KD and Usp9x KD cells.
Usp9x KD reduces myeloma cell survival and proliferation and upregulates Usp24. (A) Immunoblot confirming Usp9x and Mcl-1 KD in H929 and MM1.S myeloma cells. Usp9x or Mcl-1 were knocked down by control or specific shRNA retrovirus and were selected for puromycin resistance before cell lysates were examined for the protein indicated (left) or cell survival by measurement of annexin positivity (right). The results represent the average ± standard deviation (SD) of triplicate assays. (B) Top: equal numbers of scrambled control and Usp9x KD cells were plated on day 0, and cell numbers were assessed daily for 3 days. The numbers represent the average ± SD of triplicate cell counts. Bottom: cell cycle analysis was performed on control (red bars) and Usp9x KD (blue bars) cells obtained at day 3 following initial plating. Cell cycle phase was determined by propidium iodide staining and flow analysis, and each value represents the average ± SD of triplicate assessments. P values are shown above each pair. (C) DUB activity was assessed (as described in the legend for Figure 1) in control KD and Usp9x KD H929 cells left untreated or treated with 5 μM WP1130 for 4 hours. Usp9x, Mcl-1, PARP, and actin were also assessed by immunoblotting. HA-UbVS–labeled Usp9x and Usp24 are denoted (migration at similar molecular size). (D) Lysates from control (Con) and chronic Usp9x KD myeloma cells were subjected to immunoblotting for Usp9x, Usp24, and actin. (E) Colon cancer cell lines with (Usp9x−/−) or without (Usp9x+/+) full Usp9x gene disruption were assessed for DUB activity by HA-UbVS labeling (top) of equal protein lysates. Lysates were also probed for Usp9x, Usp24, and β-actin protein expression. (F) Equal protein lysates from myeloma cells were subjected to immunodepletion of the DUB indicated with anti-Usp9x or Usp24 (or control antibody [Ab]) before measurement of total DUB activity by HA-UbVS labeling. The resultant loss of Usp9x or Usp24 from each supernatant was determined by immunoblotting. (G) Usp9x (left) and Usp24 (right) gene expression in myeloma patient samples was assessed by quantitative polymerase chain reaction and compared with H929 control KD and Usp9x KD cells (green bars). Samples designated as high risk (red bars) or good risk (blue bars) of progressive disease based on their cytogenetic profiles are denoted. Each bar represents the average ± SD of triplicate samples. (H) Usp9x and Usp24 protein expression was assessed by immunoblotting cell lysates derived from myeloma (H929) and MCL (Z138) cell lines, normal tonsilar B cells, or samples derived from two newly diagnosed or two relapsed myeloma patients. Actin was blotted as a protein loading control. (I) Tissue microarrays from normal tissue (spleen and tonsil) and confirmed pathologic samples from MCL (left) or CLL (right) were subjected to immunostaining for Usp9x and Usp24. Examples of tumor samples with intermediate and high expression in the same specimen are shown. The Usp9x/Usp24 staining pattern in additional samples is shown in supplemental Figure 2. **P < .01; ***P < .005.
Usp9x KD reduces myeloma cell survival and proliferation and upregulates Usp24. (A) Immunoblot confirming Usp9x and Mcl-1 KD in H929 and MM1.S myeloma cells. Usp9x or Mcl-1 were knocked down by control or specific shRNA retrovirus and were selected for puromycin resistance before cell lysates were examined for the protein indicated (left) or cell survival by measurement of annexin positivity (right). The results represent the average ± standard deviation (SD) of triplicate assays. (B) Top: equal numbers of scrambled control and Usp9x KD cells were plated on day 0, and cell numbers were assessed daily for 3 days. The numbers represent the average ± SD of triplicate cell counts. Bottom: cell cycle analysis was performed on control (red bars) and Usp9x KD (blue bars) cells obtained at day 3 following initial plating. Cell cycle phase was determined by propidium iodide staining and flow analysis, and each value represents the average ± SD of triplicate assessments. P values are shown above each pair. (C) DUB activity was assessed (as described in the legend for Figure 1) in control KD and Usp9x KD H929 cells left untreated or treated with 5 μM WP1130 for 4 hours. Usp9x, Mcl-1, PARP, and actin were also assessed by immunoblotting. HA-UbVS–labeled Usp9x and Usp24 are denoted (migration at similar molecular size). (D) Lysates from control (Con) and chronic Usp9x KD myeloma cells were subjected to immunoblotting for Usp9x, Usp24, and actin. (E) Colon cancer cell lines with (Usp9x−/−) or without (Usp9x+/+) full Usp9x gene disruption were assessed for DUB activity by HA-UbVS labeling (top) of equal protein lysates. Lysates were also probed for Usp9x, Usp24, and β-actin protein expression. (F) Equal protein lysates from myeloma cells were subjected to immunodepletion of the DUB indicated with anti-Usp9x or Usp24 (or control antibody [Ab]) before measurement of total DUB activity by HA-UbVS labeling. The resultant loss of Usp9x or Usp24 from each supernatant was determined by immunoblotting. (G) Usp9x (left) and Usp24 (right) gene expression in myeloma patient samples was assessed by quantitative polymerase chain reaction and compared with H929 control KD and Usp9x KD cells (green bars). Samples designated as high risk (red bars) or good risk (blue bars) of progressive disease based on their cytogenetic profiles are denoted. Each bar represents the average ± SD of triplicate samples. (H) Usp9x and Usp24 protein expression was assessed by immunoblotting cell lysates derived from myeloma (H929) and MCL (Z138) cell lines, normal tonsilar B cells, or samples derived from two newly diagnosed or two relapsed myeloma patients. Actin was blotted as a protein loading control. (I) Tissue microarrays from normal tissue (spleen and tonsil) and confirmed pathologic samples from MCL (left) or CLL (right) were subjected to immunostaining for Usp9x and Usp24. Examples of tumor samples with intermediate and high expression in the same specimen are shown. The Usp9x/Usp24 staining pattern in additional samples is shown in supplemental Figure 2. **P < .01; ***P < .005.
WP1130 treatment resulted in complete inhibition of high-molecular-weight (HMW) DUB activity and reduced Mcl-1 protein expression in both control KD and Usp9x KD cells. These results suggested that complete ablation of Usp9x activity was necessary to suppress myeloma cell survival and reduce Mcl-1 protein levels. These results were also consistent with the possibility of expression of a WP1130-sensitive DUB with Usp9x-like molecular characteristics that compensate for Usp9x deficiency. Usp24 bears a high degree of biochemical and sequence similarity to Usp9x.18 Lysates from H929 and MM1.S chronic Usp9x KD cells showed increased Usp24 protein expression (Figure 3D) when compared with control KD cells. To determine whether these changes were an artifact of Usp9x shRNA-mediated KD, we assessed DUB activity and Usp24 expression in two colon cancer cell lines with full genetic disruption of the Usp9x gene.6 HCT116Usp9x−/− and DLD-1Usp9x−/− cells had no detectable Usp9x expression but retained HMW DUB activity that was associated with increased Usp24 protein expression (Figure 3E), suggesting that Usp24 compensates for loss of Usp9x in multiple cell types. To confirm Usp9x and Usp24 expression and activity in myeloma cells, each DUB was subjected to immunodepletion prior to assessing DUB activity in H929 and MM1.S myeloma cells. As shown in Figure 3F, immunodepletion of either DUB resulted in a partial reduction of HMW DUB activity (HA-Usp9x/Usp24) in both H929 and MM1.S cells, suggesting that both DUBs are active in myeloma cells.
To assess potential clinical relevance, 11 primary myeloma samples from patient tumors with high-risk or good-risk cytogenetics were evaluated for Usp9x and Usp24 gene expression by quantitative polymerase chain reaction and compared with control KD or Usp9x KD H929 cells (Figure 3G). Usp9x expression was elevated (2- to 100-fold) compared with the H929 myeloma cell line and was independent of the cytogenetic profile. Similarly, Usp24 expression was above H929 cellular levels in 8 of 11 samples, with all 8 co-expressing Usp9x and Usp24. Oncomine (Compendia Bioscience, Ann Arbor, MI) data acquired from Zhan et al19 showed that the expression of both Usp9x and Usp24 was increased in monoclonal gammopathy of undetermined significance (n = 44) and myeloma (n = 12) bone marrow when compared with normal bone marrow (n = 22) (supplemental Figure 1C).
Usp9x and Usp24 protein expression was compared in 2 newly diagnosed patients, 2 relapsed myeloma patients, 2 normal tonsilar B-cell preparations, and representative (H929 and Z138) cell lines (Figure 3H). All 4 myeloma samples had Usp9x protein expression levels that were greater than those detected in tonsilar cells and established cell lines. Elevated Usp24 expression was detected in Z138 cells, newly diagnosed patient samples, and drug-refractory patient samples. Direct analysis of DUB activity in tonsilar CD138+ plasma cells and CD138+ myeloma and plasma cell leukemia showed increased Usp9x/Usp24 activity in tumor vs normal plasma cells (supplemental Figure 1D). Usp24 protein was below detection in normal plasma cells and one myeloma sample. To determine whether other B-cell tumors also express these proteins, tissue microarrays assembled from patients with MCL and CLL were stained for either DUB and compared with various normal tissues. In MCL, 6 of 14 and 7 of 14 samples were positive for elevated Usp9x or Usp24, respectively, when compared with spleen or tonsil staining (supplemental Figure 2A). Three samples were positive for both DUBs. In CLL, 4 of 14 and 3 of 14 samples stained positive for elevated Usp9x and Usp24 expression, respectively, with 2 of 14 samples demonstrating dual positivity (supplemental Figure 2B). Examples of high and intermediate Usp9x and Usp24 positivity in MCL and CLL tumors are shown in Figure 3I. These results suggest that B-cell tumors commonly overexpress one or both of these DUBs.
Usp24 is essential for myeloma cell survival
To assess the role of Usp24 in myeloma, two shRNA constructs were used to execute Usp24 KD in H929 and MM1.S myeloma cell lines (note that both 24-1 and 24-2 were effective; supplemental Figure 1A). As shown in Figure 4A (left), Usp24 levels were reduced by shRNA in both cell lines and were associated with reduction in Mcl-1 protein expression. Furthermore, Usp24 KD reduced cell survival in both H929 and MM1.S cells to a level greater than that achieved by direct Mcl-1 KD (Figure 4A, right), suggesting that Usp24 plays a role in Mcl-1 stability and myeloma cell survival.
Usp24 KD reduces Mcl-1 levels and myeloma cell survival. (A) Usp24 or Mcl-1 were knocked down by control or specific shRNA retrovirus and selected for puromycin resistance before cell lysates were examined for the protein indicated (left) or cell survival by measurement of annexin positivity (right). The results represent the average ± SD of triplicate assays. (B) RPMI-8226 myeloma cell lines were subjected to siRNA-mediated KD of control (luciferase), Usp9x, or Usp24 before assessing the protein indicated by immunoblotting (left). Increase in annexin positivity was assessed as a measure of cell viability. The values represent the average ± SD of 3 replicates from 2 independent experiments. **P < .01; ***P < .005.
Usp24 KD reduces Mcl-1 levels and myeloma cell survival. (A) Usp24 or Mcl-1 were knocked down by control or specific shRNA retrovirus and selected for puromycin resistance before cell lysates were examined for the protein indicated (left) or cell survival by measurement of annexin positivity (right). The results represent the average ± SD of triplicate assays. (B) RPMI-8226 myeloma cell lines were subjected to siRNA-mediated KD of control (luciferase), Usp9x, or Usp24 before assessing the protein indicated by immunoblotting (left). Increase in annexin positivity was assessed as a measure of cell viability. The values represent the average ± SD of 3 replicates from 2 independent experiments. **P < .01; ***P < .005.
To confirm a complementary role of Usp24 and Usp9x in myeloma cell survival, small interfering RNA (siRNA) (SmartPool) was used to knock down either Usp9x or Usp24 in RPMI-8226 cells, and the effect on Mcl-1 protein levels and cell survival was examined. Both Usp9x and Usp24 siRNA reduced Mcl-1 protein expression in RPMI-8226 cells (Figure 4B, left). In addition, cell survival was reduced by KD of either DUB, with greater levels of annexin staining consistently noted in Usp24 silenced cells (Figure 4B, right). These results suggest that Usp24 plays a prominent role in myeloma cell survival and Mcl-1 regulation, and increased Usp24 expression may compensate cell survival in the absence of Usp9x. Increased ubiquitination of immunoprecipitated Mcl-1 was observed in Usp24 KD cell lysate when compared with control lysates, consistent with a role for Usp24 in Mcl-1 deubiquitination (supplemental Figure 1E).
Usp9x and Usp24 interact with Mcl-1
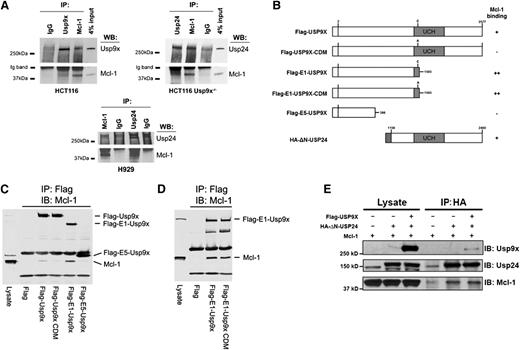
Previous studies have demonstrated that Usp9x complexes with the N-terminus of Mcl-1.5 After we observed that Usp24 upregulation modulated Mcl-1 protein expression levels in Usp9x-silenced cells (Figure 3), we wanted to determine whether Usp24 also binds Mcl-1 and determine whether this binding is constrained by the presence of Usp9x. Immunoprecipitation assays confirmed the association of endogenous Mcl-1 with Usp9x and Usp24 as demonstrated by the recovery of Mcl-1 protein after direct Usp9x or Usp24 pulldown and reciprocal recovery of either DUB by immunoprecipitation of Mcl-1 (Figure 5A). To define determinants on Usp9x that are essential for Mcl-1 binding, full-length and deletion constructs of Flag-Usp9x (Figure 5B) were co-expressed with Mcl-1 in HEK293T cells and subjected to Flag-pulldown, followed by blotting for Mcl-1. Full-length Usp9x with catalytic activity demonstrated weak but detectable Mcl-1 binding, which was absent in cells expressing a catalytic domain mutant (inactive) Usp9x protein (Figure 5C). Partial deletion of the catalytic domain and the entire C-terminus of Usp9x (Flag-E1-USP9X) resulted in increased recovery of Mcl-1 whereas expression of just the N-terminal domain of Usp9x (Flag-E5-USP9X) did not retain Mcl-1 binding (Figure 5C, right lane). Because the active site Cys (C1566) was retained in the E1 construct, we compared Mcl-1 binding to an E1 construct expressing Cys or Ala at position 1566 and did not detect an impact on Mcl-1 association (Figure 5D). These results suggest that Mcl-1 binding is mediated through primary interaction with the central core of Usp9x and can be attenuated by other domains or catalytic activity.
Usp9x and Usp24 associate with Mcl-1. (A) HCT116 Usp9x+/+ (wild-type) and Usp9x−/− cell lysates were subjected to immunoprecipitation (IP) with normal immunoglobulin G (IgG), anti-Mcl-1, anti-Usp9x, or anti-Usp24 before immunoblotting (IB) the pulldown for Mcl-1, Usp24, or Usp9x (top 2 panels). H929 cell lysates were immunoprecipitated with normal IgG, anti-Mcl-1, or anti-Usp24 and immunoblotted for Mcl-1 or Usp24 (bottom panel). (B) Organization of the Usp9x and Usp24 constructs used in pulldown experiments and summary of their Mcl-1 binding activity. The position of the catalytic domain ubiquitin C-terminal hydrolase (UCH) is shown by shading. Flag sequence and HA epitope in the constructs are positioned at the N-termini. Numbers and letters designate highlighted amino acids. (C) Flag-tagged control or Flag-tagged full-length and deletion constructs of Usp9x (illustrated in panel B) were expressed in HEK293T cells and subjected to Flag immunoprecipitation followed by immunoblotting of Mcl-1. (D) Flag-tagged control or deletion constructs of Flag-E1-Usp9x retaining the active site cysteine (C1566) or a catalytic domain mutant (CDM-C1566A) were expressed in HEK293T cells and subjected to Flag pulldown followed by Mcl-1 immunoblotting. (E) HEK293T cells were transfected with the constructs designated before the input lysates and anti-HA (Usp24) immunoprecipitates were subjected to immunoblotting with antibodies against Usp9x, Usp24, and Mcl-1. WB, western blotting.
Usp9x and Usp24 associate with Mcl-1. (A) HCT116 Usp9x+/+ (wild-type) and Usp9x−/− cell lysates were subjected to immunoprecipitation (IP) with normal immunoglobulin G (IgG), anti-Mcl-1, anti-Usp9x, or anti-Usp24 before immunoblotting (IB) the pulldown for Mcl-1, Usp24, or Usp9x (top 2 panels). H929 cell lysates were immunoprecipitated with normal IgG, anti-Mcl-1, or anti-Usp24 and immunoblotted for Mcl-1 or Usp24 (bottom panel). (B) Organization of the Usp9x and Usp24 constructs used in pulldown experiments and summary of their Mcl-1 binding activity. The position of the catalytic domain ubiquitin C-terminal hydrolase (UCH) is shown by shading. Flag sequence and HA epitope in the constructs are positioned at the N-termini. Numbers and letters designate highlighted amino acids. (C) Flag-tagged control or Flag-tagged full-length and deletion constructs of Usp9x (illustrated in panel B) were expressed in HEK293T cells and subjected to Flag immunoprecipitation followed by immunoblotting of Mcl-1. (D) Flag-tagged control or deletion constructs of Flag-E1-Usp9x retaining the active site cysteine (C1566) or a catalytic domain mutant (CDM-C1566A) were expressed in HEK293T cells and subjected to Flag pulldown followed by Mcl-1 immunoblotting. (E) HEK293T cells were transfected with the constructs designated before the input lysates and anti-HA (Usp24) immunoprecipitates were subjected to immunoblotting with antibodies against Usp9x, Usp24, and Mcl-1. WB, western blotting.
Because Usp9x and Usp24 are closely related, we constructed a Usp24 expression vector that retained a portion of its central core and the entire C-terminus of Usp24 (HA-ΔN-Usp24) and assessed its Mcl-1 interaction in the presence or absence of full-length Flag-Usp9x (Figure 5B). As shown in Figure 5E, HA-ΔN-Usp24 retained Mcl-1 binding activity. In addition, co-expression with Usp9x led to recovery of Usp9x in the Usp24/Mcl-1 complex. Together, these results suggest that Usp9x and Usp24 are capable of Mcl-1 binding.
Small-molecule–mediated Usp9x/Usp24 inhibition
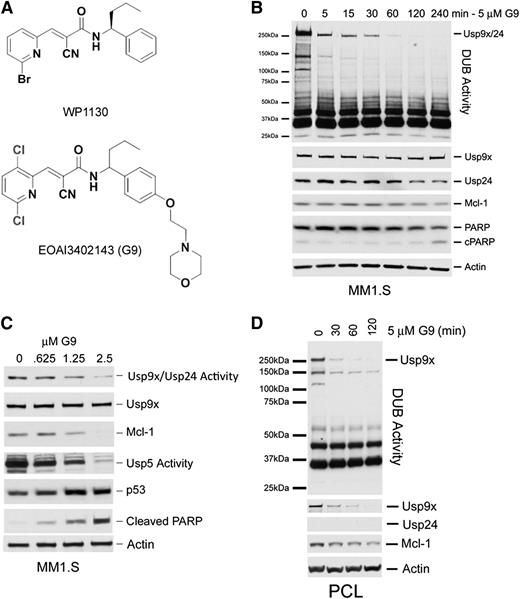
WP1130 was the first small molecule described with Usp9x inhibitory activity and has shown activity against specific tumors in vitro and in vivo.9,12,20 However, other DUBs are also WP1130 targets, with some recognized benefits and risks associated with the use of a partially selective DUB inhibitor in a clinical setting.4 The main limitation for WP1130 as a clinical candidate is its low aqueous solubility (∼2.3 μM), which limits its delivery and bioavailability. To improve the solubility while retaining the inhibitory activity of WP1130 against Usp9x, we carried out structure-activity relationship studies to map and define critical and usable chemical modifications to the parent compound. More than 220 compounds were designed, synthesized, and assessed for Usp9x catalytic domain inhibitory activity (as previously described) and compared with WP1130.21 Those retaining catalytic domain inhibition were further screened for aqueous solubility, metabolic stability, pharmacokinetics, cellular activity, and tumor vs normal cell differential apoptosis. One compound emerged (G9; Figure 6A) that was threefold more effective against Usp9x catalytic activity than WP1130 and had increased aqueous solubility and greater cellular Usp9x inhibitory activity than WP1130. G9 also demonstrated a therapeutic index of >10, had sufficient metabolic stability, and had adequate pharmacokinetics to allow assessment of its in vivo antitumor activity (supplemental Table 2, supplemental Figure 3). G9 displayed nM apoptotic activity against other myeloma cell lines and diffuse large B-cell lymphomas (supplemental Figure 4). G9 treatment of MM1.S cells resulted in the rapid onset and sustained inhibition of Usp9x and Usp24 (coincident migration of both DUBs) activity (Figure 6B). We confirmed Usp24 as a G9 target by using HCT116 Usp9x−/− cells (supplemental Figure 3D). Other DUBs, such as Usp5, were also affected by G9 in these and other cell types, as was recently reported.22 Dose-dependent inhibition of Usp9x/Usp24 and Usp5 were noted in G9-treated MM1.S cells (Figure 6C), which was associated with a reduction in Mcl-1 levels, an increase in p53 stability, and the rapid onset of apoptosis (cleaved PARP). We also noted a similar pattern of Usp9x inhibition and reduction of Mcl-1 levels in a freshly prepared G9-treated plasma cell leukemia sample derived from a newly diagnosed patient (Figure 6D). Interestingly, we did not detect Usp24 expression in this sample, but we did note loss of Usp9x protein after G9 treatment, suggesting that Usp9x may be susceptible to decay in some cell types.
Structure and activity of a novel DUB inhibitor. (A) Chemical structures of WP1130 and G9. (B) MM1.S cells were treated as described with 5 µM G9 before assessing DUB activity (top) and total protein expression of Usp9x, Usp24, Mcl-1, PARP, and β-actin. The HA-labeled Usp9x/Usp24 band is shown at the top. (C) MM1.S cells were treated with the G9 concentration indicated before assessing changes in DUB activity associated with Usp9x/Usp24 and Usp5. Lysates were also immunoblotted for Mcl-1, p53, and cleaved PARP as a measure of apoptosis. Actin was blotted as a loading control. (D) Cells derived from a plasma cell leukemia (PCL) patient were treated with G9 for the time indicated before DUB activity and specific protein levels were determined by HA-UbVS labeling and immunoblotting.
Structure and activity of a novel DUB inhibitor. (A) Chemical structures of WP1130 and G9. (B) MM1.S cells were treated as described with 5 µM G9 before assessing DUB activity (top) and total protein expression of Usp9x, Usp24, Mcl-1, PARP, and β-actin. The HA-labeled Usp9x/Usp24 band is shown at the top. (C) MM1.S cells were treated with the G9 concentration indicated before assessing changes in DUB activity associated with Usp9x/Usp24 and Usp5. Lysates were also immunoblotted for Mcl-1, p53, and cleaved PARP as a measure of apoptosis. Actin was blotted as a loading control. (D) Cells derived from a plasma cell leukemia (PCL) patient were treated with G9 for the time indicated before DUB activity and specific protein levels were determined by HA-UbVS labeling and immunoblotting.
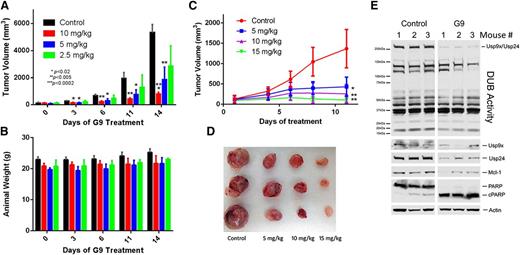
Studies of the mechanisms of action suggest that G9 inhibits Usp9x (and Usp24) activity through a covalent, slowly reversible conjugation with Cys residues essential for substrate binding (unpublished observation, M.Y. and N.D.). Furthermore, in vitro analysis demonstrated that 5 minutes of incubation with G9 is sufficient to mediate long-term Usp9x inhibition. Although G9 has a short half-life, sufficient peak levels were achievable to inhibit Usp9x activity in vivo. Animals with subcutaneous MM1.S tumors were treated with G9 doses ranging from 2.5 to 10 mg/kg intraperitoneally once per day for 2 weeks. As shown in Figure 7A, G9 doses at or above 5 mg/kg significantly suppressed tumor growth, noted as early as 3 days after initiating dosing. Animal weight was not adversely affected throughout the treatment interval (Figure 7B). Tumor regrowth was observed after discontinuation of G9 dosing (supplemental Figure 5A), which demonstrated a dose-dependent delay in resumption of tumor growth after stopping G9 treatment.
In vivo antimyeloma activity of a DUB inhibitor. (A) NSG mice with equal size-matched dorsal MM1.S tumors were treated with the indicated dose of G9 every day for 2 weeks. Tumor size was measured at the time points indicated. Each bar represents the average ± SD of tumor size from 5 animals. P values are shown within the figure. (B) Animal body weight was measured in control and G9-treated mice from Figure 7A. Body weight was measured at the interval noted, and each bar represents the average ± SD from 5 mice per group. (C) MM1.S tumors growing in NSG mice (3 per group) were treated daily with the indicated concentration of G9. Tumor volumes at each designated interval were recorded. The average tumor volume ± SD from 3 mice per group is shown. P values are the same as described in Figure 7A. (D) Tumors from Figure 7C were excised at the conclusion of the study (day12) and photographed. (E) One hour after the final G9 treatment, tumors from control and G9-treated (15 mg/kg) mice were excised, quick frozen in liquid nitrogen, and subjected to lysis. Equal protein lysates from these tumors was subjected to DUB activity assessment (top; as in Figure 1A) and immunoblotted for the protein indicated. The top band represents HA-UbVS–labeled Usp9x and Usp24 (co-migrating proteins).
In vivo antimyeloma activity of a DUB inhibitor. (A) NSG mice with equal size-matched dorsal MM1.S tumors were treated with the indicated dose of G9 every day for 2 weeks. Tumor size was measured at the time points indicated. Each bar represents the average ± SD of tumor size from 5 animals. P values are shown within the figure. (B) Animal body weight was measured in control and G9-treated mice from Figure 7A. Body weight was measured at the interval noted, and each bar represents the average ± SD from 5 mice per group. (C) MM1.S tumors growing in NSG mice (3 per group) were treated daily with the indicated concentration of G9. Tumor volumes at each designated interval were recorded. The average tumor volume ± SD from 3 mice per group is shown. P values are the same as described in Figure 7A. (D) Tumors from Figure 7C were excised at the conclusion of the study (day12) and photographed. (E) One hour after the final G9 treatment, tumors from control and G9-treated (15 mg/kg) mice were excised, quick frozen in liquid nitrogen, and subjected to lysis. Equal protein lysates from these tumors was subjected to DUB activity assessment (top; as in Figure 1A) and immunoblotted for the protein indicated. The top band represents HA-UbVS–labeled Usp9x and Usp24 (co-migrating proteins).
Additional studies were initiated to test higher doses and the impact of G9 on tumor Usp9x activity. As shown in Figure 7C, tumor growth was significantly suppressed at all doses tested, with evidence for some tumor regression at the 15-mg/kg dose. A second trial of antitumor assessment at the 15-mg/kg G9 dose level confirmed evidence for tumor regression (supplemental Figure 5B). At the conclusion of the treatment interval, tumors were excised and photographed (Figure 7D), and tumor lysates were assessed for DUB activity and blotted for several proteins. The results demonstrate that G9 suppressed Usp9x/Usp24 activity and reduced the protein expression levels of Usp9x, Usp24, and Mcl-1 in vivo (Figure 7E). PARP cleavage was also activated by G9. Together, these results suggest that G9 targets Usp9x and Usp24 (as well as additional DUBs) in vivo and is able to safely reduce the growth or regress MM1.S myeloma tumors.
Discussion
In this study, we demonstrate that WP1130 effectively inhibits Usp9x in multiple B-cell malignancies with a wide spectrum of basal Usp9x expression or enzymatic activities. However, WP1130 is not absolutely Usp9x specific, which may not be a major concern because clinical evidence suggests that highly specific inhibitors are more likely to engage multiple resistance mechanisms.23,24 In addition, emerging data on the role of other cancer-associated DUBs supports the benefit of multitarget inhibition.4 As was recently demonstrated, the WP1130 chemotype may be useful against various cancers with overexpressed or activated Usp9x.12
Mcl-1 was previously reported to be a substrate of Usp9x in some tumors.5 We sought to confirm a direct role for Usp9x as an Mcl-1 regulator by assessing the response to Usp9x KD and identified several unexpected outcomes. Although short-term selection of Usp9x KD myeloma cells (shRNA- and siRNA-mediated KD) were shown to have an impact on cell survival, this effect was not sustained and did not appear to be fully dependent on Mcl-1. However, Usp9x KD caused sustained suppression of myeloma cell growth because cells were unable to efficiently exit G2→M in the absence of Usp9x. These observations suggested that some Usp9x functions may be redundant, and cellular adaptation may ensure cell survival in the absence of Usp9x. DUB activity measurements in chronic Usp9x KD cells or those with full genetic Usp9x disruption supported evidence of a role for Usp24 in that adaptation because these DUBs are closely related and because Usp24 expression or activity was increased in Usp9x-deficient cells.18,25 Importantly, Usp24 KD was more effective in controlling both myeloma cell survival and Mcl-1 protein levels, suggesting that WP1130 activity against Mcl-1 is manifested through dual Usp9x/Usp24 inhibition. Indeed, we have accruing evidence that both DUBs are capable of Mcl-1 association, but the extent of the association may be controlled by more than recognition by the catalytic domain. Because the C-terminus of Usp9x was previously shown to bind other substrates,26 our mapping studies suggest that Mcl-1/Usp9x association may be subject to regulation by the extent of occupancy with other C-terminally bound Usp9x substrates. This may explain the previous inconsistencies associated with definition of Mcl-1 as a Usp9x substrate in cells of distinct origin. Interestingly, Mcl-1–binding maps to a domain distinct from that of other Usp9x substrates (ie, Smurf1),26 suggesting that substrate recognition is not mediated by uniform Usp9x interactions. Furthermore, the central Usp9x domain (aa 385-1593) appears critical for Mcl-1 binding, but finer mapping will be required to fully define critical contacts. This effort has been complicated by our inability to stably express catalytic domain mutants of Usp9x. Interestingly, an N-terminal deletion mutant of Usp24 retained Mcl-1 and Usp9x binding activity, suggesting the existence of a Usp9x/Usp24/Mcl-1 complex. These observations, coupled with the observed effect of Usp24 KD on myeloma cell survival, support the possibility that Usp24 plays a central role in control of both Mcl-1 and Usp9x function, and dual Usp9x/Usp24 inhibition may provide greater antitumor activity than more selective Usp9x or Usp24 inhibitors alone. Our preliminary studies have demonstrated that WP1130 blocks Usp9x activity by forming slowly reversible covalent adducts with cysteines, configuring part of a substrate-binding zinc finger motif (Cys1771, Cys1774) within the catalytic domain of Usp9x. Usp24 retains a homologous zinc finger motif (Cys1859, Cys1863), which may underlie dual Usp9x/Usp24 inhibition by WP1130. This is certainly a mechanistic departure from other DUB inhibitors that either directly target the active site Cys or interfere through binding at allosteric sites.27
To exploit the Usp9x/Usp24 dual inhibitory activity of WP1130, we improved its activity and drug-like properties. G9 was more effective than WP1130 in Usp9x inhibition and Mcl-1 destruction and had an improved therapeutic index against myeloma vs normal CD34+ cells (supplemental Figure 3C).11 Although some improvement was achieved in selectivity, we were unable to fully modify DUB selectivity with G9 because we retained Usp5 inhibition, which was previously noted as a WP1130 target.10,22 However, Usp5 was activated in myeloma cells, and its inhibition increases p53 accumulation, as we have shown in this study and as others have shown in their studies and in other tumor types.10,22,28 Because p53 is infrequently inactivated by mutation in B-cell tumors, it remains a potential effector of apoptosis. Therefore, G9-mediated inhibition of Usp5 and p53 accumulation may amplify the effects of Usp9x/Usp24 inhibition in some tumors rather than pose a disadvantage as a result of partial selection.
Although G9 showed only an acceptable-to-moderate drug metabolism and pharmacokinetics profile, this was overcome by taking advantage of the covalent character of G9, which included a dissociation of pharmacokinetics from pharmacodynamics and making quickly cleared compound more acceptable and even desirable because of a lower systemic exposure and reduced risk of idiosyncratic effects.29-32 Overall, G9 had greater in vivo antitumor activity at lower doses than WP1130 and was well tolerated in animals (based on minimal loss of body weight). To investigate the pharmacodynamics of the compound, we have shown that Usp9x inhibition in vivo is sustained for up to 24 hours after G9 intraperitoneal administration. We have seen dose-dependent tumor regrowth delays after stopping G9 treatment (supplemental Figure 5A), suggesting a slow recovery from reversible covalent inhibition. Further modifications of this current lead compound will result in an optimized candidate for detailed preclinical evaluation.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
The authors thank the patients who agreed to be part of an institutional review board–approved translational study and Dr Fred Bunz (The Johns Hopkins University) for kindly providing Usp9x−/− colon cell lines, and they acknowledge the editorial assistance of Dr Anupama Pal during preparation of this manuscript.
This work was supported by grants from the Leukemia Lymphoma Society (Translational Research Program and Therapeutics Acceleration Program).
Authorship
Contribution: L.F.P., H.S., M.E., M.Y., H.D.S., D.S., M.T., and N.J.D. designed the study; L.F.P., H.S., Y.L., H.P., M.K., M.E., S.M.C., M.Y., H.D.S., and D.S. performed the research; A.J., S.N.M., and M.T. contributed materials; L.F.P., M.E., M.Y., H.D.S., D.S., and N.J.D. analyzed the data; N.J.D. wrote the manuscript; and all authors contributed to data review and provided comments on the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nicholas J. Donato, Department of Internal Medicine/Division of Hematology/Oncology, University of Michigan School of Medicine and Comprehensive Cancer Center, 1500 E Medical Center Dr, Ann Arbor, MI, 48109; e-mail: ndonato@med.umich.edu.
References
Author notes
L.F.P. and H.S. contributed equally to this work.



![Figure 3. Usp9x KD reduces myeloma cell survival and proliferation and upregulates Usp24. (A) Immunoblot confirming Usp9x and Mcl-1 KD in H929 and MM1.S myeloma cells. Usp9x or Mcl-1 were knocked down by control or specific shRNA retrovirus and were selected for puromycin resistance before cell lysates were examined for the protein indicated (left) or cell survival by measurement of annexin positivity (right). The results represent the average ± standard deviation (SD) of triplicate assays. (B) Top: equal numbers of scrambled control and Usp9x KD cells were plated on day 0, and cell numbers were assessed daily for 3 days. The numbers represent the average ± SD of triplicate cell counts. Bottom: cell cycle analysis was performed on control (red bars) and Usp9x KD (blue bars) cells obtained at day 3 following initial plating. Cell cycle phase was determined by propidium iodide staining and flow analysis, and each value represents the average ± SD of triplicate assessments. P values are shown above each pair. (C) DUB activity was assessed (as described in the legend for Figure 1) in control KD and Usp9x KD H929 cells left untreated or treated with 5 μM WP1130 for 4 hours. Usp9x, Mcl-1, PARP, and actin were also assessed by immunoblotting. HA-UbVS–labeled Usp9x and Usp24 are denoted (migration at similar molecular size). (D) Lysates from control (Con) and chronic Usp9x KD myeloma cells were subjected to immunoblotting for Usp9x, Usp24, and actin. (E) Colon cancer cell lines with (Usp9x−/−) or without (Usp9x+/+) full Usp9x gene disruption were assessed for DUB activity by HA-UbVS labeling (top) of equal protein lysates. Lysates were also probed for Usp9x, Usp24, and β-actin protein expression. (F) Equal protein lysates from myeloma cells were subjected to immunodepletion of the DUB indicated with anti-Usp9x or Usp24 (or control antibody [Ab]) before measurement of total DUB activity by HA-UbVS labeling. The resultant loss of Usp9x or Usp24 from each supernatant was determined by immunoblotting. (G) Usp9x (left) and Usp24 (right) gene expression in myeloma patient samples was assessed by quantitative polymerase chain reaction and compared with H929 control KD and Usp9x KD cells (green bars). Samples designated as high risk (red bars) or good risk (blue bars) of progressive disease based on their cytogenetic profiles are denoted. Each bar represents the average ± SD of triplicate samples. (H) Usp9x and Usp24 protein expression was assessed by immunoblotting cell lysates derived from myeloma (H929) and MCL (Z138) cell lines, normal tonsilar B cells, or samples derived from two newly diagnosed or two relapsed myeloma patients. Actin was blotted as a protein loading control. (I) Tissue microarrays from normal tissue (spleen and tonsil) and confirmed pathologic samples from MCL (left) or CLL (right) were subjected to immunostaining for Usp9x and Usp24. Examples of tumor samples with intermediate and high expression in the same specimen are shown. The Usp9x/Usp24 staining pattern in additional samples is shown in supplemental Figure 2. **P < .01; ***P < .005.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/23/10.1182_blood-2014-10-605584/4/m_3588f3.jpeg?Expires=1767754480&Signature=PFM9hOeynSJNDhV8PRJzY9vTEoUVifAhyxOuIn77IqkaBeIZZwEzRmgBXN40ZUUBODrjlTgsaaCE~6zceYOePIG8EIJg70N6QqcqeCJYnLGnYEHm1uLXcuZ4quFfAg4A1rToEMcB0mo938-K139FZZPKcgyiCCC~k7jGeBjS3kmnskNLvk~dwKVaxSXo7iD5e6YzYt3c7dSwLw792mbZzBX19bKkZ0e9jpteVzoe51ZFIAji7TUFE0lGEmlTQiSiUx1qbYBS6gA4Fx7nFtVOdr28HIOYFIzAqj8Ty3fHkmYZx-tDE0e~akXNFzqXzoTLCzUwM6yrdRfL4z2MEtt0zA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)