Key Points
Myeloma cell–derived Runx2 promotes myeloma progression.
High levels of Runx2 expression are associated with a high-risk myeloma population.
Abstract
The progression of multiple myeloma (MM) is governed by a network of molecular signals, the majority of which remain to be identified. Recent studies suggest that Runt-related transcription factor 2 (Runx2), a well-known bone-specific transcription factor, is also expressed in solid tumors, where expression promotes both bone metastasis and osteolysis. However, the function of Runx2 in MM remains unknown. The current study demonstrated that (1) Runx2 expression in primary human MM cells is significantly greater than in plasma cells from healthy donors and patients with monoclonal gammopathy of undetermined significance; (2) high levels of Runx2 expression in MM cells are associated with a high-risk population of MM patients; and (3) overexpression of Runx2 in MM cells enhanced tumor growth and disease progression in vivo. Additional studies demonstrated that MM cell–derived Runx2 promotes tumor progression through a mechanism involving the upregulation of Akt/β-catenin/Survivin signaling and enhanced expression of multiple metastatic genes/proteins, as well as the induction of a bone-resident cell-like phenotype in MM cells. Thus, Runx2 expression supports the aggressive phenotype of MM and is correlated with poor prognosis. These data implicate Runx2 expression as a major regulator of MM progression in bone and myeloma bone disease.
Introduction
Multiple myeloma (MM) is a largely incurable B-cell malignancy characterized by the clonal expansion of malignant plasma cells in the bone marrow.1-3 A hallmark of MM is the predominant localization in the bone marrow and the propensity for progression from primary bone sites to new bone sites in both local and distant bones.2,4 Bone disease occurs in ∼90% of patients with MM5 and is the main cause of patient mortality, however, the cellular mechanisms driving MM progression in bone remain largely undefined.
Runt-related transcription factor 2 (Runx2), a member of the runt-related gene family, is a bone-specific transcription factor6,7 considered to be the master regulator of osteoblastogenesis and bone formation.6-9 Accumulating evidence has demonstrated that various solid tumors, such as breast and prostate cancers, also express Runx210-17 and that Runx2 expression is significantly correlated with the development of bone metastasis and subsequent osteolysis.10-19 Despite the evidence in solid tumors, the role of Runx2 in MM remains unclear. In this study, the regulatory roles and mechanisms of Runx2 in the promotion of MM growth, survival, and progression in bone were elucidated.
Materials and methods
Cell lines and cell culture
Mouse myeloma 5TGM1 cells were a gift from Dr Claire M. Edwards (University of Oxford, Oxford, United Kingdom). Human myeloma MM.1R cells were purchased from American Type Culture Collection. All cells were grown in RPMI 1640 medium (Sigma-Aldrich) supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 1% l-glutamine at 37°C and 5% CO2.
Generation of Runx2-overexpressing myeloma cells and luciferase labeling
Mouse Runx2 complementary DNA (cDNA) was subcloned into the pIRES2-EGFP vector (Clontech), which allowed both Runx2 and the enhanced green fluorescent protein (EGFP) to be translated from a single messenger RNA (mRNA). The empty pIRES2-EGFP vector or pIRES2-EGFP/Runx2 construct was electrotransfected into 5TGM1 mouse myeloma cells using program DN-100 on the 4D-Nucleofector system and the Amaxa SF cell line 4D-nucleofector X kit (Lonza). Transfected cells were selected with G418 (500 μg/mL) and GFP sorting by flow cytometry. Overexpression of Runx2 in pIRES2-EGFP/Runx2–transfected cells was confirmed by western blotting. 5TGM1 control and Runx2 knock-in (Runx2 k/in) cells were then transfected with lentivirus carrying luciferase and were selected with blasticidin (10 μg/mL; Sigma-Aldrich). Vector control and Runx2 k/in cells with equal luciferase expression were selected for specific experiments (supplemental Figure 1, see supplemental Data available on the Blood Web site). Both Runx2 k/in and vector control cells secreted similar levels of immunoglobulin G2bκ (IgG2bκ) into the conditioned medium (CM) (supplemental Figure 2).
Knockdown of Runx2 in MM cell lines by Runx2 shRNA
Runx2 expression was knocked down in human MM.1R or mouse 5TGM1 myeloma cells by transduction with specific Runx2 short hairpin RNA (shRNA) lentiviruses 90 and 91 or nontargeted (NT) shRNA control (Sigma-Aldrich). The cells were transduced in 96-well plates, in triplicate, according to the manufacturer’s protocol. After transduction, cells were selected with puromycin (5 μg/mL; Sigma-Aldrich) and the extent of Runx2 knockdown (k/d) was determined by western blotting. Runx2 k/d did not affect IgG2bκ secretion from MM cells (supplemental Figure 2).
Western blot analysis
Equal amounts of protein (80 μg) were subjected to 4% to 12% gradient sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gels (Bio-Rad) and transferred to nitrocellulose membranes (Schleicher and Schuell).1 Transferred proteins were probed with appropriate antibodies (supplemental Table 2) and visualized using an enhanced chemiluminescence system (Amersham Biosciences). Western blots were quantified by NIH ImageJ software version 1.45 (rsb.info.nih.gov/ij).
RNA sequencing and real-time PCR
Total RNA was isolated from 5TGM1 nontargeted control (NT) and Runx2 k/d as well as 5TGM1 control and Runx2 k/in cells using RNeasy Mini kits (Qiagen Inc). cDNA was synthesized using reverse transcriptase (Clontech). Gene expression profiles (GEPs) were generated by RNA sequencing at the University of Alabama at Birmingham (UAB) Heflin Center Genomics Core. Changes in genes of interest identified by RNA sequencing were confirmed by real-time polymerase chain reaction (PCR), using appropriate specific primers (primer sequences are listed in supplemental Table 1) and SYBR Green Supermix (Bio-Rad). Gene expression data are expressed relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) for mouse cells and 28S ribosomal RNA (rRNA) for human cells according to the comparative cycle threshold (CT) method.20
Evaluation of Runx2 k/in and Runx2 k/d cells in animal models of MM
All animal studies were performed in accordance with UAB and National Insitittues of Health (NIH) guidelines after institutional review and approval.
SCID-s.c. model.
Six-week-old male severe combined immunodeficient (SCID) (BALB/cByJ-scid/scid) mice were purchased from Harlan Laboratories Inc. NT control or Runx2 k/d MM.1R human MM cells (106 in 100 μL of phosphate-buffered saline [PBS]) were subcutaneously (s.c.) injected into the left flank of SCID mice. Mouse serum was collected biweekly for measurement of human Ig λ light chain (a soluble marker of MM.1R cells) as an indicator of whole animal tumor burden.1,21 Mice were sacrificed 4 weeks after the injection of tumor cells.
5TGM1-IV and 5TGM1-tibia models.
C57BL/KaLwRij mice were purchased from Harlan Laboratories Inc. This syngenic model of mouse MM has been reported to faithfully replicate many aspects of human MM.22-24 For IV injection, control or Runx2 k/in 5TGM1cells constitutively expressing firefly luciferase, as well as NT or Runx2 k/d 5TGM1 cells, were injected into 6-week-old male C57BL/KaLwRij mice via the tail vein. For intratibial injection, NT or Runx2 k/d 5TGM1 cells were injected directly into the bone marrow cavities of right tibias. Serum was collected biweekly, and IgG2bκ levels (a soluble marker of 5TGM1 cells) measured by enzyme-linked immunosorbent assay (ELISA). In the case of IV injection, weekly bioluminescent imaging noninvasively tracked tumor growth and the location of tumor cells in bone in vivo. At the end of all experiments, femurs and tibias were harvested, fixed, decalcified, paraffin-embedded, sectioned, and stained with hematoxylin and eosin (H&E).1,25
ELISA
The levels of human Ig λ light chain or mouse IgG2bκ in mouse sera were measured using human Ig λ or mouse IgG2bκ ELISA kits (Bethyl Laboratories Inc), respectively. The osteopontin (OPN) levels in CM of mouse 5TGM1 Runx2 k/d and Runx2 k/in cells were measured using an OPN mouse ELISA kit (Abcam). All assays were performed according to manufacturer protocols with each sample measured in duplicate.
Cytokine/chemokine array
A customized cytokine array (RayBiotech) which utilizes a chemiluminescent sandwich ELISA format and probes for 29 cytokines, chemokines, and growth factors was used according to manufacturer recommendations. Film was analyzed by densitometry using ImageJ software.
Cell proliferation and cell cycle assay
Cell proliferation was determined using a 3[4,5-dimethylthiazol-2-y]-2,5-diphenyltetrazolium (MTT) assay kit (Abnova) according to the manufacturer’s instructions. Assays were performed at 24 and 48 hours, and each sample was assayed in triplicate. For cell cycle assessment, propidium iodide staining for DNA was performed followed by flow cytometric analysis.26
Invasion assay
Runx2 k/in or Runx2 k/d 5TGM1 cell invasiveness was determined using a commercially available invasion assay (BD Biosciences). Briefly, Matrigel-coated inserts containing 8-µm pores were allowed to rehydrate in serum-free medium for 2 hours at 37°C. Subsequently, 2 × 105 cells in 500 µL of serum-free medium were added in triplicate into inserts and allowed to migrate toward complete medium in the bottom wells at 37°C and 5% CO2. Cells that invaded into bottom wells were enumerated at 24 and 48 hours in triplicate, using a Z1 Dual threshold Coulter Counter (Beckman Coulter).27
Akt inhibition
Akt signaling was inhibited using the allosteric Akt inhibitor MK2206 dissolved in dimethylsulfoxide (DMSO). Runx2 k/in 5TGM1 cells were cultured in the presence of MK2206 (2.5 µM) or DMSO for 24 hours after which cells were harvested for western blotting. For invasion assays, cells were pretreated with MK2206 (2.5 µM) for 90 minutes. Cells were then isolated and seeded in serum-free medium containing the inhibitor into invasion assay inserts as described.
Gelatin zymography
Matrix metallopeptidase 9 (MMP-9) activity in the CM of Runx2 k/in, Runx2 k/d, and control cells was measured by gelatin zymography as previously described.25 Briefly, cells were incubated in serum-free medium for 48 hours after which the CM was collected and concentrated (10-fold) using a Spin-X UF concentrator (Corning). Equal amounts of protein (80 μg) were loaded without heating and analyzed by SDS-PAGE using 10% polyacrylamide gels copolymerized with gelatin (Bio-Rad). Electrophoresis was carried out at 10 mA for 2 hours. After washing, the gel was stained with Coomassie blue R-250 and proteolytic activity visualized.25
Microarray analysis of Runx2 gene expression in clinical myeloma samples
GEP data obtained from 22 normal healthy subjects (normal plasma cells [NPCs]), 44 patients with monoclonal gammopathy of undetermined significance (MGUS), and 351 newly diagnosed MM patients in the total therapy 2 (TT2) trial were collected from a publicly available website28-30 and Runx2 gene expression in the 3 patient groups analyzed. In addition, Runx2 gene expression was examined in MM patient populations identified as high-risk and low-risk by the 70-gene model.28,29
Human bone marrow Runx2 immunohistochemistry
All biopsy procedures and immunohistochemistry protocols were approved by the UAB Institutional Review Board. Paraffin-embedded bone marrow core biopsy specimens from 14 normal bone marrow donors, 11 patients with MGUS, and 35 multiple myeloma patients, obtained from the Department of Pathology at UAB, were stained for Runx2. Staining densities were determined by 2 independent readers, including a board-certified hematopathologist as described.31
Statistical analysis
Statistical comparisons between 2 experimental groups were analyzed by the Student t test. For comparisons among multiple groups, analysis of variance (ANOVA) followed by a post hoc Bonferroni correction was used. The Wilcoxon rank-sum test was used for the statistical comparison of survival rate. Data were considered significantly different when P < .05 and are reported as such.
Results
Runx2 expression in myeloma cells does not affect cell proliferation in vitro but increases invasiveness of these cells
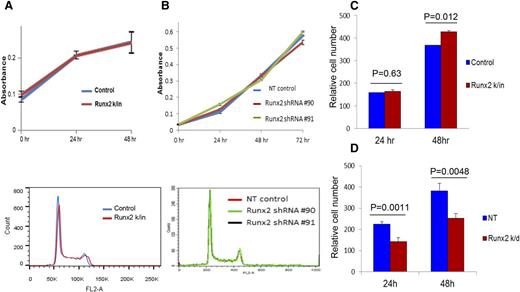
MTT and cell cycle assays were performed on Runx2 k/in, Runx2 k/d, and control 5TGM1 cells to determine the effects of Runx2 expression on tumor cell proliferation in vitro. No differences in cell proliferation were observed (Figure 1A-B). However, the invasion of Runx2 k/d cells through Matrigel was significantly reduced compared with control cells, whereas Runx2 k/in cells had significantly increased invasion (Figure 1C-D).
Runx2 expression in MM cells does not affect cell proliferation but promotes tumor cell invasion in vitro. (A) MTT assay (top) and cell cycle flow cytometry (bottom) of Runx2 k/in and control 5TGM1 cells. (B) MTT assay (top) and cell cycle flow cytometry (bottom) of Runx2 k/d (Runx2 shRNA #90 and #91 transfectants) and NT control 5TGM1cells. No differences in cell proliferation were observed. Each sample was analyzed in triplicate and each assay was performed at least twice. (C) Invasion assay using Runx2 k/in and control 5TGM1 cells. (D) Invasion assay using Runx2 k/d (#90) and NT control 5TGM1 cells. Each sample was analyzed in triplicate and each assay was performed 3 times. Error bars represent mean ± SEM of each group. Significant differences between groups are indicated by P value.
Runx2 expression in MM cells does not affect cell proliferation but promotes tumor cell invasion in vitro. (A) MTT assay (top) and cell cycle flow cytometry (bottom) of Runx2 k/in and control 5TGM1 cells. (B) MTT assay (top) and cell cycle flow cytometry (bottom) of Runx2 k/d (Runx2 shRNA #90 and #91 transfectants) and NT control 5TGM1cells. No differences in cell proliferation were observed. Each sample was analyzed in triplicate and each assay was performed at least twice. (C) Invasion assay using Runx2 k/in and control 5TGM1 cells. (D) Invasion assay using Runx2 k/d (#90) and NT control 5TGM1 cells. Each sample was analyzed in triplicate and each assay was performed 3 times. Error bars represent mean ± SEM of each group. Significant differences between groups are indicated by P value.
Runx2 overexpression in myeloma cells promotes tumor growth and progression in vivo
In vivo studies were conducted to determine whether MM-derived Runx2 affects tumor growth via regulation of the tumor microenvironment. First, luciferase-expressing vector control cells or Runx2 k/in 5TGM1 cells (Figure 2A) were injected into C57BL/KaLwRij mice via the tail vein (106 cells per mouse, n = 8 per group). Tumor growth was evaluated by measuring the levels of IgG2bκ in mouse sera, and dissemination of tumor cells was tracked by bioluminescent imaging. Mice injected with Runx2 k/in 5TGM1 cells had significantly higher IgG2bκ in the serum compared with mice injected with vector control cells at 4 weeks (Figure 2B). This difference was even more dramatic at 6 weeks (Figure 2B), suggesting that enhanced Runx2 in MM cells promotes tumor growth in vivo. Bioluminescent imaging confirmed these observations and localized MM cells in bone (Figure 2C). In fact, tumor cells were detected in bone in the Runx2 k/in group by bioluminescent imaging as early as 2 weeks after cell injection, whereas the vector control group tumors were not observed in bone until 4 weeks (data not shown). Mice were sacrificed at week 6 and histologic evaluation demonstrated that the tibia/femurs from both groups had tumors in bone at this time point, but that the mice injected with Runx2 k/in cells had significantly larger tumors. These results demonstrate that the enhancement of Runx2 expression in myeloma cells promotes disease progression in vivo.
Overexpression of Runx2 in MM cells promotes tumor progression in vivo. (A) Expression of Runx2 in Runx2 k/in 5TGM1 cells was analyzed by western blot. Runx2 k/in cells show enhanced expression of Runx2 compared with vector control cells. (B) IgG2bκ in serum (marker of total tumor burden) was measured by ELISA at 0, 4, and 6 weeks after IV injection of vector control (blue bar) or Runx2 k/in 5TGM1 cells (red bar) in C57BL/KaLwRij mice (n = 8 per group). Error bars represent mean ± SEM of each group. Significant differences between groups are indicated by P value. (C) Bioluminescent imaging 4 weeks after IV injection of both control and Runx2 k/in 5TGM1 cells (8 mice per group). (i) Robust tumor bioluminescence is observed in mice bearing Runx2 k/in 5TGM1 cells compared with mice bearing control 5TGM1 tumors. (ii) Tumors in mice bearing control 5TGM1cells are detectable by bioluminescence; however, the maximal signal in the control group is 14× less than the Runx2 k/in group as indicated by the different scale bar.
Overexpression of Runx2 in MM cells promotes tumor progression in vivo. (A) Expression of Runx2 in Runx2 k/in 5TGM1 cells was analyzed by western blot. Runx2 k/in cells show enhanced expression of Runx2 compared with vector control cells. (B) IgG2bκ in serum (marker of total tumor burden) was measured by ELISA at 0, 4, and 6 weeks after IV injection of vector control (blue bar) or Runx2 k/in 5TGM1 cells (red bar) in C57BL/KaLwRij mice (n = 8 per group). Error bars represent mean ± SEM of each group. Significant differences between groups are indicated by P value. (C) Bioluminescent imaging 4 weeks after IV injection of both control and Runx2 k/in 5TGM1 cells (8 mice per group). (i) Robust tumor bioluminescence is observed in mice bearing Runx2 k/in 5TGM1 cells compared with mice bearing control 5TGM1 tumors. (ii) Tumors in mice bearing control 5TGM1cells are detectable by bioluminescence; however, the maximal signal in the control group is 14× less than the Runx2 k/in group as indicated by the different scale bar.
Runx2 k/d in myeloma cells inhibits tumor growth and progression in vivo
The impact of Runx2 on MM progression in bone was further examined with Runx2 k/d 5TGM1 cells in 3 different animal models. 5TGM1 Runx2 k/d cells were first tested by IV injection (2 × 106 cells per mouse, n = 10 per group). In contrast to 5TGM1 Runx2 k/in, knockdown of Runx2 expression in 5TGM1 cells (Figure 3A) significantly prevented tumor dissemination. Mice bearing Runx2 k/d tumors had significantly reduced tumor burden by serum IgG2bκ levels than those with NT tumors (Figure 3B). H&E staining demonstrated that all mice injected with NT control cells had tumors in tibiae/femurs, whereas only 20% of mice injected with Runx2 k/d cells had detectable tumors in bone (Figure 3C). In a separate 5TGM1-IV injection experiment (n = 10 per group), the survival of mice bearing NT and Runx2 k/d tumors was examined. The survival rates of mice bearing Runx2 k/d 5TGM1 cells were significantly increased compared with control mice (Figure 3D).
Runx2 knockdown inhibits MM progression in vivo. (A) Expression of Runx2 in 2 Runx2 k/d 5TGM1 cell lines (Runx2 shRNA #90– and Runx2 shRNA #91–transfected 5TGM1 cells) was probed by western blot. Both Runx2 shRNA #90 and #91 decreased the expression of Runx2 compared with NT control. (B) Serum IgG2bκ was measured by ELISA 6 weeks after IV injection of NT control or Runx2 k/d 5TGM1 cells. Error bars represent mean ± SEM (n = 10 animals per group). Significant differences between groups are indicated by P value. (C) H&E-stained bone sections from mice injected IV with either NT control or Runx2 k/d 5TGM1 cells. Tumors were present in mice injected with NT cells, but not in the mice injected with Runx2 k/d cells (original magnification, ×100). Inset, Abundant myeloma cells in NT-bearing mice compared with Runx2 k/d 5TGM1 cell-injected mice. (D) Survival was significantly increased in mice injected IV with 5TGM1 Runx2 k/d cells (clone #90 and #91) compared with those injected with NT 5TGM1 cells. (E) Six weeks after intratibial injection of Runx2 k/d or NT 5TGM1 cells, levels of serum IgG2bκ were measured by ELISA. Error bars represent mean ± SEM (n = 10 animals per group). Significant differences between groups are indicated by P value. (F) Western blot shows reduction of Runx2 expression in MM.1R cells transduced with Runx2 shRNA compared with wild-type (WT) or NT control cells. (G) Six weeks after s.c. injection of NT or Runx2 k/d human MM.1R cells, serum human Ig λ light chain was measured by ELISA. Error bars represent mean ± SEM (n = 10 animals per group). Significant differences between groups are indicated by P value.
Runx2 knockdown inhibits MM progression in vivo. (A) Expression of Runx2 in 2 Runx2 k/d 5TGM1 cell lines (Runx2 shRNA #90– and Runx2 shRNA #91–transfected 5TGM1 cells) was probed by western blot. Both Runx2 shRNA #90 and #91 decreased the expression of Runx2 compared with NT control. (B) Serum IgG2bκ was measured by ELISA 6 weeks after IV injection of NT control or Runx2 k/d 5TGM1 cells. Error bars represent mean ± SEM (n = 10 animals per group). Significant differences between groups are indicated by P value. (C) H&E-stained bone sections from mice injected IV with either NT control or Runx2 k/d 5TGM1 cells. Tumors were present in mice injected with NT cells, but not in the mice injected with Runx2 k/d cells (original magnification, ×100). Inset, Abundant myeloma cells in NT-bearing mice compared with Runx2 k/d 5TGM1 cell-injected mice. (D) Survival was significantly increased in mice injected IV with 5TGM1 Runx2 k/d cells (clone #90 and #91) compared with those injected with NT 5TGM1 cells. (E) Six weeks after intratibial injection of Runx2 k/d or NT 5TGM1 cells, levels of serum IgG2bκ were measured by ELISA. Error bars represent mean ± SEM (n = 10 animals per group). Significant differences between groups are indicated by P value. (F) Western blot shows reduction of Runx2 expression in MM.1R cells transduced with Runx2 shRNA compared with wild-type (WT) or NT control cells. (G) Six weeks after s.c. injection of NT or Runx2 k/d human MM.1R cells, serum human Ig λ light chain was measured by ELISA. Error bars represent mean ± SEM (n = 10 animals per group). Significant differences between groups are indicated by P value.
Next, Runx2 k/d 5TGM1 cells were tested in the intratibial model to determine whether Runx2 expression in MM cells influences tumor cell survival and growth in bone. NT control or Runx2 k/d 5TGM1 (105) cells were injected into the right tibia of C57BL/KaLwRij mice (n = 7 per group). The levels of serum IgG2bκ demonstrated that Runx2 knockdown significantly diminished tumor burden by ∼70% (Figure 3E). Subsequently, Runx2 k/d MM.1R human myeloma cells were tested in the SCID-s.c. model (Figure 3F) (n = 8 per group). The level of human Ig λ light-chain in mouse serum demonstrated that tumor growth was significantly inhibited in mice injected with human Runx2 k/d MM.1R cells compared with mice bearing NT control tumors (Figure 3G). These results demonstrate that MM cell-derived Runx2 plays an important role in supporting tumor growth and bone homing in vivo, and that suppressing Runx2 in MM cells inhibits MM proliferation and progression.
Runx2 activates Akt /β-catenin/survivin signaling pathway in MM cells
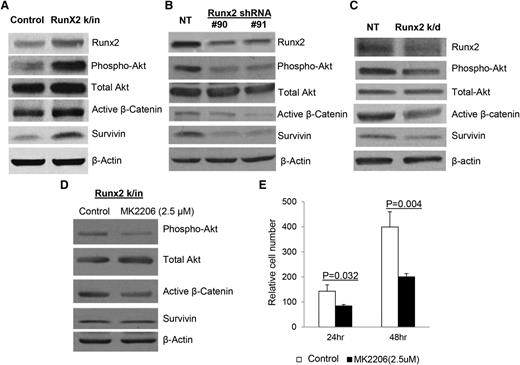
The activities of multiple signaling pathways in both Runx2 k/in and Runx2 k/d MM cells were measured by western blot to ascertain which are involved in Runx2-promoted MM growth. Interestingly, Runx2 expression did not affect bone morphogenetic protein 2 (BMP-2) and transforming growth factor-β (TGF-β) signaling in MM cells (data not shown), the two signaling pathways commonly activated by Runx2 in solid tumors.32,33 However, overexpression of Runx2 in MM cells resulted in increased Akt phosphorylation in addition to increased levels of active β-catenin (a downstream target of Akt34 ) and survivin (a downstream target of β-catenin34,35 ) (Figure 4A). In contrast, knockdown of Runx2 in mouse or human MM cells downregulated Akt signaling, active β-catenin, and survivin (Figure 4B-C). To demonstrate the functional role of Akt, Runx2 k/in 5TGM1 cells were treated with the specific allosteric Akt inhibitor MK2206 (2.5 μM) for 24 hours.36 As expected, Akt inhibition resulted in significantly decreased Akt phosphorylation, as well as decreased active β-catenin and survivin (Figure 4D). The inhibition of Akt had functional consequences as MK2206 treatment (2.5 μM) resulted in significantly decreased invasion of Runx2 k/in cells (Figure 4E). These data implicate Akt /β-catenin/survivin signaling as a functional mediator of Runx2-promoted MM tumor growth and invasion.
Akt/β-catenin/survivin signaling pathway is upregulated by Runx2 in MM cells. (A) Expression of Runx2, phosphorylated Akt, total Akt, active β-catenin, survivin, and β-actin in Runx2 k/in 5TGM1 cells and control cells analyzed by western blot. (B) Expression of Runx2, phosphorylated Akt, total Akt, active β-catenin, survivin, and β-actin in Runx2 k/d 5TGM1 cells (Runx2 shRNA #90 and #91) compared with NT control cells. (C) Expression of Runx2, phosphorylated Akt, total Akt, active β-catenin, survivin, and β-actin in NT control and Runx2 k/d MM.1R cells analyzed by western blot. (D) Runx2 k/in cells were cultured with the Akt inhibitor MK2206 (2.5 μM) or without (DMSO control) and analyzed for phosphorylated Akt, total Akt, active β-catenin, survivin, and β-actin by western blot. (E) Invasion assays were performed with Runx2 k/in cells treated with DMSO (Control) (□) or MK2206 (2.5 μM) (▪) for 24 or 48 hours. Each sample was analyzed in triplicate and each assay was performed 3 times. Error bars represent mean ± SEM of each group. Significant differences between groups are indicated by P value.
Akt/β-catenin/survivin signaling pathway is upregulated by Runx2 in MM cells. (A) Expression of Runx2, phosphorylated Akt, total Akt, active β-catenin, survivin, and β-actin in Runx2 k/in 5TGM1 cells and control cells analyzed by western blot. (B) Expression of Runx2, phosphorylated Akt, total Akt, active β-catenin, survivin, and β-actin in Runx2 k/d 5TGM1 cells (Runx2 shRNA #90 and #91) compared with NT control cells. (C) Expression of Runx2, phosphorylated Akt, total Akt, active β-catenin, survivin, and β-actin in NT control and Runx2 k/d MM.1R cells analyzed by western blot. (D) Runx2 k/in cells were cultured with the Akt inhibitor MK2206 (2.5 μM) or without (DMSO control) and analyzed for phosphorylated Akt, total Akt, active β-catenin, survivin, and β-actin by western blot. (E) Invasion assays were performed with Runx2 k/in cells treated with DMSO (Control) (□) or MK2206 (2.5 μM) (▪) for 24 or 48 hours. Each sample was analyzed in triplicate and each assay was performed 3 times. Error bars represent mean ± SEM of each group. Significant differences between groups are indicated by P value.
Runx2 elicits a bone-resident cell-like phenotype in myeloma cells
Because Runx2 is a master regulator of osteoblastogenesis,37 we hypothesized that enhanced Runx2 expression in MM cells may result in increased expression of osteogenic genes downstream of Runx2. To test this hypothesis, GEPs of Runx2 k/d, Runx2 k/in, and control 5TGM1 cells were analyzed by RNA sequencing. Compared with control cells, the expression of multiple osteogenic genes was significantly downregulated in Runx2 k/d 5TGM1 cells, including genes expressed by osteoblasts (eg, OPN), osteoclasts (eg, receptor activator of nuclear factor κB [RANK]), and osteocytes (eg, dentin matrix protein 1 [DMP1]) (Table 1). In addition, Runx2 k/d cells had reduced levels of several proteolytic enzymes secreted by osteoclasts (eg, cathepsin K [CtsK] and MMP-9) and factors secreted by resident bone marrow stromal cells, such as CD44 and α4β1 integrin, which are important molecules for adhesion to the bone matrix (Table 1). Conversely, in 5TGM1 Runx2 k/in cells, the expression of these molecules was markedly increased.
Bone-related genes are regulated by Runx2 in MM cells
| Classes . | Gene symbol . | Gene name . | Fold change . | |
|---|---|---|---|---|
| Runx2 k/d . | Runx2 k/in . | |||
| Osteoblast | BGLAP | Osteocalcin (OCN) | −3.25 | 1.22 |
| SPP1 | Osteopontin (OPN) | −2.09 | 4.32 | |
| Osteoclast | CTSK | Cathepsin K | −15.12 | 2.73 |
| RANK | Receptor activator of nuclear factor κB | −7.45 | 2.27 | |
| MMP9 | Matrix metallopeptidase 9 | −8.93 | 4.12 | |
| Osteocyte | DMP1 | Dentin matrix protein 1 | −2.02 | 7.36 |
| Adhesion molecule | CD44 | CD44 | −6.64 | 1.26 |
| ITGA4 | Integrin α4 | −5.23 | 1.26 | |
| Classes . | Gene symbol . | Gene name . | Fold change . | |
|---|---|---|---|---|
| Runx2 k/d . | Runx2 k/in . | |||
| Osteoblast | BGLAP | Osteocalcin (OCN) | −3.25 | 1.22 |
| SPP1 | Osteopontin (OPN) | −2.09 | 4.32 | |
| Osteoclast | CTSK | Cathepsin K | −15.12 | 2.73 |
| RANK | Receptor activator of nuclear factor κB | −7.45 | 2.27 | |
| MMP9 | Matrix metallopeptidase 9 | −8.93 | 4.12 | |
| Osteocyte | DMP1 | Dentin matrix protein 1 | −2.02 | 7.36 |
| Adhesion molecule | CD44 | CD44 | −6.64 | 1.26 |
| ITGA4 | Integrin α4 | −5.23 | 1.26 | |
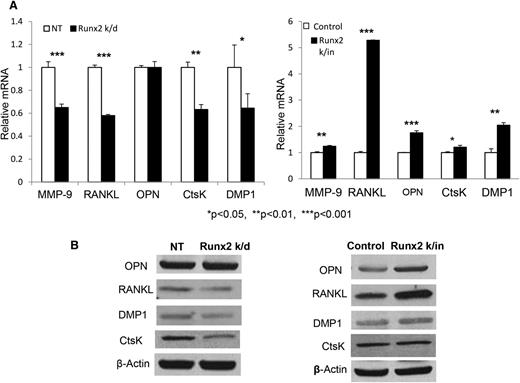
Real-time PCR confirmed that downregulation of Runx2 in 5TGM1 cells resulted in significantly decreased expression of CtsK, MMP-9, RANK ligand (RANKL), and DMP1, whereas Runx2 overexpression significantly enhanced expression of these genes, compared with control cells (Figure 5A). Western blot analysis revealed that Runx2 overexpression stimulated the increased expression of RANKL, CtsK, and DMP1 at the protein level (Figure 5B). Taken together, these results support the hypothesis that MM cells express bone-related genes in a Runx2-dependent fashion that mimics bone marrow resident cells and likely contributes to tumor survival and growth in the bone microenvironment.
MM cell–derived Runx2 upregulates expression of bone-related genes at the mRNA and protein level. (A) Real-time PCR of MMP9, OPN, RANKL, CtsK, and DMP1 expression levels in Runx2 k/d (left panel) and Runx2 k/in 5TGM1 (right panel) cells compared with respective control cells (NT or control). Error bars represent mean ± SEM. *Significant differences with corresponding P value. (B) OPN, RANKL, DMP1, CtsK, and β-actin protein expression in Runx2 k/d and NT control cells (left panel) and in Runx2 k/in and vector control 5TGM1 cells (right panel) were evaluated by western blot.
MM cell–derived Runx2 upregulates expression of bone-related genes at the mRNA and protein level. (A) Real-time PCR of MMP9, OPN, RANKL, CtsK, and DMP1 expression levels in Runx2 k/d (left panel) and Runx2 k/in 5TGM1 (right panel) cells compared with respective control cells (NT or control). Error bars represent mean ± SEM. *Significant differences with corresponding P value. (B) OPN, RANKL, DMP1, CtsK, and β-actin protein expression in Runx2 k/d and NT control cells (left panel) and in Runx2 k/in and vector control 5TGM1 cells (right panel) were evaluated by western blot.
Runx2 enhances the secretion of soluble factors related to tumor progression in myeloma cells
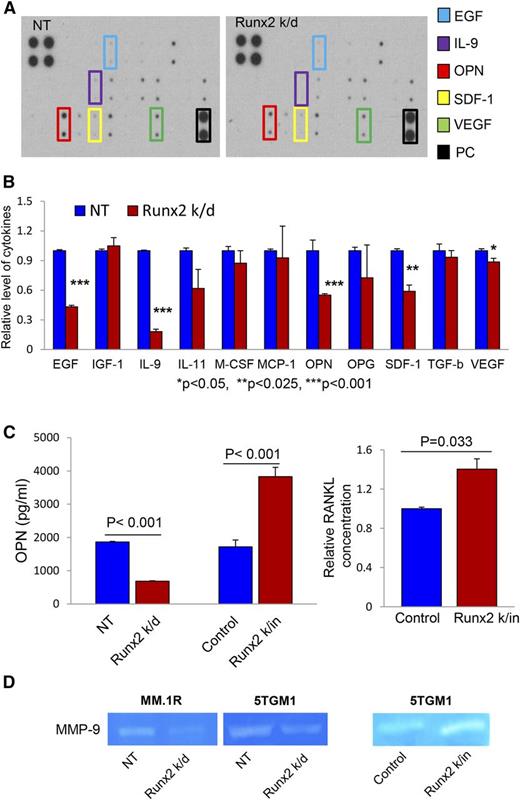
MM cell-secreted soluble factors have been suggested to play essential roles in the alteration of the bone marrow microenvironment, myeloma bone disease, and progression.38-40 A cytokine array was performed on CM of Runx2 k/d and NT 5TGM1 cells to determine the effects of decreased Runx2 expression on the secretion of soluble factors by myeloma cells. Of the 29 tested cytokines/chemokines/growth factors, epidermal growth factor (EGF), interleukin-9 (IL-9), stromal cell–derived factor-1 (SDF-1), vascular endothelial growth factor (VEGF), and OPN were significantly reduced in Runx2 k/d CM compared with control cells (Figure 6A-B). Given the profound reduction of OPN in the CM, OPN secretion was further analyzed by ELISA. Consistent with the cytokine array, Runx2 k/d led to significantly reduced (63%) secretion of OPN, whereas overexpression of Runx2 resulted in significantly increased secretion (200%) compared with controls (Figure 6C). Similarly, MM cell-derived Runx2 significantly enhanced RANKL secretion by MM cells (Figure 6C).
Runx2 regulates the secretion of soluble factors related to myeloma progression. (A) Cytokine array analysis of CM from NT control and Runx2 k/d 5TGM1 cells. Specific cytokines of interest (EGF, IL-9, OPN, SDF-1, VEGF) are identified by colored boxes. PC, cytokine array positive control. (B) The relative levels of cytokines in NT control (blue bars) and Runx2 k/d 5TGM1 cells (red bars) were normalized by comparison with the positive control signal. Levels of OPN, EGF, IL-9, SDF-1, and VEGF were significantly decreased in CM of Runx2 k/d cells. *Significant differences with corresponding P value. (C) OPN (left panel) ELISA of CM from Runx2 k/d and k/in 5TGM1 cells (red bars) compared with appropriate controls (blue bars) and RANKL (right panel) ELISA of CM from Runx2 k/in 5TGM1 cells (red bar) compared with control 5TGM1 cells (blue bar). Error bars represent mean ± SEM. Each sample was analyzed in duplicate. Significant differences between groups are indicated by P value. (D) Gelatin proteolytic analysis (zymogram) of MMP-9 activity in CM of Runx2 k/d MMIS and 5TGM1 cells compared with NT controls and Runx2 k/in 5TGM1 cells compared with vector control.
Runx2 regulates the secretion of soluble factors related to myeloma progression. (A) Cytokine array analysis of CM from NT control and Runx2 k/d 5TGM1 cells. Specific cytokines of interest (EGF, IL-9, OPN, SDF-1, VEGF) are identified by colored boxes. PC, cytokine array positive control. (B) The relative levels of cytokines in NT control (blue bars) and Runx2 k/d 5TGM1 cells (red bars) were normalized by comparison with the positive control signal. Levels of OPN, EGF, IL-9, SDF-1, and VEGF were significantly decreased in CM of Runx2 k/d cells. *Significant differences with corresponding P value. (C) OPN (left panel) ELISA of CM from Runx2 k/d and k/in 5TGM1 cells (red bars) compared with appropriate controls (blue bars) and RANKL (right panel) ELISA of CM from Runx2 k/in 5TGM1 cells (red bar) compared with control 5TGM1 cells (blue bar). Error bars represent mean ± SEM. Each sample was analyzed in duplicate. Significant differences between groups are indicated by P value. (D) Gelatin proteolytic analysis (zymogram) of MMP-9 activity in CM of Runx2 k/d MMIS and 5TGM1 cells compared with NT controls and Runx2 k/in 5TGM1 cells compared with vector control.
MMP-9 is a protease normally released by osteoclasts during bone resorption41,42 and is an important regulator of the tumor microenvironment during tumor metastasis.25 Because Runx2 k/in upregulates MMP-9 expression (Figure 5A), the effect of Runx2 expression in MM cells on MMP-9 enzyme activity was examined by gelatin zymography. The enzymatic activity of secreted MMP-9 was significantly increased by overexpression of Runx2 in 5TGM1 cells whereas downregulation of Runx2 in either MM.1R or 5TGM1 cells resulted in significantly decreased MMP-9 activity (Figure 6D).
Runx2 expression is enhanced in primary myeloma cells and high level of MM cell-derived Runx2 is correlated with high-risk disease
To determine the clinical relevance of Runx2 expression, bone marrow biopsies from 14 normal bone marrow donors, 11 MGUS patients, and 35 MM patients treated at the UAB hospital were immunostained for Runx2. The average age of normal, MGUS, and MM patients was 56.1, 67.1, and 60.7 years, respectively. Significant increases in Runx2 expression were observed in both the nuclei and cytoplasm of MM cells compared with those in the plasma cells of normal and MGUS bone marrow (Figure 7A). Interestingly, although Runx2 expression appeared elevated in MGUS, no significant difference in Runx2 expression between MGUS patients and normal bone marrow donors was observed (Figure 7B). In addition, although MM occurs more frequently in men than women, no significant difference in Runx2 expression levels was observed between genders.
Runx2 expression in plasma cells of normal bone marrow donors, MGUS, and MM patients. (A) Runx2 staining of plasma cells in normal bone marrow, MGUS, and MM patient biopsies. Strong Runx2 expression (brown color) was seen in the nuclei and cytoplasm of MM cells, whereas light staining was found in plasma cells of normal bone marrow donors and MGUS patients as indicated by arrows (original magnification, ×400). Not all positively stained cells are shown. (B) Runx2 staining density in cytoplasm and nuclei of plasma cells in normal (blue bars), MGUS (red bars), and MM patient (green bars) groups. Error bars represent mean ± SEM. Significant differences between groups are indicated by P value. (C) Runx2 mRNA expression was measured by gene microarray and compared between normal (NPC), MGUS, and MM patient groups. The number of patients in each group (n) is shown. (D) Runx2 expression was compared between low-risk and high-risk MM patients by gene expression profiling (GEP). The number of patients in each group (n) is shown. Significant differences between groups are indicated by P value.
Runx2 expression in plasma cells of normal bone marrow donors, MGUS, and MM patients. (A) Runx2 staining of plasma cells in normal bone marrow, MGUS, and MM patient biopsies. Strong Runx2 expression (brown color) was seen in the nuclei and cytoplasm of MM cells, whereas light staining was found in plasma cells of normal bone marrow donors and MGUS patients as indicated by arrows (original magnification, ×400). Not all positively stained cells are shown. (B) Runx2 staining density in cytoplasm and nuclei of plasma cells in normal (blue bars), MGUS (red bars), and MM patient (green bars) groups. Error bars represent mean ± SEM. Significant differences between groups are indicated by P value. (C) Runx2 mRNA expression was measured by gene microarray and compared between normal (NPC), MGUS, and MM patient groups. The number of patients in each group (n) is shown. (D) Runx2 expression was compared between low-risk and high-risk MM patients by gene expression profiling (GEP). The number of patients in each group (n) is shown. Significant differences between groups are indicated by P value.
Analysis of the GEP demonstrated that the expression of Runx2 mRNA was significantly increased in primary MM cells compared with MGUS and NPCs from healthy bone marrow donors (Figure 7C; P < .0001). In addition, Runx2 expression in high-risk and low-risk MMs indicated that high-risk myeloma patients exhibited significantly higher levels of Runx2 than low-risk patients (Figure 7C). These results suggest that the increased expression of Runx2 is significantly associated with a more aggressive MM phenotype.
Discussion
Runx2 is well-known as a bone-specific transcription factor6,7 essential for bone development.6-9 Recent studies demonstrate that Runx2 is also expressed in many cancer cells,15,43-47 including myeloma cells.48 Aberrant Runx2 expression in carcinomas is associated with bone metastasis,18,49 disease progression, and poor prognosis.14 However, the function of MM cell-derived Runx2 in MM progression has not been studied. Here, Runx2 expression at the mRNA and protein levels in bone marrow plasma cells of normal bone marrow donors, patients with MGUS, and those with newly diagnosed MM were analyzed. MM cells express significantly higher levels of Runx2 mRNA and protein than plasma cells from normal bone marrow donors and MGUS patients. To determine whether this enhanced Runx2 plays a role in MM progression, Runx2 was either overexpressed or knocked down in 5TGM1 mouse myeloma cells as well as in MM.1R human myeloma cells (Runx2 k/d only) and evaluated in 3 distinct animal models. Following IV injection, Runx2 k/in MM cells were detected in bone earlier and developed into larger tumors than control cells. In contrast, only 20% of mice injected with Runx2k/d MM cells formed tumors in bone and these tumors were significantly smaller than those in the control group. These data demonstrate that Runx2 expression in MM cells is associated with a more aggressive phenotype of MM, and that inhibiting MM-Runx2 significantly decreases tumor survival and growth in bone.
The signaling pathways in MM cells altered by Runx2 expression were analyzed to determine the mechanism(s) underlying Runx2 regulation of MM progression. Interestingly, Akt signaling (and not BMP or TGF-β signaling) as well as two downstream Akt targets, β-catenin and survivin, were positively regulated by Runx2 in MM cells. In addition, when Akt activity was specifically inhibited by the small molecule MK2206, both active β-catenin and survivin were significantly decreased in Runx2 k/in cells, as was the invasion of these cells. Akt signaling regulates cell proliferation, survival, cell motility, and angiogenesis.50 Survivin, a member of the inhibitor of apoptosis family,51 is upregulated by active β-catenin.51,52 Disruption of pathways that induce survivin has been shown to lead to increased apoptosis and decreased tumor growth.52 These results directly implicate an Akt/β-catenin/survivin signaling pathway downstream of Runx2 that mediates MM growth, survival, and progression in bone. Interestingly, canonical β-catenin signaling is a major mediator of Runx2 activation in osteoblasts.53 Our data demonstrate that in MM cells enhanced Runx2 expression stimulates β-catenin signaling. The molecules and mechanisms responsible for this process are the focus of ongoing investigation.
Because Runx2 is a master regulator of bone remodeling (directly regulating osteoblast differentiation and indirectly regulating osteoclast differentiation), we hypothesized that Runx2 may also upregulate bone cell gene expression in MM cells, thereby facilitating MM cells survival, dissemination, and progression in the bone microenvironment. Such a phenotype has been suggested for breast cancer cells.54 Indeed, RNASeq analyses of Runx2 k/in or Runx2 k/d cells revealed that multiple osteogenic genes were significantly increased in Runx2 k/in and decreased in Runx2 k/d MM cells. The osteogenic genes regulated included osteocalcin, OPN, DMP1, as well as osteoclast markers RANK, MMP-9, and CtsK. These changes were independently confirmed using other biochemical approaches. Many of these molecules including OPN, RANKL, and MMP-9, are known to promote and support the bone metastasis of solid tumors.25,55-58 The expression of RANKL and MMP-9 by tumor cells has been suggested to promote bone metastasis and tumor growth in bone via the stimulation of osteoclastic bone resorption38 which releases cytokines and growth factors from the bone matrix that support further tumor cell homing and growth in bone.59,60
OPN is a CD44 ligand secreted by osteoblasts and bone marrow stromal cells.61,62 MM cell-secreted OPN localizes to the surface of bone marrow cells and binds to the bone matrix, potentially playing a role in attracting MM cells to bone.61-65 High plasma levels of OPN have also been found in breast cancer,66,67 prostate cancer,68 and MM patients69,70 where elevated expression is associated with poor prognosis.67 These data support the idea that increased Runx2 expression promotes MM cell survival, growth and progression in bone through upregulation of multiple pro-metastatic molecules in MM cells.
In conclusion, the studies presented here demonstrate that Runx2 expression is significantly enhanced in MM cells. Increased Runx2 expression is associated with an aggressive bone phenotype in vivo as well as in the malignant plasma cells of MM patients, suggesting an important role for Runx2 in MM progression. Several potential mechanisms by which Runx2 expression in MM cells promotes tumor progression in bone have been uncovered. The activation of Akt/β-catenin/survivin signaling, upregulation of multiple metastatic genes, induction of a bone resident cell-like phenotype in MM cells, as well as the production and secretion of numerous cytokines and growth factors are impacted by Runx2 expression. Collectively, these Runx2-mediated effects have the potential to modify the tumor-bone microenvironment and support MM cell growth in bone. Therefore, the targeting of Runx2 expression in MM cells may represent a new therapeutic strategy for the treatment of aggressive MM.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs John Kappes and Qun Dai for providing lentivirus and helping with luciferase labeling of 5TGM1 Runx2 k/in and control cells, and Drs Claire M. Edwards and Ralph D. Sanderson for MM cells. The authors also thank Dr Michael Crowley and David Crossman from University of Alabama at Birmingham’s Heflin Center for DNAA sequencing and gene profile analysis, and Ms Enid Keyser and Mr Marion Spell from University of Alabama at Birmingham’s Comprehensive Flow Cytometry Core for helping with flow cytometry analysis.
This work was supported by National Institutes of Health (NIH) National Cancer Institute grant R01CA151538 (Y.Y.), a Multiple Myeloma Research Foundation Senior Award (Y.Y.), an International Myeloma Foundation Senior Award (Y.Y.), and 2014 University of Alabama at Birmingham Center for Metabolic Bone Disease (CMBD) pilot grant (Y.Y.). University of Alabama at Birmingham’s Comprehensive Flow Cytometry Core was supported by an NIH National Institute of Arthritis and Musculoskeletal and Skin Diseases grant P30 AR48311.
Authorship
Contribution: T.N.T. and M.L. designed and performed in vivo and in vitro experiments, analyzed and interpreted data, and wrote the manuscript; Q.P. and P.D.R. designed and carried out in vivo and in vitro experiments; D.P. collected bone marrow specimens of patients and evaluated immune staining data; J.L. evaluated clinical data; F.Z. analyzed gene array data; and A.J., L.J.S., and Y.Y. designed experiments, analyzed and interpreted data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Yang Yang, Department of Pathology, University of Alabama at Birmingham, WTI Building Room 320A, 1824 6th Ave South, Birmingham, AL 35294; e-mail: yangyang@uab.edu.
References
Author notes
T.N.T. and M.L. contributed equally to this work.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal