Key Points
ATL clones are preserved in a rare CD4+CD45RA+ TSCM population.
ATL-TSCM cells unidirectionally produce conventional CD45RO+ ATL cells and show a high potency of repopulating identical ATL clones in vivo.
Abstract
Adult T-cell leukemia (ATL) is a peripheral CD4+ T-cell neoplasm caused by human T-cell leukemia virus type 1 (HTLV-1). Despite several investigations using human specimens and mice models, the exact origin of ATL cells remains unclear. Here we provide a new insight into the hierarchical architecture of ATL cells. HTLV-1–infected cells and dominant ATL clones are successfully traced back to CD45RA+ T memory stem (TSCM) cells, which were recently identified as a unique population with stemlike properties, despite the fact that the majority of ATL cells are CD45RA–CD45RO+ conventional memory T cells. TSCM cells from ATL patients are capable of both sustaining themselves in less proliferative mode and differentiating into other memory T-cell populations in the rapidly propagating phase. In a xenograft model, a low number of TSCM cells efficiently repopulate identical ATL clones and replenish downstream CD45RO+ memory T cells, whereas other populations have no such capacities. Taken together, these findings demonstrate the phenotypic and functional heterogeneity and the hierarchy of ATL cells. TSCM cells are identified as the hierarchical apex capable of reconstituting identical ATL clones. Thus, this is the first report to demonstrate the association of a T-cell malignancy with TSCM cells.
Introduction
Adult T-cell leukemia (ATL) is a peripheral CD4+ T-cell neoplasm caused by human T-cell leukemia virus type 1 (HTLV-1).1,2 HTLV-1 is primarily transmitted during the neonatal period through breast feeding from HTLV-1 carriers, and approximately 5% of HTLV-1 carriers develop ATL decades after the initial infection. A recent report showed that multiple subclones with various chromosomal gain or loss originate as a result of clonal evolution,3 suggesting that longitudinal accumulations of genetic abnormalities in the host genome are required for the development of ATL.
These abnormalities have been thought to accumulate in some specific populations with longevity such as hematopoietic stem cells (HSCs) and to consequently transform normal stem cells into cancer stem cells such as leukemia-initiating cells (LICs) in myeloid malignancies.4,5 However, because lymphocytes generally have a long life span, it remains uncertain whether lymphoid malignancies develop in particular populations or randomly in common mature populations.6
The origin of ATL cells is currently unclear. Although several studies have pointed out close immunophenotypic and functional resemblances between ATL cells and regulatory T cells,7-9 a recent paper noted that such features can be acquired or induced by virus molecules but not inherited from the initially infected T cells.10 In Tax-transgenic mice, CD117+ hematopoietic progenitor cells recapitulated T-cell lymphoma.11 These findings raise the possibility that the initial ATL clones might originate in a clinically disregarded population rather than the major population with conventional phenotypes.
Human CD4+ T cells are very heterogeneous and are conventionally classified to CD45RA+ naïve T (TN) cells and CD45RO+ memory T cells, which include both central memory T (TCM) cells and effector memory T (TEM) cells, classified based on the expression of CCR7. In addition, a recent report identified a novel rare CD45RA+ subpopulation, termed T memory stem (TSCM) cells. This population possesses stem cell–like properties including enhanced self-renewal capacity and multipotency to generate TCM and TEM cells,12 and it appears to support long-lived T-cell memory against viruses.13 Because many cancers are organized hierarchically and are sustained by a rare subpopulation with self-renewal capacity at the apex of the hierarchy,14 we investigated the phenotypic and functional heterogeneity and the hierarchy of ATL cells and whether the TSCM population is involved in the development of ATL.
Materials and methods
Clinical samples
This study was approved by the institutional review board of the Graduate School of Medicine, Kyoto University, and abided by the tenets of the Declaration of Helsinki. All specimens from healthy individuals (HIs), HTLV-1 carriers, and ATL patients were collected after written informed consent was obtained. The diagnosis of ATL was based on the presence of anti–HTLV-1 antibodies, expansion of CD3+CD4+CD25+ T cells, presence of clonal expansion proven by Southern blot hybridization analysis to detect the HTLV-1 genome, and classification criteria proposed by Shimoyama.15 In cases positive for anti–HTLV-1 antibody but without clonal expansion, patients were classified as HTLV-1 carriers. The details of each patient are shown in supplemental Table 1, available on the Blood Web site.
Flow cytometric analysis and sorting
Peripheral blood mononuclear cells (PBMCs) or bone marrow mononuclear cells (BMMCs) were purified using Ficoll-Paque PLUS (GE Healthcare Bio-sciences AB). We isolated CD4+ T-cell subpopulations according to a previously described protocol12,16 with minor modifications. Because CD3 often declines aberrantly and CD25 is a crucial marker in ATL and CD62L is typically positive in the CD45RA+CCR7+ population, lineage markers such as CD11b, CD14, and CD56 were used instead of CD3 to exclude other CD4+ populations including dendritic cells, monocytes, and natural killer cells, and CD62L was skipped instead of adding CD25. Thus PBMCs were stained with anti-CD45RO, anti-CCR7, anti-CD95, anti-CD122, anti-CD25, anti-CD4, and anti-CD45RA, in addition to a cocktail including anti-CD11b, anti-CD14, and anti-CD56. For analyses of human cells repopulating in mice, anti-human CD45 and anti-mouse CD45.1 were used. To isolate lineage–CD34+ cells, BMMCs were stained with anti-CD34, anti-CD38, and a lineage cocktail including anti-CD3, anti-CD4, anti-CD8, anti-CD11b, anti-CD14, anti-CD19, anti-CD20, anti-CD56, and anti-Glycophorin A. The list of antibodies is shown in supplemental Table 2. Analyses and sorting were performed with FACS Aria II Special Order System and Diva software (Becton Dickinson Biosciences) or FlowJo software (Tomy Digital Biology).
Polymerase chain reaction (PCR) for HTLV-1 provirus, inverse long PCR, and ATL clone–specific PCR
Genomic DNA was isolated with a QIAamp DNA Mini or Micro Kit (QIAGEN, Tokyo, Japan). Qualitative and quantitative PCR were performed for the pX region, which is relatively conserved in the HTLV-1 genome, and RNase P as the internal control, with Ex-Taq Hot Start Version or SYBR Premix Ex Taq II and the TaKaRa Thermal Cycler Dice (TP800) (Takara, Shiga, Japan). After normalizing the value of pX to that of RNase P (pX/RNase P) in each sample, HTLV-1 proviral load (PVL) was described as the relative value of pX/RNase P to that in CD3+CD4+CD25+ cells from Pt-14 because this case has the highest number of white blood cells and single provirus integration proven by Southern blot hybridization analysis so that the copy number of pX in these cells is estimated to be 100 copies per 100 cells. Inverse long PCR was performed to identify ATL clone among numerous HTLV-1–infected cells as previously described.17 To determine the individual integration site, the cases presenting single band proceeded to direct sequencing with a 3130xl Genetic Analyzer (Applied Biosystems) according to the manufacturer’s instructions. Subsequently, to amplify each ATL clone–specific sequence, we constructed 2 forward primers located at the 3′ long terminal repeat of provirus and reverse primers located in the human genome adjacent to the provirus integration site in each case. Primers and PCR conditions used in these assays are shown in supplemental Table 3.
In vitro assay
Each sorted population was labeled with carboxyfluorescein succinimidyl ester (CFSE; Biotium) as previously described.18 In brief, cells were labeled with 5 μM CFSE for 10 minutes at room temperature in phosphate-buffered saline supplemented with 5% heat-inactivated fetal bovine serum (Sigma-Aldrich), and washed 3 times. Subsequently, 20 000 cells were suspended in Iscove’s modified Dulbecco’s medium (GIBCO) supplemented with 20% fetal bovine serum, 100 μg/mL primocin (InvivoGen), and 25 ng/mL recombinant human interleukin 7 (IL-7) (PeproTech). Cells were then plated in 96-well U-bottom plates and incubated at 37°C in a humidified 5% CO2 incubator. Half of the culture medium was replaced every 4 to 5 days. Two weeks after starting cultures, cells were harvested and stained with anti-CCR7 and anti-CD45RA, and then analyzed.
Mice and xenogeneic transplantation assay
NOD.Cg-PrkdcscidIl2rgtm1Wjl /SzJ (NSG) mice19 and NOD.Cg-PrkdcscidIl2rgtm1Sug /Jic (NOG) mice20 were purchased from Charles River (Yokohama, Japan) and the Central Institute for Experimental Animals (Chiba, Japan), respectively. NOD.Cg-Rag1tm1MomIl2rgtm1Wjl /SzJ (NRG) mice21 were obtained from The Jackson Laboratory (Bar Harbor, ME). These mice were maintained under specific pathogen-free conditions in our institute of laboratory animals, and all experiments were approved by the animal research committee of Kyoto University. Each sorted population was suspended in Hank’s balanced salt solution (GIBCO) and inoculated intraperitoneally into 6- to 10-week-old NOG, NSG, or NRG mice. NOG and NSG mice were irradiated (150 cGy) 1 day before inoculation. Peripheral blood cells were analyzed at indicated time points for the repopulation and phenotype of repopulating human cells. Finally, mice were euthanized 6 to 12 weeks after transplantation, and cells from peripheral blood and spleen were analyzed with regard to chimerisms, phenotype, and clonality.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 6 (GraphPad Software, Inc). Three or more groups were compared with the Kruskal-Wallis test followed by Dunn’s multiple comparisons test. P < .05 was considered to indicate statistical significance.
Results
No existence of HTLV-1–infected cells in hematopoietic stem and progenitor populations
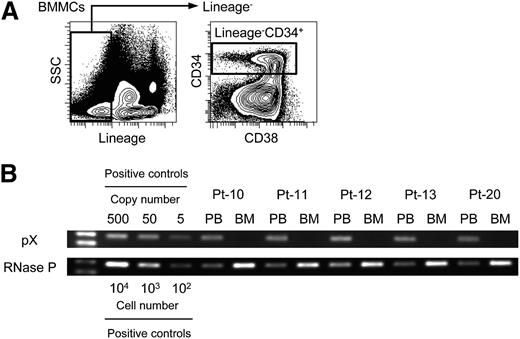
We first investigated whether hematopoietic stem and progenitor cells (HSPCs) were infected with HTLV-1. We obtained BMMCs from 5 ATL patients (supplemental Table 1) and sorted more than 1 × 104 lineage–CD34+ cells that contain HSPCs but not mature hematopoietic cells including T, B, natural killer, myeloid, and erythroid cells (Figure 1A). Sorting purity was >96.3% for all cases. We then performed PCR, which can successfully amplify ≥5 copies of the pX region. PCR results demonstrated that amplification of pX was detected only in PBMCs but not in lineage–CD34+ cells in all tested samples (Figure 1B). This result indicates that HTLV-1–infected cells do not exist in HSPCs, which is consistent with the previous report.22
No infection with HTLV-1 in phenotypically sorted hematopoietic stem and progenitor cells. (A) Representative data of sorting lineage–CD34+ cells in BMMCs from an ATL patient are shown. Lineage markers include CD3, CD4, CD8, CD11b, CD14, CD19, CD20, CD56, and CD235. (B) Data of PCR targeting pX as a proviral region and RNase P as an internal control are shown. The same amount of genomic DNA purified from PBMCs (0.6 ng) and lineage–CD34+ cells (6 ng) sorted from BMMCs of each ATL patient are used as a template. Numbers of patients (Pt) are correspondent to those in supplemental Table 1. Positive controls indicate a plasmid DNA-cloning HTLV-1 provirus or genomic DNA of PBMCs from a HI. The left lane shows the size marker.
No infection with HTLV-1 in phenotypically sorted hematopoietic stem and progenitor cells. (A) Representative data of sorting lineage–CD34+ cells in BMMCs from an ATL patient are shown. Lineage markers include CD3, CD4, CD8, CD11b, CD14, CD19, CD20, CD56, and CD235. (B) Data of PCR targeting pX as a proviral region and RNase P as an internal control are shown. The same amount of genomic DNA purified from PBMCs (0.6 ng) and lineage–CD34+ cells (6 ng) sorted from BMMCs of each ATL patient are used as a template. Numbers of patients (Pt) are correspondent to those in supplemental Table 1. Positive controls indicate a plasmid DNA-cloning HTLV-1 provirus or genomic DNA of PBMCs from a HI. The left lane shows the size marker.
Identification of phenotypic subpopulations in CD4+ T cells in ATL
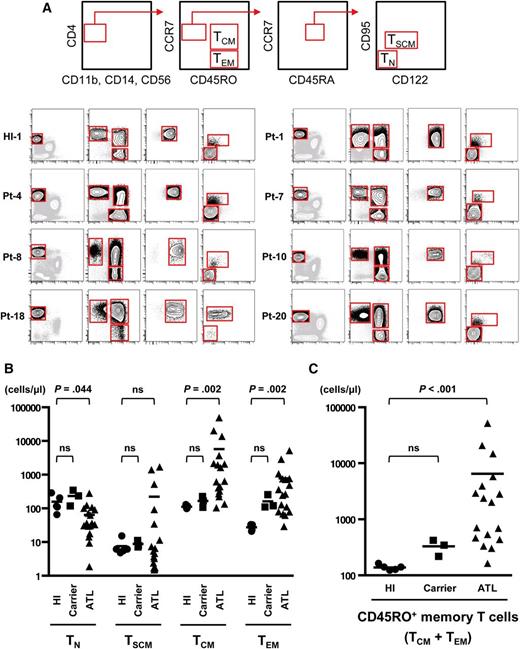
To investigate the heterogeneity of CD4+ T cells in ATL patients, we adopted a multicolor staining as described in Materials and methods and successfully divided CD4+ T cells from PBMCs of HIs into 4 subpopulations including TN (CD4+CCR7+CD45RA+CD45RO–CD95−CD122−), TSCM (CD4+CCR7+CD45RA+CD45RO–CD95+CD122dim), TCM (CD4+CCR7+CD45RA−CD45RO+), and TEM (CD4+CCR7−CD45RA−CD45RO+) in agreement with previous studies (Figure 2A and supplemental Figure 1).13
Identification and absolute numbers of 4 subpopulations—TN, TSCM, TCM, and TEM—divided from the CD4+ T cells of HTLV-1 carriers and ATL patients. (A) Top panels show a scheme to isolate the TN, TSCM, TCM, and TEM populations from PBMCs. After excluding dendritic cells, monocytes, and natural killer cells, we gated CD4+ cells and then separated into 4 subpopulations. The lower panels show representative data from a HI, a HTLV-1 carrier, and ATL patients. Additional data from other samples are presented in supplemental Figure 1. (B-C) Absolute numbers of each population (TN, TSCM, TCM, and TEM) or the CD45RO+ memory T-cell population (TCM + TEM) in peripheral blood from HIs (n = 5), HTLV-1 carriers (n = 3), and ATL patients (n = 17) were calculated with PBMC counts and fluorescence-activated cell sorting (FACS) analysis. Each value is presented in supplemental Table 4. P values by Dunn’s multiple comparisons test after Kruskal-Wallis test are presented. Bars indicate means.
Identification and absolute numbers of 4 subpopulations—TN, TSCM, TCM, and TEM—divided from the CD4+ T cells of HTLV-1 carriers and ATL patients. (A) Top panels show a scheme to isolate the TN, TSCM, TCM, and TEM populations from PBMCs. After excluding dendritic cells, monocytes, and natural killer cells, we gated CD4+ cells and then separated into 4 subpopulations. The lower panels show representative data from a HI, a HTLV-1 carrier, and ATL patients. Additional data from other samples are presented in supplemental Figure 1. (B-C) Absolute numbers of each population (TN, TSCM, TCM, and TEM) or the CD45RO+ memory T-cell population (TCM + TEM) in peripheral blood from HIs (n = 5), HTLV-1 carriers (n = 3), and ATL patients (n = 17) were calculated with PBMC counts and fluorescence-activated cell sorting (FACS) analysis. Each value is presented in supplemental Table 4. P values by Dunn’s multiple comparisons test after Kruskal-Wallis test are presented. Bars indicate means.
First we examined whether TSCM can be infected by HTLV-1. Several receptors such as GLUT1, Nrp-1, HSPGs, and Syndecan-4 are reported to be involved in HTLV-1 binding and entry, and are especially induced by stimulation.23-25 Consistent with previous reports, all receptors were hardly or faintly expressed without activation but clearly induced on populations phenotypically identical to TSCM, TCM, and TEM after activation, except for GLUT124 (supplemental Figure 2A-C). The coculture assay of CD4+ T cells with HTLV-1–producing cell line MT-2 illustrated that HTLV-1 was integrated to TSCM as well as TCM and TEM (supplemental Figure 3A-B), indicating that all the memory T-cell populations including TSCM have susceptibility to HTLV-1 infection.
Next we applied the same gating method to PBMCs from 3 HTLV-1 carriers and 17 ATL patients and found that all 4 populations were separable across all disease phases (Figure 2A, supplemental Figure 1, and supplemental Table 4). Numbers of TCM or TEM cells were increased in ATL patients (Figure 2B), especially in the chronic phase and acute phase (supplemental Figure 4A). Numbers of entire CD45RO+ memory T cells (TCM + TEM) were increased in ATL patients and even in HTLV-1 carriers (Figure 2C) consistently with the clinical aspect that the majority of ATL cells express CD45RO, and intriguingly more increased as disease stages worsen (supplemental Figure 4B), suggesting that expansion of conventional memory T cells already starts in a low-grade phase and accelerates with disease progression. In contrast, numbers of TN cells were decreased in ATL patients (Figure 2B), especially in the acute phase (supplemental Figure 4A). Numbers of TSCM cells were mostly comparable between HIs and ATL patients, although they were increased in some acute-type ATL cases. We were also concerned that tax expression might drive the expansion of TSCM, TCM, and TEM populations in patients. However, tax expression level was very low in many cases and not correlated to cell numbers of any population (supplemental Figure 4C). Finally, we analyzed CD25 expression, which is a key marker of ATL cells, to roughly estimate the occupancy of ATL cells in each population (supplemental Figure 5A and supplemental Table 5). Proportions of CD25+ cells were significantly increased in TCM and TEM populations, but not in TSCM and TN populations of ATL patients and HTLV-1 carriers (supplemental Figure 5B). Therefore, additional genetic analyses were required to acquire conclusive evidence with regard to the existence of ATL cells in the TSCM population.
Phenotypic heterogeneity of HTLV-1–infected CD4+ T cells across all disease phases
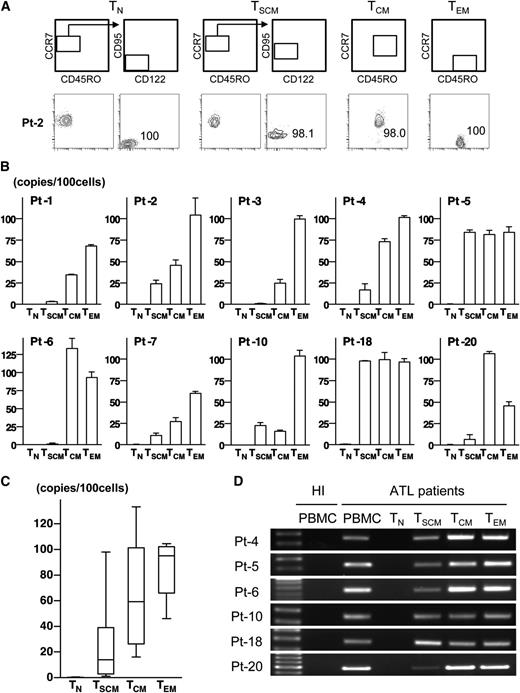
To examine the exact distribution of HTLV-1–infected cells in CD4+ subpopulations, we sorted these 4 subpopulations and measured PVL. To minimize the influence of contaminations, we only analyzed cases in which ≥97% purity was achieved with reanalysis of sorted cells (Figure 3A and supplemental Figure 6). We also strictly checked for contamination of the TSCM population with CD45RO+ cells. With the exception of 1 case (Pt-18), which had 0.2% contamination, the remaining cases had no evidence of CD45RO+ cells contamination (supplemental Figure 6). The majority of TCM and TEM cells were infected with HTLV-1 (TCM: PVL mean 64.2 copies/100 cells [SD 40.4 copies/100 cells]; TEM: PVL mean 85.9 copies/100 cells [SD 20.7 copies/100 cells]), consistent with a previous study reporting the correlation of CD4+ memory T-cell number and PVL (Figure 3B-C).26 Strikingly, the PVL of TSCM cells was clearly measurable in all tested cases including not only ATL patients but also HTLV-1 carriers (PVL range, 1.0-98.0 copies/100 cells, mean 26.9 copies/100 cells [SD 35.0 copies/100 cells]). The PVL of TN cells was barely detectable in only half of the tested cases at the level that is explicable as contamination (supplemental Table 6). These results indicate that the pool of HTLV-1–infected cells is composed not only of TCM and TEM cells but also TSCM cells.
Existence of HTLV-1–infected cells and ATL clones not only in TCM and TEM populations but also in the TSCM population. (A) Representative data of postsorting samples. Values indicate the purity of each sorted population. (B) The mean value of PVL in each population from HTLV-1 carriers and ATL patients are presented. Error bars indicate 1 standard deviation (SD) in triplicate. (C) Data described in (B) are summarized with box and whisker plots. The boxes represent the 25% to 75% percentiles, whiskers represent the range, and lines in the box represent the median values of the distribution. (D) Data of ATL clone–specific PCR using genomic DNA from 1 × 103 cells of each population are shown. Data from PBMCs of each patient or a HI are presented as positive controls or negative controls. The left lane shows the size marker.
Existence of HTLV-1–infected cells and ATL clones not only in TCM and TEM populations but also in the TSCM population. (A) Representative data of postsorting samples. Values indicate the purity of each sorted population. (B) The mean value of PVL in each population from HTLV-1 carriers and ATL patients are presented. Error bars indicate 1 standard deviation (SD) in triplicate. (C) Data described in (B) are summarized with box and whisker plots. The boxes represent the 25% to 75% percentiles, whiskers represent the range, and lines in the box represent the median values of the distribution. (D) Data of ATL clone–specific PCR using genomic DNA from 1 × 103 cells of each population are shown. Data from PBMCs of each patient or a HI are presented as positive controls or negative controls. The left lane shows the size marker.
Existence of ATL clones not only in TCM and TEM populations but also in the TSCM population
ATL is generally composed of only one or a few dominant clones among numerous HTLV-1–infected cells, raising the possibility that the distribution of ATL clones may differ from that of HTLV-1–infected cells. We then investigated HTLV-1 integration sites. Inverse long PCR showed the single product in all tested cases excluding 1 case (Pt-7), indicating that Pt-7 was supposed to be oligoclonal but all other cases had monoclonal expansion (supplemental Figure 7A). Based on sequencing data of these single products (supplemental Figure 7B), we constructed ATL clone–specific PCR, which amplified the unique flanking region of the HTLV-1 integration site in each case (supplemental Table 3). We then examined the presence of the ATL clone in each sorted population from ATL patients. In all tested cases, each targeted site was amplified not only in TCM and TEM cells but also in TSCM cells, whereas no amplification was detected in TN cells (Figure 3D). Even in TSCM cells from Pt-6 with a low value of PVL (1.3 copies/100 cells), the ATL clone–specific HTLV-1 integration site was clearly amplified. This result indicates that ATL clones are also phenotypically heterogeneous and clearly exist in the TSCM population.
Unidirectional differentiation of TSCM cells to TCM and TEM cells in ATL
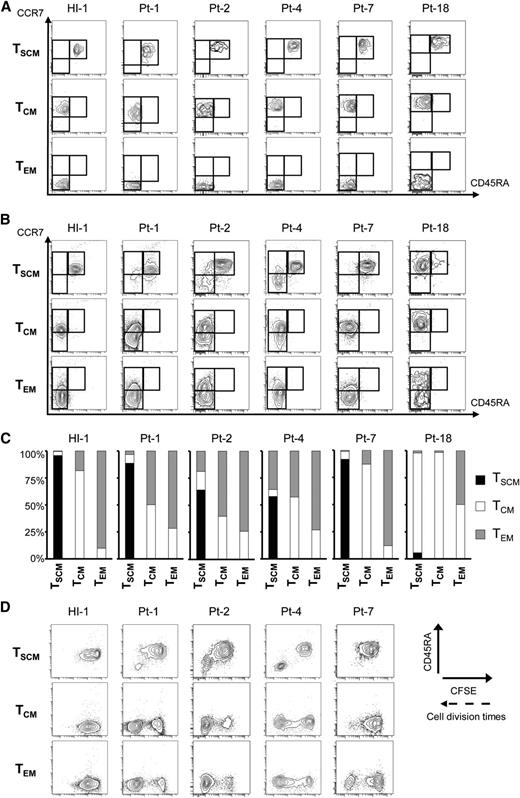
The successful demonstration of phenotypic heterogeneity of HTLV-1–infected cells and ATL clones encouraged us to investigate the possibility of functional hierarchy, namely that TSCM cells are upstream of TCM and TEM cells in HTLV-1 carriers and ATL patients. We highly purified each memory T-cell population (Figure 4A) and performed an in vitro culture assay with IL-7, which promotes survival but not differentiation of CD4+ T cells.27 In a case of HI, each sorted population sustained its original phenotype. In contrast, in HTLV-1 carriers and ATL patients, TSCM cells sustained themselves and concurrently generated TCM and/or TEM cells (Figure 4B-C). In Pt-18, TSCM cells, most of which were thought to be HTLV-1–infected cells according to their PVL (98.0 copies/100 cells), sustained themselves in a small proportion (5.7% of total) and markedly generated TCM cells. Also in Pt-4, TSCM cells with an intermediate level of PVL (17.1 copies/100 cells) produced TCM and TEM cells, and these populations were expanded up to 42% of the total cells after culture. Conversely, the opposite induction from TCM or TEM cells to the TSCM population was never observed in all tested cases (Figure 4B-C). We additionally evaluated the correlation between the propagating capacity and population alterations. TCM and/or TEM cells in HTLV-1 carriers and ATL patients proliferated more rapidly than those in a HI. More importantly, rapidly proliferating TSCM cells no longer sustained themselves and concurrently generated CD45RA– memory T cells (Figure 4D). For instance, in Pt-1, a small number of TSCM cells proliferated rapidly and differentiated into TCM and TEM cells with high propagating capacity. These data are consistent with the increase of PVL that was 3.4 copies per 100 cells in TSCM, 34.3 copies per 100 cells in TCM, and 68.0 copies per 100 cells in TEM cells (supplemental Table 6). Such an increase in PVL from TSCM to TCM and TEM cells was validated in many other cases, regardless of disease phases (Figure 3B). These data demonstrate the existence of unidirectional alteration from TSCM to TCM and TEM cells, which contributes to expansion of the HTLV-1–infected T-cell pool.
Unidirectional generation from HTLV-1–infected TSCM cells to TCM and TEM cells with rapid proliferation capacity. Data from culture of sorted TSCM, TCM, and TEM cells with 25 ng/mL recombinant human IL-7 and CFSE dilution for 2 weeks are presented. (A) FACS analyses of sorted samples in each case before in vitro culture. (B) FACS plots show sorted populations from a HI, HTLV-1 carriers, and ATL patients alter their phenotype. (C) The proportions of resulting phenotypic TSCM, TCM, and TEM cells shown in (B) are summarized. (D) The correlations of phenotypic alterations based on CD45RA level with proliferation ability are presented. The proliferation ability of each sorted population is assessed by the intensity of CFSE.
Unidirectional generation from HTLV-1–infected TSCM cells to TCM and TEM cells with rapid proliferation capacity. Data from culture of sorted TSCM, TCM, and TEM cells with 25 ng/mL recombinant human IL-7 and CFSE dilution for 2 weeks are presented. (A) FACS analyses of sorted samples in each case before in vitro culture. (B) FACS plots show sorted populations from a HI, HTLV-1 carriers, and ATL patients alter their phenotype. (C) The proportions of resulting phenotypic TSCM, TCM, and TEM cells shown in (B) are summarized. (D) The correlations of phenotypic alterations based on CD45RA level with proliferation ability are presented. The proliferation ability of each sorted population is assessed by the intensity of CFSE.
High repopulating capacity of TSCM cells in ATL
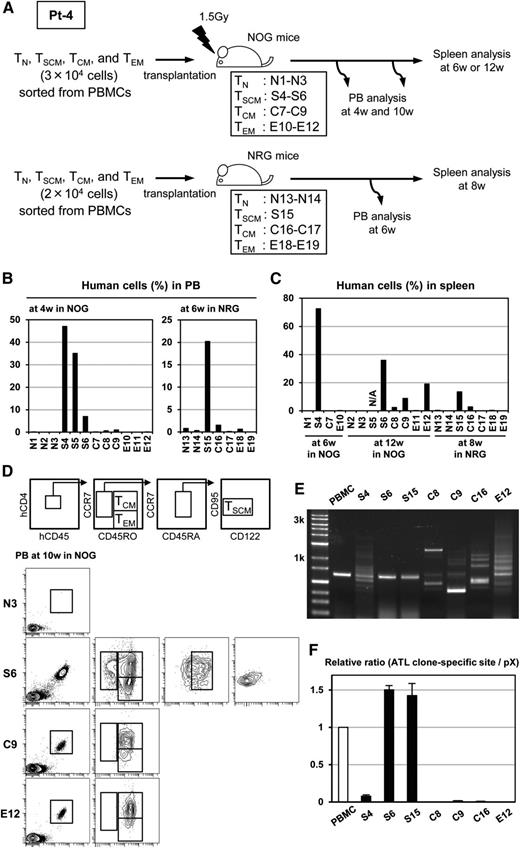
To validate the hierarchy demonstrated in the in vitro assay and to further assess the tumorigenicity of TSCM cells from ATL patients in vivo, we conducted a xenogeneic transplantation assay. We inoculated 2 to 3 × 104 cells of sorted TN, TSCM, TCM, or TEM cells from Pt-4 intraperitoneally into immunodeficient mice and evaluated the engraftment capability of each population and the phenotype and clonal architecture of repopulating cells in mice (Figure 5A). The proportions of human CD45+ cells in the peripheral blood of mice at 4 weeks after transplantation were markedly higher in NOG mice inoculated with the TSCM population than with other populations (TN: mean 0.01% [SD 0.02%]; TSCM: mean 29.0% [SD 19.6%]; TCM: mean 0.5% [SD 0.5%]; and TEM: mean 0.03% [SD 0.06%]; each N = 3) (Figure 5B, left); this was reconfirmed in another independent arm with NRG mice at 6 weeks after transplantation (Figure 5B, right). We also observed that TSCM achieved the highest proportions of human CD45+ cells in spleens at all time points (Figure 5C). These results suggest that TSCM cells possess an enhanced repopulating capacity, a key feature of stem cells. At 10 weeks after transplantation, the phenotype of repopulating human cells in NOG mice was analyzed. Transplanted TSCM cells reproduced themselves and simultaneously generated TCM and TEM populations, whereas transplanted TCM or TEM cells did not generate the TSCM population (Figure 5D). This result clearly indicates that TSCM cells can unidirectionally replenish TCM and TEM populations, implying that TSCM cells could be the hierarchical apex of heterogeneous ATL cells. Transplanted TEM cells generated TCM population in mice, suggesting that a more flexible differentiation pathway between TEM and TCM may exist in ATL cells. We were concerned that the possibility that many HTLV-1–infected clones other than the ATL clone might expand in immunodeficient mice. Therefore, we investigated the clonality of repopulating human cells. Human CD45+ cells were collected from spleens at the time points described in Figure 5C and analyzed with inverse long PCR. As expected, many clones were detected in mice transplanted with TCM and TEM cells. However, the single clone was clearly detected in S6 and S15 mice transplanted with TSCM cells, and this clone was identical to the primary ATL clone detected in the patient (Figure 5E). Furthermore, this result was validated by measuring the compensated value of the identical ATL clone to the HTLV-1 provirus. The proportions were comparable between repopulating cells in S6 or S15 mice and PBMCs in the patient. Also in the S4 mouse that was transplanted with TSCM cells and in which multiple HTLV-1–infected clones were propagated, the identical ATL clone filled a part of the HTLV-1–infected cell pool. In contrast, this was not the case in C8, C9, C16, and E12 mice (Figure 5F). In addition, we performed a xenotransplantation assay with another case and attempted to reconfirm the capacity of TSCM cells to repopulate the identical ATL clone. We inoculated 3 × 104 cells of each sorted population and 3 × 105 TCM cells from Pt-6 into NSG mice (supplemental Figure 8A). Human cells were repopulated in one (N20) of 2 mice transplanted with TN cells and both (S22 and S23) mice transplanted with TSCM cells (supplemental Figure 8B). Strikingly, even in cases (C30 and C31) inoculated with a tenfold higher number of TCM cells, TCM cells failed to be repopulated in mice. The identical ATL clone was clearly detected in 1 (S22) of 2 mice transplanted with TSCM cells (supplemental Figure 8C). Notably, PVL in the TSCM populations was very low, 17.1 copies per 100 cells in Pt-4 and only 1.3 copies per 100 cells in Pt-6 (supplemental Table 6), which means the actual inoculated numbers of HTLV-1–infected TSCM cells were estimated to be approximately 3400 cells or 5100 cells in Pt-4 and only 390 cells in Pt-6. Nevertheless, only TSCM cells could efficiently reconstitute identical ATL clones in immunodeficient mice, indicating the high repopulating capacity of ATL-TSCM cells. Collectively, these results suggest that only TSCM cells, not TCM and TEM cells, are competent to preserve ATL clones and to give rise to ATL.
Better engraftment, replenishment of TCM and TEM cells, and high capacity to repopulate the identical ATL clone by TSCM in ATL. (A) The scheme of 2 independent experiments is shown. Mice numbers are indicated in boxes. N, S, C, and E at the head of the numbers are correspondent to mice transplanted with TN, TSCM, TCM, and TEM population, respectively. The proportions of human CD45+ cells in peripheral blood (B) and in spleen (C) are presented. The data of the spleen from S5 mouse are not available owing to death before the indicated time. (D) Representative FACS analyses data on the phenotypic alteration of human CD45+ cells in the peripheral blood from N3, S6, C9, and E12 mice at 10 weeks after transplantation are depicted. (E) Inverse long PCR shows diversity of HTLV-1–infected clones in human CD45+ cells from the spleens of recipient mice. Data from PBMCs of the patient are presented together. The left lane shows the size marker. (F) Quantitative PCR targeting the ATL clone–specific site and the pX region is performed. Value of the ATL clone–specific site normalized to the value of the pX region is displayed as the relative ratio to that of PBMCs of the patient.
Better engraftment, replenishment of TCM and TEM cells, and high capacity to repopulate the identical ATL clone by TSCM in ATL. (A) The scheme of 2 independent experiments is shown. Mice numbers are indicated in boxes. N, S, C, and E at the head of the numbers are correspondent to mice transplanted with TN, TSCM, TCM, and TEM population, respectively. The proportions of human CD45+ cells in peripheral blood (B) and in spleen (C) are presented. The data of the spleen from S5 mouse are not available owing to death before the indicated time. (D) Representative FACS analyses data on the phenotypic alteration of human CD45+ cells in the peripheral blood from N3, S6, C9, and E12 mice at 10 weeks after transplantation are depicted. (E) Inverse long PCR shows diversity of HTLV-1–infected clones in human CD45+ cells from the spleens of recipient mice. Data from PBMCs of the patient are presented together. The left lane shows the size marker. (F) Quantitative PCR targeting the ATL clone–specific site and the pX region is performed. Value of the ATL clone–specific site normalized to the value of the pX region is displayed as the relative ratio to that of PBMCs of the patient.
Discussion
In this study, we have provided the new concept of the hierarchical architecture of ATL. First, we examined how upstream ATL clones can be phenotypically traced. Several recent studies have challenged the conventional belief that mature lymphocytes are the origin of lymphoid malignancies. For instance, tumor development appears to be initiated in HSPCs in certain mature lymphoid malignancies, including chronic lymphocytic leukemia (CLL),28,29 angioimmunoblastic T-cell lymphoma,30 and hairy cell leukemia.31 Moreover, the identification of the TSCM population with stem cell–like features12,13 has produced the new concept that the TSCM population is a reservoir of another retrovirus, HIV-1.32 Based on these findings, we investigated the phenotypic architecture and the distribution of genetic marks including HTLV-1 DNA, a definite first hit for ATL, and ATL clone–specific provirus integration sites, in CD4+ T cells from HTLV-1 carriers and ATL patients. Our data have clearly shown the presence of ATL clones in the TSCM population as well as in the TCM and TEM populations, indicating that ATL clones are very heterogeneous and may originate from CD45RA+CD45RO– TSCM cells despite the fact that the majority of ATL cells are usually CD45RO+.
We then examined whether TSCM cells are the functional apex of ATL. Our in vitro culture data demonstrated that the movement between TSCM cells and other populations including TCM and TEM cells, in HTLV-1 carriers and ATL patients, is unidirectional. Intriguingly, less divided TSCM cells sustained the TSCM population, whereas rapidly propagating TSCM cells differentiated into TCM and TEM cells. Considering the concept of LICs in myeloid malignancies that LICs are generally quiescent and are able to reproduce themselves and differentiate into progenitor cells which explosively propagate to shape leukemia,14,33 TSCM cells in ATL seem to behave as stem cells, whereas TCM and TEM cells may resemble progenitors.
These findings should be validated using in vivo xenograft models,4,34 however, it should be heeded that HTLV-1–infected cells rather than ATL clones could generate xenografts. In fact, although a number of xenograft models of ATL have been reported,35-38 identical ATL clones engrafted in only a few cases.36,38 In the present study, we also observed multiple nonidentical clones when transplanting TCM and TEM cells. Intriguingly, only TSCM cells were capable of repopulating identical ATL clones efficiently in immunodeficient mice, and TCM and TEM cells hardly engraft, even when increased cell numbers were transplanted. In addition, TSCM cells were able to unidirectionally regenerate TCM and TEM cells in vivo. Of note, a low number of HTLV-1–infected TSCM cells successfully repopulated identical ATL clones. Compared with previously reported xenograft models in which a minimum of 1 million PBMCs or CD4+ T cells were inoculated, our results have clearly demonstrated the high efficiency of repopulating capacity in TSCM cells. In addition, several reports have illustrated the existence of preleukemia stem cells. For instances, in CLL, only HSCs can be repopulated and cause monoclonal B lymphocytosis, which is a preleukemia phase of CLL in a xenogeneic model.28 In acute myeloid leukemia, the clone that has only a first hit is detected in HSCs, but the clone with a secondary hit is generated in progenitor levels.39 Given these reports, it is noteworthy in understanding clonal evolution of ATL that the repopulating capacity of TSCM could be observed in the early phase of ATL such as smoldering cases, although unfortunately, we could not perform xenotransplantation assay of acute-type ATL cases because of the sample limitation. Taken together, our data suggest that TSCM cells could be a candidate of ATL-initiating or preleukemia stem cells.
The current study has not focused on whether the TSCM population is a direct target for HTLV-1 infection. Although our in vitro data showed the susceptibility of TSCM cells to HTLV-1 infection, further studies such as detailed phenotypic and genetic analyses in a HTLV-1 infection model using humanized mice40 will be required to delineate the exact nature of HTLV-1 infection and HTLV-1–infected cells.
Collectively, our results have demonstrated the phenotypic and functional heterogeneity and the existence of a hierarchy in ATL cells. TSCM cells have been identified as the hierarchical apex capable of reconstituting the identical ATL clones. This report is the first demonstration of the association between a T-cell malignancy and TSCM cells. A recent report revealed the long-term persistence of quasispecies of HIV-1 and HIV-1 persistence in TSCM cells after long-term antiretroviral therapy.32 Taken together, TSCM cells may be exploited as a venue for clonal evolution and a reservoir for ATL cells. Thus, eradication of ATL-TSCM cells may be a promising strategy to cure ATL.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to Drs Yoshitomo Maesako, Takashi Akasaka, and Hitoshi Ohno (Tenri Hospital); Akihito Yonezawa and Kazunori Imada (Kokura Memorial Hospital); and Takashi Miyoshi and Kenichi Nagai (Kansai Electric Power Hospital) for supplying clinical samples.This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (M.K., M.H.).
Authorship
Contribution: Y.N. and M.K. designed, performed, and analyzed all experiments and wrote the manuscript; M.H. collected samples and wrote the manuscript; Y.S., N.S., and S.F. performed some of the experiments; and N.K. and A.T.-K. supervised the project.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Masakatsu Hishizawa, Department of Hematology and Oncology, Graduate School of Medicine, Kyoto University, 54 Shogoin-Kawahara-cho, Sakyo-ku, Kyoto 606-8507, Japan; e-mail: hishiza@kuhp.kyoto-u.ac.jp.
References
Author notes
Y.N. and M.K. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal