Key Points
IgH-V(D)J NGS-MRD detection pretransplant identifies a cohort at low risk for relapse, for which treatment modification could be considered.
Positive NGS-MRD was highly predictive of relapse and survival as early as 30 days after HCT.
Abstract
Positive detection of minimal residual disease (MRD) by multichannel flow cytometry (MFC) prior to hematopoietic cell transplantation (HCT) of patients with acute lymphoblastic leukemia (ALL) identifies patients at high risk for relapse, but many pre-HCT MFC-MRD negative patients also relapse, and the predictive power MFC-MRD early post-HCT is poor. To test whether the increased sensitivity of next-generation sequencing (NGS)–MRD better identifies pre- and post-HCT relapse risk, we performed immunoglobulin heavy chain (IgH) variable, diversity, and joining (V[D]J) DNA sequences J NGS-MRD on 56 patients with B-cell ALL enrolled in Children’s Oncology Group trial ASCT0431. NGS-MRD predicted relapse and survival more accurately than MFC-MRD (P < .0001), especially in the MRD negative cohort (relapse, 0% vs 16%; P = .02; 2-year overall survival, 96% vs 77%; P = .003). Post-HCT NGS-MRD detection was better at predicting relapse than MFC-MRD (P < .0001), especially early after HCT (day 30 MFC-MRD positive relapse rate, 35%; NGS-MRD positive relapse rate, 67%; P = .004). Any post-HCT NGS positivity resulted in an increase in relapse risk by multivariate analysis (hazard ratio, 7.7; P = .05). Absence of detectable IgH-V(D)J NGS-MRD pre-HCT defines good-risk patients potentially eligible for less intense treatment approaches. Post-HCT NGS-MRD is highly predictive of relapse and survival, suggesting a role for this technique in defining patients early who would be eligible for post-HCT interventions. The trial was registered at www.clinicaltrials.gov as #NCT00382109.
Introduction
Response to therapy, measured through detection of minimal residual disease (MRD) by PCR techniques or multichannel flow cytometry (MFC) has become an essential part of determining disease risk and directing therapeutic approach for children and adults with acute lymphoblastic leukemia (ALL).1,2 In patients at very high risk for treatment failure and deemed eligible for allogeneic hematopoietic cell transplantation (HCT), detection of MRD just prior to starting the transplant preparative regimen has been shown to be highly prognostic.3,4 Investigators from the ALL Relapse Berlin-Frankfurt-Muenster group showed a risk of relapse at 5 years of 55% in children with MRD ≥10−4, and Children’s Oncology Group (COG)/Pediatric Blood and Marrow Transplant Consortium (PBMTC) investigators have shown that relapse rates are increased by 3.3-fold, to 60% at 2 years in children with the presence of ≥0.1% marrow MRD by flow pre-HCT. These high relapse rates have resulted in decreases in survival to as low as 20% to 35% in children who are in clinical remission but with MRD levels ≥10−4 by polymerase chain reaction (PCR) or >0.1% by flow cytometry detected pre-HCT. In contrast, patients with low or absent MRD pre-HCT achieve survival rates of 60% to 80%. These large differences in survival have prompted clinicians to give additional courses of chemotherapy after achieving remission prior to transplant in order to eliminate or minimize pre-HCT MRD, although efficacy of this practice has not been demonstrated.
Although absence of detectable MRD pre-HCT defines a lower-risk population, relapse rates of 15% to 25% persist in this better-risk group. Because levels of MRD right up to the limits of detection of flow cytometry and PCR seem to be important in defining relapse risk, logically, measurement of MRD at even lower levels could possibly either more accurately predict relapse or define a level below which relapse would not occur. Recently, next-generation sequencing (NGS) techniques measuring immunoglobulin heavy chain (IgH)–variable, diversity, and joining (V[D]J) or T-cell receptor clonal rearrangements as a method of detecting MRD have been introduced.5 These approaches expand the sensitivity of MRD detection from 1 blast cell in 104 to 105 cells offered by PCR and flow techniques currently used by large cooperative groups to as high as 1 in 107 cells and have been shown to be predictive of relapse in children with ALL receiving standard chemotherapy.6,7 In order to assess whether the increased sensitivity of NGS-MRD detection could improve our ability to predict low or absent relapse after transplant, we tested banked pre-HCT bone marrow (BM) samples for MRD in a recently closed COG/PBMTC phase 3 trial and compared the predictive power for relapse and survival of IgH-V(D)J deep-sequence (NGS-MRD) detection with standardized flow cytometric MRD data gathered prospectively for the trial. We also looked at the predictive power for relapse of IgH-V(D)J NGS-MRD tested on post-HCT samples and compared this approach directly with MFC-MRD.
Materials and methods
Patients and study design
In order to test the hypothesis that NGS-MRD could improve predictive ability in patients with ALL undergoing HCT, we identified a cohort of uniformly treated patients who enrolled on COG ASCT0431 (PBMTC ONC051 conducted from 2007 to 2011) who had pretransplant ALL blast samples stored as part of COG study AALL05B1 (biology banking study) or other up-front or relapsed COG trials. Patients were ages 1 to 21 with ALL in first complete remission (CR1) and second complete remssion (CR2) and were transplanted utilizing related or unrelated BM, peripheral blood stem cells, or cord blood. Details regarding eligibility and approach were published previously.8 All patients had consented through their parent trials to allow additional biology study performance; all trials were approved through the National Cancer Institute Institutional Review Board and local institutional review boards as applicable. Research was conducted in accordance with the Declaration of Helsinki.
Sample availability and determination of informative ALL blast clonal sequences
All patients were in morphologic remission (<5% blasts) at the time of the pre-HCT BM samples. Results of pre- or post-HCT BM samples for MRD were blinded to treating physicians. Because of the logistics of pretransplant BM assessments sometimes being obtained at outside centers referring to transplant programs, submission of the pre-HCT sample for flow cytometry MRD and biobanking was at the discretion of the participating center. Thus, 74% and 78% of patients had pre-HCT and post-HCT BM samples available for analysis. Secondary encoding of patient identification by the biorepository ensured that neither the study committee nor the laboratory team knew the patient identity or outcome data.
We limited the NGS-MRD analysis to patients with available pre-HCT blast samples in the COG ALL biobank from whom definitive ALL blast clonal sequences could be identified. There were 79 of 143 ASCT0431 participants for whom a pre-HCT blast sample and study BM sample(s) (pre- or post-HCT) was available for laboratory analysis (supplemental Figure 1, flow diagram; see the Blood Web site). Sixty-six patients had B-lineage leukemia, from which 56 patients had clearly detectable IgH-V(D)J complementarity determining region 3 (CDR3) tagging sequences; 2 patients had IgH-DJ clones, and 8 were noninformative.
For 15 of the 56 patients (27%) with IgH-V(D)J determined tagging sequences, a pre-HCT BM sample was not available, leaving 41 patients for the primary analysis of pre-HCT NGS-MRD detection. There were a total of 125 post-HCT MRD assessments available for analysis in 53 of the 56 IgH-V(D)J tagging sequence patients (supplemental Figure 1).
Immunosequencing (IgH-V[D]J NGS-MRD testing)
DNA was extracted from BM cells at all available time points for each patient and was submitted to Adaptive Biotechnologies for sequencing using the ImmunoSeq IgH assay. Details of the assay have been published previously.5 DNA samples were amplified in a single reaction using 128 forward primers, positioned in the framework 2 region of each IgH-V segment, and 9 reverse primers for the 9 IgH-J segments. Any possible IgH-V(D)J rearrangement, including rearrangements using pseudogene V or J segments, can be amplified using this multiplex PCR. Assay sensitivity for detection of any possible V-J combination was confirmed by synthesizing all 1152 possible V-J combinations9 ; an equimolar mix of these synthetic templates was used to titer the IgH primer concentrations in the multiplex PCR for minimal PCR bias. As previously observed for T cell receptor gamma, amplification bias from V and J primers was independent, and residual bias in the assay was consistent, allowing us to computationally remove residual amplification bias of specific clones by adjusting observed frequencies for the V and J primers used to amplify the clone.
In order to define the tumor-tagging IgH-CDR3 sequence(s) for a patient, 120 ng of DNA, representing 40 000 haploid genomes, was used to prepare and sequence an IgH library from a diagnostic time point, when the patient had active disease. This library was sequenced to an average depth of 5 to 10× per unique clone, and the tumor tagging clones were defined as all clones exceeding 5% of the total sequence count in the diagnostic library. In order to track these clones in the pre-HCT samples, a deeper library was analyzed; 3 μg of DNA, representing 1 million haploid genomes, was used to prepare and sequence an IgH library from each available pre-HCT sample. The cumulative frequency of the tumor clones in the pre-HCT sample was used to calculate tumor burden as a fraction of B cells, assuming that all tumor-tagging CDR3 sequences were present in the same cell.
To compare tumor burden from immunosequencing with tumor burden from MFC, we converted the tumor burden from fraction of B cells to the fraction of nucleated cells. In order to do this, an aliquot containing an average of 100 unique synthetic templates was spiked into the pre-HCT library multiplex PCR reactions. The observed depth of coverage per synthetic template provides a direct estimate of the total amplification factor for the library (reads per unique input template) and, thus, an estimate of the total number of amplified input templates. Constraining analysis to the tumor-tagging clones, this allowed us to directly estimate the frequency of tumor genomes among the input genomes.
MFC-MRD detection
All MFC-MRD was performed at a single COG reference laboratory and transplant center personnel were blinded to MRD results. MRD was measured on BM aspirates using 6-color flow cytometry, the current standard of care MRD detection method for COG trials.10 Samples were stained with 2 different 6-color antibody combinations: CD20-FITC/CD10-PE/CD38-PerCPCy5.5/CD58-APC/CD19-PECy7/CD45-APCH7 and CD9/CD13+33/CD34/CD10/CD19/CD45. A third tube contained SYTO-16 to identify all nucleated cells using a method previously described by Dworzak.11 CD19 in this tube was used to express B cells as a percent of all nucleated cells; MRD identified in either of the 2 test tubes was expressed as a percent of B cells and the third tube used to calculate MRD as a percent of nucleated cells. Finally, mononuclear cells were estimated on a display of CD45/side scatter to exclude granulocytes, and MRD ultimately expressed as a percent of mononuclear cells, so as to better match the denominator of the Ficoll-Hypaque–separated banked cells used for NGS-MRD detection.12
Statistical analysis
The primary outcome was time to relapse from HCT, and differences in relapse rates were assessed using the likelihood ratio test from Cox regression of relapse rates comparing appropriate nested models. Nonparametric estimation of relapse risk, accounting for treatment related mortality as a competing risk was done using the Aalen-Johansen method.13 All reported relapse probabilities are cumulative incidence, accounting for treatment related mortality. To provide simple graphical comparisons of outcome probabilities over time by acute graft-versus-host disease (aGVHD) or post-HCT MRD status, the landmark analysis curve approach14 was used in which outcome probabilities were computed from day a given landmark time forward with status determined and fixed at that landmark time. For aGVHD, day +55 was chosen because 93% of those who developed aGVHD had experienced peak grade by this point, and determination of risk classification at this time would allow sufficient time for implementing further treatment strategies. For post-HCT MRD, landmark times were chosen after peri-engraftment, 3-month, and 8-month MRD samples were taken. The use of landmark curves avoids bias that would occur comparing Kaplan-Meier estimates starting at HCT based on aGVHD or post-HCT MRD status, events that occur after HCT. Reported hazard ratios and P values are from the Cox regression analysis with aGVHD or/and post-HCT MRD as a time-dependent covariates and uses all the available data.
Results
Predictive power of pretransplant IgH-V(D)J NGS-MRD for relapse and survival after total body irradiation (TBI)–based myeloablative HCT
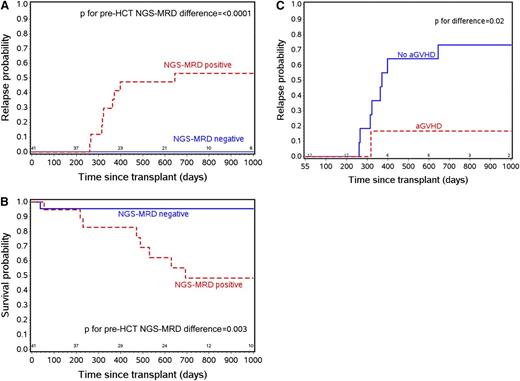
A total of 41 patients were eligible for pre-HCT NGS-MRD analysis. Table 1 shows the distributions of patient and disease characteristics in the full ASCT0431 population and in the subset of B-cell patients with clones detected using the IgH-V(D)J NGS-MRD assay. There were no statistical differences in clinical characteristics of patients detected by IgH-V(D)J NGS-MRD and the remainder of the study population. The actuarial median follow-up for this group of patients is 24 months (interquartile range, 18-37) compared with the 26 months (23-38) for the full study. Of note, none of the 22 pre-HCT IgH-V(D)J NGS-MRD negative patients relapsed, whereas 9 of the 19 MRD positive patients relapsed. Figure 1 shows the estimated relapse and survival probabilities by time from transplant. The 2-year relapse probability was 0% and 53% in pre-HCT NGS-MRD negative and positive patients, respectively (P < .0001). This led to improved 2-year probability of OS of 96% vs 48% in pre-HCT NGS-MRD negative vs positive patients, respectively (Figure 1B; P = .003).
Patient characteristics for all ASCT0431 patients and patients included in the pre-HCT NGS-MRD statistical analysis
| . | All patients . | Tagging sequence determined by IgH-V(D)J in pre-HCT blast sample . | ||
|---|---|---|---|---|
| N . | Percentage . | N . | Percentage . | |
| Total | 143 | 100 | 41 | 100 |
| Age | ||||
| 1-9 | 76 | 53 | 23 | 56 |
| 10+ | 67 | 47 | 18 | 44 |
| Sex | ||||
| Female | 59 | 41 | 19 | 46 |
| Male | 84 | 59 | 22 | 54 |
| Relapse risk group assessment | ||||
| High-risk CR1 | 49 | 34 | 17 | 41 |
| High-risk CR2 | 69 | 48 | 14 | 34 |
| Intermediate-risk CR2 | 25 | 17 | 10 | 24 |
| Stem cell source/HLA matching | ||||
| Matched sibling | 78 | 55 | 24 | 59 |
| Other related donor | 4 | 3 | 2 | 5 |
| Unrelated donor, BM or PBSC | 34 | 24 | 9 | 22 |
| Unrelated donor, cord blood | 27 | 19 | 6 | 15 |
| Relapse risk group/stem cell source | ||||
| High-risk CR1 sib | 29 | 20 | 11 | 27 |
| High-risk CR1 noncord nonsib | 17 | 12 | 5 | 12 |
| High-risk CR1 cord | 3 | 2 | 1 | 2 |
| High-risk CR2 sib | 24 | 17 | 3 | 7 |
| High-risk CR2 noncord nonsib | 21 | 15 | 6 | 15 |
| High-risk CR2 cord | 24 | 17 | 5 | 12 |
| Intermediate-risk CR2 sib | 25 | 17 | 10 | 24 |
| Grade of acute GVHD | ||||
| None | 88 | 62 | 25 | 61 |
| 1 | 20 | 14 | 8 | 20 |
| 2 | 19 | 13 | 5 | 12 |
| 3 | 9 | 6 | 2 | 5 |
| 4 | 7 | 5 | 1 | 2 |
| Chronic GVHD | ||||
| No | 107 | 75 | 29 | 71 |
| Yes | 36 | 25 | 12 | 29 |
| MFC-MRD status pretransplant* | N = 105 | N = 40 | ||
| Negative | 75 | 71 | 28 | 70 |
| Positive | 30 | 29 | 12 | 30 |
| . | All patients . | Tagging sequence determined by IgH-V(D)J in pre-HCT blast sample . | ||
|---|---|---|---|---|
| N . | Percentage . | N . | Percentage . | |
| Total | 143 | 100 | 41 | 100 |
| Age | ||||
| 1-9 | 76 | 53 | 23 | 56 |
| 10+ | 67 | 47 | 18 | 44 |
| Sex | ||||
| Female | 59 | 41 | 19 | 46 |
| Male | 84 | 59 | 22 | 54 |
| Relapse risk group assessment | ||||
| High-risk CR1 | 49 | 34 | 17 | 41 |
| High-risk CR2 | 69 | 48 | 14 | 34 |
| Intermediate-risk CR2 | 25 | 17 | 10 | 24 |
| Stem cell source/HLA matching | ||||
| Matched sibling | 78 | 55 | 24 | 59 |
| Other related donor | 4 | 3 | 2 | 5 |
| Unrelated donor, BM or PBSC | 34 | 24 | 9 | 22 |
| Unrelated donor, cord blood | 27 | 19 | 6 | 15 |
| Relapse risk group/stem cell source | ||||
| High-risk CR1 sib | 29 | 20 | 11 | 27 |
| High-risk CR1 noncord nonsib | 17 | 12 | 5 | 12 |
| High-risk CR1 cord | 3 | 2 | 1 | 2 |
| High-risk CR2 sib | 24 | 17 | 3 | 7 |
| High-risk CR2 noncord nonsib | 21 | 15 | 6 | 15 |
| High-risk CR2 cord | 24 | 17 | 5 | 12 |
| Intermediate-risk CR2 sib | 25 | 17 | 10 | 24 |
| Grade of acute GVHD | ||||
| None | 88 | 62 | 25 | 61 |
| 1 | 20 | 14 | 8 | 20 |
| 2 | 19 | 13 | 5 | 12 |
| 3 | 9 | 6 | 2 | 5 |
| 4 | 7 | 5 | 1 | 2 |
| Chronic GVHD | ||||
| No | 107 | 75 | 29 | 71 |
| Yes | 36 | 25 | 12 | 29 |
| MFC-MRD status pretransplant* | N = 105 | N = 40 | ||
| Negative | 75 | 71 | 28 | 70 |
| Positive | 30 | 29 | 12 | 30 |
One patient with deep-sequence MRD did not have flow MRD.
PBSC, peripheral blood stem cells.
Estimated relapse and survival probabilities by time from transplant. (A) Relapse risk by NGS-MRD status. (B) Overall survival (OS) by NGS-MRD status. (C) Relapse risk by aGVHD status among NGS-MRD positive patients.
Estimated relapse and survival probabilities by time from transplant. (A) Relapse risk by NGS-MRD status. (B) Overall survival (OS) by NGS-MRD status. (C) Relapse risk by aGVHD status among NGS-MRD positive patients.
There was no trend in relapse rates among pre-HCT NGS-MRD positive patients by quantity of leukemia cells detected as a percentage of total nucleated cells in the marrow samples (hazard ratio [HR] = 1.0 for upper vs lower than the median, P = .99), with relapse occurring frequently even at the lowest levels of detection (<10−6).
Instead, the occurrence of aGVHD was important in defining relapse risk of pre-HCT NGS-MRD positive patients. Among the 19 pre-HCT MRD positive patients, the estimated 2-year relapse probabilities were 73% for patients with no aGVHD by day +55 and 17% for those who experienced aGVHD by day +55 (P = .02; Figure 1C). The association of aGVHD with decreased relapse risk was described in the parent trial report,15 but it is notable how important aGVHD was for decreasing relapse specifically in pre-HCT NGS-MRD positive patients. Also of note, there was no difference in the aGVHD occurrence between NGS-MRD negative (41% aGVHD) and positive (37% aGVHD) patients.
Comparison of NGS-MRD with standardized MFC-MRD
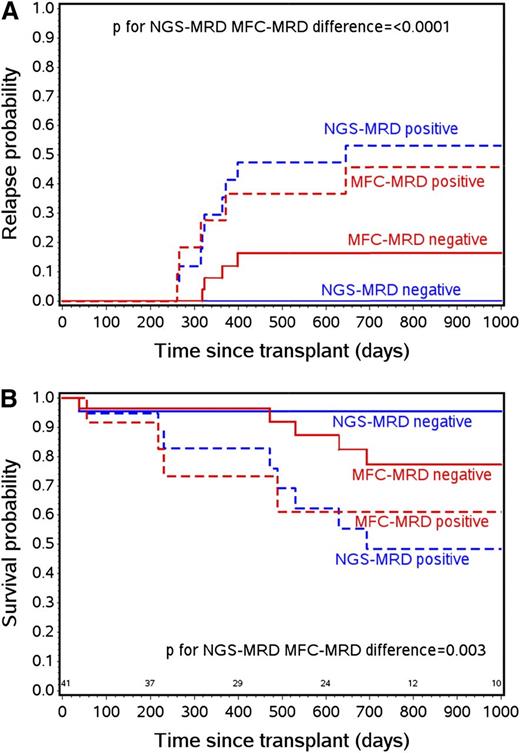
Forty of the 41 IgH-NGS-MRD informative patients also had pre-HCT MRD status determination by MFC. NGS-MRD predicted relapse significantly better than flow cytometry MRD (P < .0001), whereas MFC-MRD did not improve relapse prediction after adjusting for deep sequence MRD (P = .67). Similarly, NGS-MRD was better at predicting OS compared with flow MRD (P = .003). Figure 2A-B show probability curves of patients who were MRD negative or positive by flow cytometry or deep sequencing. The 2-year relapse probabilities were 53% and 0% among NGS-MRD positive and negative patients, respectively (P < .0001), compared with 46% and 16% among flow MRD positive and negative populations (P = .02). In addition, NGS-MRD negativity predicted a rate of 0% relapse and 96% OS in this cohort.
Probability curves of patients who were MRD negative or positive by flow cytometry or deep sequencing. (A) Relapse risk by pre-HCT NGS-MRD compared with MFC-MRD status. (B) OS by pre-HCT NGS-MRD compared with NGS-MRD status.
Probability curves of patients who were MRD negative or positive by flow cytometry or deep sequencing. (A) Relapse risk by pre-HCT NGS-MRD compared with MFC-MRD status. (B) OS by pre-HCT NGS-MRD compared with NGS-MRD status.
Effect of post-HCT NGS-MRD detection on risk of relapse
Between 1 and 4 post-HCT BM MRD assessments per patient performed at days +30, +100, or 8 to 12 months were available for analysis. In 53 of the 56 patients with IgH-V(D)J tagged tumors, there were a total of 125 post-HCT samples available for NGS-MRD assessment (Table 2). Eleven relapses occurred in the 15 MRD+ patients (73%) with any positive post-HCT MRD, compared with 5 of 38 patients (13%) with consistently negative post-HCT MRDs (HR = 14.5, P < .0001). Risk for death was also increased in post-HCT NGS-MRD patients (HR = 6.0, P = .005). NGS-MRD libraries prepared at the 30-day time point contained significantly fewer total sequences than any other time points, reflecting lymphopenia as would be expected early after transplant. NGS-MRD negative patients at this time point who later relapse may reflect incomplete sampling of a hypoplastic marrow.
Post-HCT NGS-MRD correlations
| . | . | . | Relapse . | |
|---|---|---|---|---|
| Number of post-HCT NGS-MRD . | Positive, n . | Patients, n . | N . | % . |
| No. of tests | ||||
| None | — | 3 | 0 | 0 |
| 1 | 0 | 6 | 1 | 17 |
| 1 | 1 | 2 | 1 | 50 |
| 2 | 0 | 15 | 4 | 27 |
| 2 | 1 | 4 | 2 | 50 |
| 2 | 2 | 1 | 1 | 100 |
| 3-4 | 0 | 17 | 0 | 0 |
| 3-4 | 1 | 5 | 4 | 80 |
| 3-4 | 2+ | 3 | 3 | 100 |
| . | . | . | Relapse . | |
|---|---|---|---|---|
| Number of post-HCT NGS-MRD . | Positive, n . | Patients, n . | N . | % . |
| No. of tests | ||||
| None | — | 3 | 0 | 0 |
| 1 | 0 | 6 | 1 | 17 |
| 1 | 1 | 2 | 1 | 50 |
| 2 | 0 | 15 | 4 | 27 |
| 2 | 1 | 4 | 2 | 50 |
| 2 | 2 | 1 | 1 | 100 |
| 3-4 | 0 | 17 | 0 | 0 |
| 3-4 | 1 | 5 | 4 | 80 |
| 3-4 | 2+ | 3 | 3 | 100 |
| NGS-MRD status pre- and post-HCT* . | . | . | . | |
|---|---|---|---|---|
| Pre-HCT . | Positive post-HCT . | . | . | . |
| Negative | 0 | 19 | 0 | 0 |
| Negative | 1+ | 2 | 0 | 0 |
| Positive | 0 | 7 | 1 | 14 |
| Positive | 1+ | 10 | 8 | 80 |
| NGS-MRD status pre- and post-HCT* . | . | . | . | |
|---|---|---|---|---|
| Pre-HCT . | Positive post-HCT . | . | . | . |
| Negative | 0 | 19 | 0 | 0 |
| Negative | 1+ | 2 | 0 | 0 |
| Positive | 0 | 7 | 1 | 14 |
| Positive | 1+ | 10 | 8 | 80 |
| Post-HCT deep-sequence and flow MRD . | . | . | . | |
|---|---|---|---|---|
| Positive NGS-MRD . | Positive MFC-MRD* . | . | . | . |
| 0 | 0 | 35 | 5 | 14 |
| 0 | 1+ | 3 | 0 | 0 |
| 1+ | 0 | 11 | 7 | 64 |
| 1+ | 1+ | 3 | 3 | 100 |
| Post-HCT deep-sequence and flow MRD . | . | . | . | |
|---|---|---|---|---|
| Positive NGS-MRD . | Positive MFC-MRD* . | . | . | . |
| 0 | 0 | 35 | 5 | 14 |
| 0 | 1+ | 3 | 0 | 0 |
| 1+ | 0 | 11 | 7 | 64 |
| 1+ | 1+ | 3 | 3 | 100 |
Among patients with at least 1 post-HCT assessment (3 patients had a pre- but no post-HCT NGS-MRD performed, and 1 patient had post-HCT NGS-MRD but no post-HCT MFC-MRD).
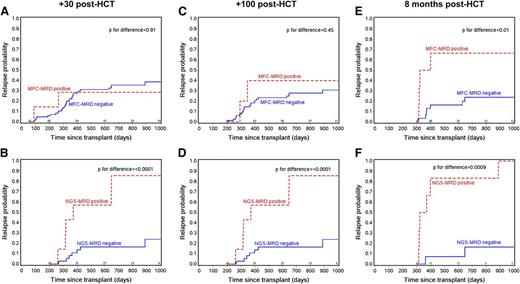
In spite of lymphopenia, the ability to predict relapse is higher using NGS-MRD compared with MFC-MRD, especially early after HCT. Figure 3A-C and D-F show landmark curves for relapse probability as a function of time post-HCT (1, 3, and 8 months) for MFC-MRD (n = 112 with at least 1 post-HCT MRD time point) and NGS-MRD (n = 53) cohorts, respectively. At day +30, rates of relapse of MFC-MRD positive vs negative patients overlap, and post-HCT NGS-MRD positive patients have estimated relapse probability of 67% compared with post-HCT NGS-MRD negative patients with relapse rates of 25% (P = .01). The better predictive power of post-HCT NGS-MRD continues vs MFC-MRD at day 100 and 8 months. Multivariate analysis showed that the presence of detectable MRD after transplant led to an increase in relapse (HR = 7.7; P = .05) independent of other factors, including pre-HCT MRD and aGVHD status.
Landmark curves for relapse probability as a function of time post-HCT for MFC-MRD and NGS-MRD cohorts. (A) Relapse probability by day +30 MFC-MRD status. (B) Relapse probability by day +30 NGS-MRD status. (C) Relapse probability by day +100 MFC-MRD status. (D) Relapse probability by day +100 NGS-MRD status. (E) Relapse probability by 8-month MFC-MRD status. (F) Relapse probability by 8-month NGS-MRD status.
Landmark curves for relapse probability as a function of time post-HCT for MFC-MRD and NGS-MRD cohorts. (A) Relapse probability by day +30 MFC-MRD status. (B) Relapse probability by day +30 NGS-MRD status. (C) Relapse probability by day +100 MFC-MRD status. (D) Relapse probability by day +100 NGS-MRD status. (E) Relapse probability by 8-month MFC-MRD status. (F) Relapse probability by 8-month NGS-MRD status.
Pre- and post-HCT NGS-MRD detection correlations
None of the pre-HCT NGS-MRD negative patients relapsed. One patient relapsed out of the 7 who were pre-HCT NGS-MRD positive with only negative post-HCT MRDs, compared with 8 relapses in the 10 patients who were pre-HCT MRD positive and had at least 1 positive post-HCT MRD (P = .001; Table 2).
Comparison of post-HCT V(D)J NGS-MRD with standardized MFC-MRD with patients using both techniques
There were 117 post-HCT MRD assessments in 52 patients done by both deep-sequencing and flow cytometry methods with 14 patients (27%) with at least 1 positive post-HCT NGS-MRD compared with 6 patients (12%) with at least 1 positive post-HCT MFC-MRD. Like pre-HCT MRD, by Cox regression analysis, deep-sequencing post-HCT MRD predicted relapse better than flow cytometry MRD (P < .0001), whereas flow cytometry MRD did not significantly add to the deep sequence MRD relapse prediction (P = .09). The improved predictive ability of deep sequence MRD is primarily because of higher sensitivity. There were 7 relapses among the 11 patients who were positive post-HCT by NGS-MRD and negative by flow MRD, but none of the patients positive by flow and negative by NGS-MRD relapsed. All 3 patients positive post-HCT by both methods relapsed (Table 2).
Discussion
Our first goal with this study was to determine if low or absent levels of IgH-V(D)J NGS-MRD could better define patients at low risk of relapse after transplant. In patients where CDR3 tags were detected in the active disease sample, we were able to detect verified blast percentages as low as 4.2 × 10−7 in BM samples. This high level of sensitivity meant that pre-HCT NGS-MRD negativity was strongly correlated with relapse-free survival in our cohort. The fact pre-HCT NGS-MRD negative patients did not relapse and had an event-free survival of 96% opens the possibility of altering therapeutic approaches for these patients.
There are important implications to our ability to define, prior to the procedure, a population with ALL slated for allogeneic HCT who are at very low risk of relapse. Children and adults with high-risk ALL able to tolerate myeloablative therapy have been noted to have superior survival when TBI-based preparative regimens are used compared with chemotherapy regimens, and TBI is currently recommended in American Society for Blood and Marrow Transplantation guidelines for ALL HCT.16-22 Concern has been raised about use of these intensive approaches because of known significant late effects of TBI or cranial irradiation in growing children, and adults have a higher risk of secondary malignancies after TBI-based regimens.23-25 One multicenter trial of non-TBI containing reduced intensity conditioning in children ineligible for TBI noted that children with ALL who were MRD negative pre-HCT had promising rates of survival.26 It is possible that TBI could be eliminated for children and adults who are NGS-MRD negative pre-HCT with no increase in relapse. In addition, there is evidence that younger patients with late-relapsing (relapse occurring >36 months after diagnosis) B-ALL who become MRD negative by PCR or flow cytometry after their first round of reinduction chemotherapy have high rates of cure with chemotherapy alone.27,28 It is possible that achievement of NGS-MRD negative status at some time after first or subsequent reinduction rounds of chemotherapy in young patients with relapsed ALL may define a larger population that could be cured by chemotherapy alone, thus avoiding the potential toxicities of allogeneic HCT.
The present work demonstrates that next generation sequencing approaches can identify a subset of patients at significant risk of relapse who were previously defined as MRD negative by flow. Our results suggest a note of caution about abandoning allogeneic HCT for high-risk ALL patients. Even patients at the lowest levels of disease burden measured by NGS-MRD (10−6 to 10−7) benefitted from a graft-versus-leukemia effect, as patients with low levels of disease not getting aGVHD relapsed. This observation confirms earlier work by our group and others showing that the allogeneic effect is important in curing a large percentage of patients with high-risk ALL undergoing HCT.3,29-33
Although numbers are small, it is worth noting that patients in our trial below the level of detection of IgH NGS-MRD did not seem to need aGVHD to achieve cure. If such a result were upheld in larger studies, it would have several important implications. Perhaps, as has been seen in some studies with AML,34 patients with this level of MRD may be cured by an allogeneic effect not associated with GVHD, or may not need immunologic graft-versus-leukemia at all. It may be that such patients have reached a level of disease control where either an autologous transplant or chemotherapy alone could be sufficient for cure. We do not know if IgH-V(D)J NGS-MRD negative patients have become truly negative or if there is a hidden threshold of cells below which cure is inevitable that we simply cannot measure. In addition, MRD detection is only one aspect of disease risk, and there may be different clinical or biological subsets of ALL that require different thresholds of MRD for cure. Answers to these questions could significantly change the way we treat very high-risk patients with ALL for whom allogeneic transplantation is currently recommended therapy.
The second goal of our study was to define the predictive power of post-HCT NGS-MRD detection. Very little is published looking at the predictive power of post-HCT MRD in ALL. Balduzzi et al showed that 5-year event-free survival of patients with post-HCT MRD detectable by PCR was surprisingly high, at 40%.35 They noted, however, that early detection of MRD (in the first 100 days) was associated with an increase in risk of failure (risk ratio = 2.5; CI, 1.05-5.75; P = .04), but if the MRD was detected after 6 months, the increase in risk was more pronounced (risk ratio = 7.8; CI, 2.2-27.8; P = .002). We show here a similar finding, with a poor ability of MFC-MRD to predict relapse when detected early after HCT, but a higher risk of relapse associated with later time points. This lack of specificity in MFC-MRD early after transplant could be because of low-level clones associated with intense B-cell regeneration post-HCT, as described by Fronkova et al.36 In a larger study, Bader et al showed greater specificity of identifying patients at risk for relapse post-HCT when higher levels of disease were detected at both early and late time points (≥10−4 using PCR techniques).37 Similarly, when we used NGS-MRD techniques, even at the earliest time point of day 30 after HCT, positive results were able identify a probability of relapse approaching 70%, a threshold at which intervention to prevent relapse may be warranted. NGS-MRD may be better than MFC-MRD for defining true ALL blasts vs clonal cells associated with B-cell regeneration.
Logan et al described a single center experience testing the prognostic significance of deep sequence MRD in adult ALL patients undergoing HCT.38 The 29 patients in this series were more heterogeneous in remission state and treatment approach than our population, including CR3 and refractory patients treated by various approaches including reduced intensity, non-TBI myeloablative and TBI-based approaches. The most important difference between our approach and Logan et al is that they combined a variety of clonotypes, including IgH-V(D)J (15 patients) as well as patients with, IgH-(D)J, IgK, T-cell receptor (TCR) β, TCRδ, and TCRζ. Because of the possibility that differing rearrangements may represent biological differences in relapse risk (pro-pre-B vs pre-B-cell, B vs T-cell, etc.), and because we had a large number of IgH-V(D)J patients, we chose to focus on this specific clonotype. In addition, our patients were uniformly treated as part of a large multicenter study and were children, as opposed to adults, with a better predicted outcome with HCT. In spite of the heterogeneity and small numbers in Logan et al, they observed predictive power for relapse with pre-HCT levels above 10−4 and post-HCT levels above 10−6. Differing significantly from our outcome, their “good-risk” patients with pre-HCT NGS-MRD below 10−4 pre-HCT had disease-free survival just above 30%, whereas those with higher MRD did not survive. Although both studies find improved survival in NGS-MRD negative patients, the qualitative differences could reflect either technical differences in sensitivity of detecting NGS-MRD or clinical differences in the outcomes being analyzed; because Logan et al did not compare their deep sequencing approach with any other method of MRD detection, it is difficult to judge the comparative applicability of their approach.
In conclusion, our study suggests a role for IgH-V(D)J NGS-MRD detection in clinical management of patients with ALL undergoing allogeneic HCT. We restricted our analysis to patients with B-cell precursor ALL with IgH-V(D)J rearrangements and had sufficient number to demonstrate that (1) patients with no detectable IgH-V(D)J NGS-MRD pre-HCT have a very low risk of relapse; (2) those with IgH-V(D)J NGS-MRD detected pre-HCT who did not get GVHD or who had any detectable disease post-HCT had high rates of relapse; and (3) IgH-V(D)J NGS-MRD detection was superior to flow cytometry–based MRD for prediction of relapse or nonrelapse in both the pre- and post-HCT settings. Additional studies will be needed to appropriately characterize deep sequencing MRD approaches for patients with T-cell ALL and non–IgH-V(D)J recombinations of B-cell ALL. In addition, our trial included CR1 and CR2 patients, but no patients in CR3 and no patients treated with non-TBI myeloablative or reduced intensity regimens. Further study of different populations of patients with ALL undergoing HCT with differing approaches will assist in more precisely defining the role of this technique for MRD detection compared with other methods in determining risk and guiding therapy.
Presented in abstract form at the 55th annual meeting of the American Society of Hematology, New Orleans, LA, December 10, 2013.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by grants from the National Institutes of Health National Cancer Institute (N01 HC-45220/HHSN268200425220C, U10 CA098543, and R01CA1116660). PBMTC activities were supported by a grant from the National Institutes of Health National Cancer Institute/National Heart, Lung, and Blood Institute (2U01HL069254) and a consortium grant from the St. Baldrick’s Foundation.
Authorship
Contribution: M.A.P. and S.A.G. designed the study, analyzed the data, wrote the manuscript, and are responsible for the work as a whole; B.L. assisted with study design and performed the data analysis; C.C. performed the NGS-MRD analysis; J.M.G.-F. biobanked and processed baseline and BM samples; M.B. performed the MFC-MRD analysis; C.C., B.L., D.A.W., K.R.S., N.B., I.K., J.M.G.-F., D.W., C.D., M.K., and M.B. contributed to the study design and wrote/edited the manuscript.
Conflict-of-interest disclosure: C.D., I.K., and D.W. are employed by and C.C. and M.K. have ownership interests in Adaptive Biotechnologies, which has obtained Clinical Laboratory Improvement Amendments certification to provide residual disease testing described herein as a service, under the brand name ClonoSeq. To mitigate the potential conflict of interest, these authors were blind to the relapse status of the patients when collecting and interpreting the deep-sequence MRD data. The final freeze of the deep-sequence MRD data was made before B.L. linked these data to the clinical outcomes for analysis at COG. The remaining authors declare no competing financial interests.
Correspondence: Michael A. Pulsipher, University of Utah School of Medicine/Huntsman Cancer Institute, Division of Hematology and Hematological Malignancies, 30 North 1900 East, Room 5C402, Salt Lake City, UT 84132; e-mail: michael.pulsipher@hsc.utah.edu.