Key Points
Parmodulins are a new class of PAR1 inhibitors that target the cytosolic face of PAR1 to block signaling through Gαq, but not Gα12/13.
Unlike vorapaxar, which causes endothelial injury, parmodulins selectively block proinflammatory, but not cytoprotective, signaling.
Abstract
Protease-activated receptor-1 (PAR1) couples the coagulation cascade to platelet activation during myocardial infarction and to endothelial inflammation during sepsis. This receptor demonstrates marked signaling bias. Its activation by thrombin stimulates prothrombotic and proinflammatory signaling, whereas its activation by activated protein C (APC) stimulates cytoprotective and antiinflammatory signaling. A challenge in developing PAR1-targeted therapies is to inhibit detrimental signaling while sparing beneficial pathways. We now characterize a novel class of structurally unrelated small-molecule PAR1 antagonists, termed parmodulins, and compare the activity of these compounds to previously characterized compounds that act at the PAR1 ligand–binding site. We find that parmodulins target the cytoplasmic face of PAR1 without modifying the ligand-binding site, blocking signaling through Gαq but not Gα13 in vitro and thrombus formation in vivo. In endothelium, parmodulins inhibit prothrombotic and proinflammatory signaling without blocking APC-mediated pathways or inducing endothelial injury. In contrast, orthosteric PAR1 antagonists such as vorapaxar inhibit all signaling downstream of PAR1. Furthermore, exposure of endothelial cells to nanomolar concentrations of vorapaxar induces endothelial cell barrier dysfunction and apoptosis. These studies demonstrate how functionally selective antagonism can be achieved by targeting the cytoplasmic face of a G-protein–coupled receptor to selectively block pathologic signaling while preserving cytoprotective pathways.
Introduction
Protease-activated receptor 1 (PAR1) is a widely expressed G-protein–coupled receptor (GPCR) that detects proteases in the extracellular environment with high sensitivity. It is activated by an intramolecular signaling mechanism whereby cleavage of the N-terminus of the receptor by a protease exposes a tethered ligand that interacts with a shallow, extended binding site on the extracellular face of the receptor.1 The effective concentration of the tethered ligand is high, owing to entropy considerations and has been estimated to be ∼400 μM, making the ligand difficult to block using soluble antagonists.1 The molecular pharmacology of PAR1 is further complicated by the fact that PAR1 is a functionally-biased receptor that activates different signaling cascades depending on the proteases by which it is cleaved and the cell type on which it resides. The signaling bias of PAR1 is particularly prominent in endothelial cells, in which activation of PAR1 by thrombin or metalloproteases results in loss of barrier function and apoptosis,2-4 whereas activation of PAR1 by activated protein C (APC) is protective against loss of barrier function and apoptosis induced by chemokines and toxins.5-11
PAR1 has been the focus of several drug development programs, owing to the importance of platelet PAR1 in myocardial infarction and stroke. Two fundamental problems in developing small-molecule inhibitors of PAR1 have been (1) identifying compounds capable of competing with the tethered ligand at its binding site and (2) blocking the detrimental effects of PAR1 signaling while preserving its cytoprotective signaling. Several small-molecule inhibitors of PAR1 that effectively compete with the ligand binding site have been developed.12-16 The most well-characterized of these orthosteric inhibitors is vorapaxar, which recently received Food and Drug Administration approval for the prevention of thrombotic cardiovascular events in high-risk patients. Vorapaxar has been evaluated in 2 large, randomized phase 3 clinical studies in the settings of acute coronary syndromes (TRACER)17 and in secondary prevention of atherothrombotic events (TRA-2P).18 Its use was associated with decreased cardiovascular death or ischemic events in the secondary prevention trial.18 However, vorapaxar recipients had a significantly increased incidence of major bleeding, including intracranial hemorrhage, in both studies. Meta-analyses of clinical trials evaluating vorapaxar and a second orthosteric PAR1 antagonist, atopaxar, confirm an association of these inhibitors with bleeding risk.19,20
Like vorapaxar and atopaxar, the vast majority of small-molecule GPCR antagonists in clinical use target the ligand-binding pocket. However, selective control of downstream G-protein–coupled signaling pathways may be achieved by targeting alternative binding sites.21 This approach is particularly relevant for GPCRs such as PAR1, which mediates signaling through pathologic and protective pathways that are distinct and separable. We now describe a new class of PAR1 antagonists termed parmodulins. These compounds act at the cytosolic face of PAR1 and selectively inhibit some, but not all, Gα-mediated signaling cascades in platelets and endothelial cells. We find that orthosteric antagonists such as vorapaxar inhibit cytoprotective signaling and elicit endothelial injury, even at nanomolar concentrations achieved in plasma with dosing regimens described in clinical studies. In contrast, parmodulins inhibit proinflammatory signaling without blocking APC-mediated protective effects or eliciting apoptosis. These studies demonstrate that functionally selective compounds that target the cytosolic surface represent an important alternative to orthosteric antagonists for GPCRs with functional signaling bias.
Methods
Reagents and compounds
Parmodulins were either synthesized at the Broad Institute or purchased from ChemBridge. Vorapaxar and atopaxar were obtained from Axon Medchem BV. SCH79797 and probenecid were obtained from Tocris. The PAR1 agonist SFLLRN, the PAR4 agonist AYPGKF, and BMS-200261 were obtained from Sigma-Aldrich. Fura-2 was obtained from Life Technologies. Tissue-culture reagents were obtained from Lonza. COS-7 cells, human umbilical vein endothelial cells (HUVECs), and human microvascular endothelial cells (HMVECs) were obtained from ATCC. High-affinity thrombin-receptor–activating peptide (haTRAP, Ala-Phe[p-F]-Arg-ChA-HArg-[(3)H]Tyr-NH[2])14 was synthesized by the Tufts Analytical Core Facility. Human APC was obtained from Haematological Technologies. Radiolabeled [3H]haTRAP was obtained from PerkinElmer, Inc. Glass microfiber filters were purchased from Whatman. Rat anti-mouse GPIbβ antibody conjugated to DyLight 649 was obtained from Emfret.
PAR receptor constructs
AP-PAR1 and AP-PAR4 constructs used for transfection were a generous gift from Dr Shaun R. Coughlin, University of California–San Francisco. A construct encoding for a PAR1 ΔH8 mutant, in which amino acid residues S376-L386 are replaced with an A-A-A linker, was a generous gift of Dr. Athan Kuliopulos, Sackler School of Graduate Biomedical Sciences, Tufts University. A detailed description of chimeric construct generation and expression in COS-7 cells is provided in the supplemental Methods, available on the Blood Web site.
Ca2+ flux measurements
Ca2+ flux was evaluated using fluorometry in COS-7 cells expressing human PAR1, PAR4, PAR1:PAR4 chimeras, the ΔH8 PAR1 mutant, or washed platelets. Detailed methods are provided in the supplemental Methods.
Platelet aggregation
Aggregation of platelet-rich plasma or washed platelets was initiated by the indicated concentrations of agonists and measured using a ChronoLog 680 Aggregation System as described previously.22 Experiments using epinephrine were performed in the presence of 50 U/mL hirudin to prevent thrombin formation. Parmodulins were used at the lowest concentration, resulting in >90% inhibition of the aggregation of washed platelets in response to 5 μM SFLLRN. Studies assessing reversibility of inhibition are described in the supplemental Methods.
RhoA-GTP assay
Washed platelets (2 × 108/mL in HEPES-Tyrode buffer) or HUVECs plated in 6-well platelets were incubated with the indicated parmodulins or orthosteric inhibitors and then activated with 10 μM SFLLRN for 1 minutes before lysis. Five μL lysate was kept for analysis of total protein expression and the remaining lysate used to precipitate GTP-bound protein using agarose beads conjugated to GST-Rhotekin-RBD for RhoA (Cytoskeleton). Total and precipitated GTPase protein was measured by immunoblot analysis and visualized using enhanced chemoluminescence. Gels representative of 3 to 5 trials for each condition are presented.
Equilibrium-binding studies
Platelet membranes were prepared from outdated human donor platelets supplied by the blood bank at Beth Israel Deaconess Medical Center and BloodSource as previously described.22 Binding studies were performed in triplicate in 1.5 mL Eppendorf tubes at a final assay volume of 200 μL. Bovine serum albumin (0.1%) was included in the incubation buffer and filter plates were presoaked in 0.1% polyethyleneimine to reduce binding of [3H]haTRAP to tubes and pipette tips. [3H]haTRAP (25 nM) was mixed with the indicated concentration of compounds or vehicle in binding buffer (50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM ethylene glycol tetraacetic acid (EGTA), 0.1% bovine serum albumin). The reaction was initiated with the addition of membranes (0.4 mg/mL) to the reaction mixture. The tubes were capped and mixed gently in a water-bath shaker for 1 hour at room temperature. The reaction was stopped via vacuum filtration using Whatman GF/B filters presoaked for at least 1 hour in 0.1% polyethyleneimine. Filters were rapidly washed 4 times with 300 μL of ice-cold 50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM EGTA. The filters were then added to individual scintillation vials. Five mL of Ultima Gold Scintillation cocktail was added to each vial, and the plates were counted in a scintillation counter. Binding of [3H]haTRAP to platelet membranes was analyzed using nonlinear regression to obtain apparent Kd and βmax.
Apoptosis assay
The detection of apoptotic HUVECs was analyzed using a modified version of the Vybrant apoptosis kit #4 (Life Technologies). HUVECs were seeded onto glass coverslips in 24-well plates and grown until they were confluent. Cells were treated with the indicated concentrations of antagonists followed by the addition of Yo-Pro-1 as directed by the manufacturer. After Yo-Pro-1 labeling, cells were fixed with 4% paraformaldehyde in phosphate-buffered saline. Cells were washed in phosphate-buffered saline and subsequently stained with 300 nM 4,6 diamidino-2-phenylindole (DAPI). Coverslips were mounted onto glass slides with Aqua Poly/Mount (Polysciences, Inc.) and allowed to cure overnight. Micrographs were captured using a 20X Olympus Plan Achromat Objective, 0.4 NA, 1.2 mm WD fitted onto an Olympus Bx62 microscope with attached Hamamatsu Orca AG camera. The microscope, filters, and camera were controlled by Slidebook. Images were exported into ImageJ for analysis of cells stained with Yo-Pro-1 divided by the total cell count. Data were expressed as % apoptotic cells.
Endothelial exocytosis assays
HUVECs or human aortic endothelial cells were seeded in 6-well plates and grown until they were confluent. Cells were washed twice and incubated in EBM2 serum-free medium for 2 hours at 37°C. After preincubation of HUVECs with vorapaxar (0.3 µM), parmodulin 2 (10 µM) or vehicle (dimethyl sulfoxide) for 30 minutes, cells were exposed to buffer or SFLLRN (10 µM) for 1 hour at 37°C. The supernatants from endothelial cells were collected and centrifuged for 3 minutes at 1500g to remove cell debris. von Willebrand factor (VWF) antigen levels were quantified using a sandwich enzyme-linked immunosorbent assay (ELISA) as described previously.23
Transendothelial electric resistance
Transendothelial electric resistance (TER) was measured using an electric cell-substrate impedance sensing system (Applied BioPhysics Inc.). Values were pooled at discrete time points and were either plotted vs time or reported as bar graphs at the time point of maximal response to a given stimulus, as described elsewhere in detail.24 Each condition’s end-point resistance was divided by its starting resistance to give the normalized TER. Confluence was determined by the monolayer achieving manufacturer-recommended electric criteria (resistance >1800 ohms and capacitance <10 nF).
Transfection of siRNA
HUVECs were grown on glass coverslips until they were 70% confluent. Transfections were performed with Lipofectamine RNAiMAX (Life Technologies) according to the manufacturer’s instructions. Validated siRNA for PAR1 was purchased from Life Technologies (Cat. #4390824). Confirmation of knockdown was completed using TaqMan gene expression assay (ID #Hs00169258_m1) according to the manufacturer’s instructions. Isolation of total RNA was completed using the PureLink RNA mini kit (Life Technologies) and cDNA synthesis was completed using the Superscript Vilo cDNA synthesis kit (Life Technologies) according to the manufacturer’s instructions.
Laser-induced injury
The effect of parmodulin 2 on thrombus formation was evaluated by monitoring platelet accumulation after laser-induced injury of cremaster arterioles as previously described19 and detailed the supplemental Methods.
Tail-tip amputation assay
Hemostasis was evaluated using a tail-tip amputation assay as detailed in the supplemental Methods.
Results
Identification of 2 classes of PAR1 antagonists
In performing high-throughput screening for inhibitors of platelet activation induced by the PAR1-activating peptide, SFLLRN, we previously identified 4 structurally unrelated inhibitors of SFLLRN-induced dense granule release (Figure 1A).22,25-27 We compared the activity of these compounds with that of orthosteric antagonists of PAR1. Orthosteric antagonists included vorapaxar, atopaxar, SCH79797, and BMS-200261 (Figure 1B). To determine which PAR1 inhibitors targeted extracellular binding sites and which targeted the cytosolic face of PAR1, we evaluated their activity in COS-7 cells expressing either wild-type human PAR1 or a PAR1/PAR4CT chimera consisting of amino acids 1 to 365 of PAR1 and 334 to 385 of the C-terminus of PAR4. The PAR4 cytoplasmic tail was used because none of these compounds inhibits platelet activation mediated through PAR4.22,25-27 Expression levels of receptors were quantified using an alkaline phosphatase assay (supplemental Figure 1). All compounds inhibited [Ca2+]i flux stimulated by the PAR1-specific agonist SFLLRN when tested in COS-7 cells overexpressing PAR1 (Figure 1C). All orthosteric antagonists also inhibited activation of the PAR1/PAR4CT chimera (Figure 1D). In contrast, none of the compounds that we identified in our screens inhibited activation through the cytoplasmic tail chimera (Figure 1D). We termed these novel PAR1 inhibitors parmodulins.
Identification of 2 classes of PAR1 antagonists. (A) Structure of parmodulins. (B) Structure of orthosteric inhibitors. The effect of vehicle (N/A), 10 μM parmodulin 1 (PM1), 3 μM parmodulin 2 (PM2), 10 μM parmodulin 3 (PM3), 10 μM parmodulin 4 (PM4), 0.3 μM vorapaxar (V), 0.3 μM atopaxar (A), 3 μM SCH79797 (S), or 3 μM BMS-200261 (B) on SFLLRN-induced [Ca2+]i was evaluated in COS-7 transfected with (C) PAR1 or (D) PAR1/PAR4CT, in which the cytoplasmic tail of PAR1 (white) is replaced with that of PAR4 (gray). Increase in [Ca2+]i flux after stimulation with 5 μM SFLLRN was evaluated. Data are presented as means ± standard error of the mean (SEM) (n = 6).
Identification of 2 classes of PAR1 antagonists. (A) Structure of parmodulins. (B) Structure of orthosteric inhibitors. The effect of vehicle (N/A), 10 μM parmodulin 1 (PM1), 3 μM parmodulin 2 (PM2), 10 μM parmodulin 3 (PM3), 10 μM parmodulin 4 (PM4), 0.3 μM vorapaxar (V), 0.3 μM atopaxar (A), 3 μM SCH79797 (S), or 3 μM BMS-200261 (B) on SFLLRN-induced [Ca2+]i was evaluated in COS-7 transfected with (C) PAR1 or (D) PAR1/PAR4CT, in which the cytoplasmic tail of PAR1 (white) is replaced with that of PAR4 (gray). Increase in [Ca2+]i flux after stimulation with 5 μM SFLLRN was evaluated. Data are presented as means ± standard error of the mean (SEM) (n = 6).
Mechanism of action of parmodulins
Orthosteric PAR1 inhibitors and parmodulins were subsequently tested in platelets. All compounds blocked SFLLRN-induced platelet aggregation (Figure 2A-B). Inhibition of platelet aggregation by parmodulins was reversed after platelets were washed (Figure 2A), whereas washing did not reverse inhibition of platelet aggregation by orthosteric inhibitors (Figure 2B). Aggregometry tracings demonstrated that parmodulins failed to inhibit platelet shape change even at concentrations that totally blocked platelet aggregation, whereas orthosteric inhibitors blocked both shape change and aggregation (supplemental Figure 2A). To evaluate the activity of PAR1 antagonists on downstream signaling, we tested their effect on Gαq-mediated [Ca2+]i flux, which is required for aggregation,28 and on Gα13-mediated activation of RhoA, which is required for shape change.29 All compounds inhibited SFLLRN-stimulated [Ca2+]i flux (Figure 2C), indicating inhibition of Gαq-mediated signaling. However, although orthosteric inhibitors completely blocked activation of Gα13-mediated activation of RhoA, PAR1-mediated GDP/GTP exchange at RhoA could be detected in the presence of parmodulins (Figure 2D). Densitometry of multiple samples confirmed this result (supplemental Figure 2B). These studies demonstrate that parmodulins inhibit PAR1 activation via a mechanism that is distinct from that of orthosteric inhibitors and involves selective inhibition of Gα subunits.
Parmodulins inhibit signaling mediated through platelet Gαq, but not Gα13. Platelets were incubated in the presence (black line) or absence (dark gray line) of (A) 10 μM parmodulin 1 (PM1), 3 μM parmodulin 2 (PM2), 10 μM parmodulin 3 (PM3), or 10 μM parmodulin 4 (PM4); or (B) 0.3 μM vorapaxar, 0.3 μM atopaxar, 1 μM SCH79797, or 3 μM BMS-200261, and subsequently stimulated with 5 μM SFLLRN. Reversibility was assessed after washing platelets incubated with PAR1 antagonists and exposing them to 5 μM SFLLRN (light gray line). (C) Platelets were incubated with parmodulins or orthosteric PAR1 agonists as described in (A) and (B), respectively, and evaluated for [Ca2+]i flux after incubation with 5 μM SFLLRN. Data are presented as means ± SEM (n = 4). (D) Platelets were incubated with the indicated PAR1 antagonists at the concentrations described in the previous legends and subsequently exposed to vehicle (–) or 10 μM SFLLRN (+). Activation of RhoA after exposure to SFLLRN was evaluated.
Parmodulins inhibit signaling mediated through platelet Gαq, but not Gα13. Platelets were incubated in the presence (black line) or absence (dark gray line) of (A) 10 μM parmodulin 1 (PM1), 3 μM parmodulin 2 (PM2), 10 μM parmodulin 3 (PM3), or 10 μM parmodulin 4 (PM4); or (B) 0.3 μM vorapaxar, 0.3 μM atopaxar, 1 μM SCH79797, or 3 μM BMS-200261, and subsequently stimulated with 5 μM SFLLRN. Reversibility was assessed after washing platelets incubated with PAR1 antagonists and exposing them to 5 μM SFLLRN (light gray line). (C) Platelets were incubated with parmodulins or orthosteric PAR1 agonists as described in (A) and (B), respectively, and evaluated for [Ca2+]i flux after incubation with 5 μM SFLLRN. Data are presented as means ± SEM (n = 4). (D) Platelets were incubated with the indicated PAR1 antagonists at the concentrations described in the previous legends and subsequently exposed to vehicle (–) or 10 μM SFLLRN (+). Activation of RhoA after exposure to SFLLRN was evaluated.
The observation that parmodulins act by a common mechanism involving the cytosolic face of PAR1 to selectively inhibit downstream signaling was unexpected because these compounds were all identified in a screen of ligand-induced activation.22,25,26 Equilibrium-binding studies were therefore performed using parmodulins 1 and 2 to determine whether they affect agonist association with the ligand binding site of PAR1. [3H]haTRAP, a high-affinity PAR1 orthosteric ligand, was used to monitor binding to PAR1 on platelet membranes.14 [3H]haTRAP bound platelet membranes with a Kd of 6.87 nM ± 0.9 and a Bmax of 7.73 ± 0.47 pmol/mg. Incubation of platelet membranes with parmodulin 1 or parmodulin 2 did not significantly affect binding of [3H]haTRAP to platelet membranes (Figure 3A), consistent with experiments demonstrating activity at the intracellular face of PAR1. In contrast, the orthosteric inhibitor SCH79797 (1 μM) blocked binding of [3H]haTRAP (Figure 3A). These studies support the hypothesis that parmodulin 1 and parmodulin 2 act outside the ligand-binding site without substantially altering the PAR1 ligand–binding site.
Parmodulins bind outside the ligand-binding pocket and act at the intracellular face of PAR1. (A) Equilibrium radioligand-binding studies were performed as described in “Methods.” The effect of vehicle (closed triangles), parmodulin 1 (open diamonds), parmodulin 2 (closed squares), and SCH79797 (open circles) on bound (B) and free (F) [3H]TRAP after incubation with platelet membranes was evaluated. The effect of vehicle (N/A), 10 μM parmodulin 1 (PM1), 3 μM parmodulin 2 (PM2), and 1 μM SCH79797 (S) on agonist-induced [Ca2+]i flux was evaluated in COS-7 transfected with (B) wild-type (WT) PAR1, (C) WT PAR4, (D) the ΔH8 PAR1 mutant, (E) a chimeric PAR1 in which il2 was replaced with il2 of PAR4, and (F) a chimeric PAR1 in which il3 was replaced with il3 of PAR4. [Ca2+]i flux was evaluated after stimulation with either 5 μM SFLLRN (B,D-F) or 300 μM AYPGKF (C). Data are presented as means ± SEM (n = 3-6). Results were analyzed using a Bonferroni multiple comparison test as described in supplemental Table 1 (*P < .05).
Parmodulins bind outside the ligand-binding pocket and act at the intracellular face of PAR1. (A) Equilibrium radioligand-binding studies were performed as described in “Methods.” The effect of vehicle (closed triangles), parmodulin 1 (open diamonds), parmodulin 2 (closed squares), and SCH79797 (open circles) on bound (B) and free (F) [3H]TRAP after incubation with platelet membranes was evaluated. The effect of vehicle (N/A), 10 μM parmodulin 1 (PM1), 3 μM parmodulin 2 (PM2), and 1 μM SCH79797 (S) on agonist-induced [Ca2+]i flux was evaluated in COS-7 transfected with (B) wild-type (WT) PAR1, (C) WT PAR4, (D) the ΔH8 PAR1 mutant, (E) a chimeric PAR1 in which il2 was replaced with il2 of PAR4, and (F) a chimeric PAR1 in which il3 was replaced with il3 of PAR4. [Ca2+]i flux was evaluated after stimulation with either 5 μM SFLLRN (B,D-F) or 300 μM AYPGKF (C). Data are presented as means ± SEM (n = 3-6). Results were analyzed using a Bonferroni multiple comparison test as described in supplemental Table 1 (*P < .05).
To better define the molecular basis of the effect of parmodulins at PAR1, their activity was evaluated in studies using mutant PAR1 and PAR1/PAR4 chimeric receptors (see supplemental Figure 1 for receptor expression levels). Parmodulin 1, parmodulin 2, and the orthosteric inhibitor SCH79797 blocked [Ca2+]i flux stimulated by SFLLRN when tested in COS-7 cells overexpressing PAR1 (Figure 3B), but they failed to block activation induced by the PAR4-specific agonist AYPGKF in COS-7 cells overexpressing PAR4 (Figure 3C and supplemental Table 1). To assess the role of helix 8 in parmodulin activity, a PAR1 mutant in which residues 376 to 386 had been replaced with 3 alanine residues (ΔH8 PAR1) was tested.30 Neither parmodulin 1 nor parmodulin 2 significantly blocked activation of the ΔH8 PAR1 mutant (Figure 3D and supplemental Table 1). In contrast, SCH79797 inhibited this mutant as effectively as it inhibited wild-type PAR1. Activation of a chimera in which intracellular loop 2 of PAR1 was replaced by that of PAR4 (PAR1/PAR4 il2) was significantly inhibited by parmodulins (Figure 3E and supplemental Table 1). Yet parmodulins did not inhibit activation of a chimera in which intracellular loop 3 of PAR1 was replaced by that of PAR4 (PAR1/PAR4 il3; Figure 3F and supplemental Table 1), implicating intracellular loop 3 in parmodulin activity. SCH79797 inhibited activation-induced [Ca2+]i flux in both intracellular loop chimeras effectively (Figure 3E-F). These studies indicate a role for il3 and helix 8 in conferring sensitivity to these two structurally unrelated parmodulins.
Parmodulins demonstrate selective antagonism in endothelial cells
Activation of PAR1 by thrombin or SFLLRN stimulates a dramatic phenotypic change in endothelial cells characterized by exocytosis, loss of barrier function, and contraction. We compared the ability of parmodulins and orthosteric inhibitors to block PAR1-mediated proinflammatory signaling in endothelial cells. All PAR1 antagonists blocked PAR1-stimulated Gαq-mediated activation as detected by monitoring [Ca2+]i (Figure 4A-B). The potency of inhibition of endothelial-cell PAR1 was roughly equivalent to the inhibition of platelet PAR1.12-16,22,25-27 Parmodulin 2 was the most potent and selective parmodulin and demonstrated relatively little off-target activity when tested in GPCR profiling studies (supplemental Figure 3A). In addition, selectivity was evaluated in endothelial cells, in which parmodulin 2 inhibited PAR1-mediated secretion of VWF, but not secretion induced by histamine, DDAVP, epinephrine, or PMA (supplemental Figure 3B). Inhibition of PAR1 signaling in endothelial cells was reversible (supplemental Figure 4A) and selective for Gαq over Gα12/13 (Figure 4A and supplemental Figure 4B). Parmodulin 2 and vorapaxar inhibited PAR1-mediated endothelial cell Weibel-Palade body exocytosis, as indicated by secretion of VWF (Figure 4C and supplemental Figure 4C). Parmodulin 2 and vorapaxar also inhibited thrombin-induced endothelial cell contraction (Figure 4D). Further, parmodulin 2 and vorapaxar inhibited PAR1-mediated loss of barrier function, as evidenced by a transendothelial cell resistance assay (Figure 4E-F).
Parmodulin 2 and vorapaxar inhibit proinflammatory signaling in endothelial cells. HUVECs were incubated with the indicated concentrations of (A) parmodulins or (B) orthosteric inhibitors for 30 minutes before exposure to SFLLRN. Increase in [Ca2+]i flux after stimulation with 5 μM SFLLRN was evaluated and compared with vehicle controls that were not exposed to PAR1 antagonists. Data are presented as means ± SEM (n = 3-6). (C) HUVECs were incubated with either 3 μM parmodulin 2 (PM2) or 0.3 μM vorapaxar before exposure to vehicle (black) or SFLLRN (red). Release of von Willebrand factor (VWF) into supernatants was quantified by ELISA. Data are presented as means ± SEM (n = 4). ***P < .001. (D) Immunofluorescence microscopy of HUVECs incubated with vehicle, parmodulin 2, or vorapaxar, exposed to buffer or 1 U/ml thrombin, and subsequently stained with PE-phalloidin and DAPI. (E) Representative tracings of real-time monitoring of transendothelial electric resistance (TER) of HMVECs incubated with vehicle, 10 μM parmodulin 2, or 0.3 μM vorapaxar and subsequently exposed to SFLLRN (arrow). (F) Quantification of inhibition of SFLLRN- and thrombin-induced barrier dysfunction in HMVECs by parmodulin 2 and vorapaxar. *P < .05, **P < .01, ***P < .001.
Parmodulin 2 and vorapaxar inhibit proinflammatory signaling in endothelial cells. HUVECs were incubated with the indicated concentrations of (A) parmodulins or (B) orthosteric inhibitors for 30 minutes before exposure to SFLLRN. Increase in [Ca2+]i flux after stimulation with 5 μM SFLLRN was evaluated and compared with vehicle controls that were not exposed to PAR1 antagonists. Data are presented as means ± SEM (n = 3-6). (C) HUVECs were incubated with either 3 μM parmodulin 2 (PM2) or 0.3 μM vorapaxar before exposure to vehicle (black) or SFLLRN (red). Release of von Willebrand factor (VWF) into supernatants was quantified by ELISA. Data are presented as means ± SEM (n = 4). ***P < .001. (D) Immunofluorescence microscopy of HUVECs incubated with vehicle, parmodulin 2, or vorapaxar, exposed to buffer or 1 U/ml thrombin, and subsequently stained with PE-phalloidin and DAPI. (E) Representative tracings of real-time monitoring of transendothelial electric resistance (TER) of HMVECs incubated with vehicle, 10 μM parmodulin 2, or 0.3 μM vorapaxar and subsequently exposed to SFLLRN (arrow). (F) Quantification of inhibition of SFLLRN- and thrombin-induced barrier dysfunction in HMVECs by parmodulin 2 and vorapaxar. *P < .05, **P < .01, ***P < .001.
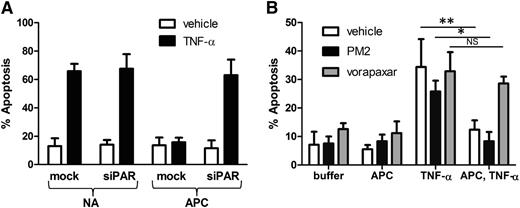
In contrast to the proinflammatory signaling elicited by thrombin, cleavage of endothelial cell PAR1 by APC results in a cytoprotective program that prevents barrier dysfunction, exocytosis, and apoptosis induced by chemokines such as tumor necrosis factor-α (TNF-α) or toxins.5-8,31,32 A potential liability of inhibition at PAR1 is interference with cytoprotective endothelial signaling stimulated by APC. We compared the ability of orthosteric inhibitors and parmodulins to block APC-mediated cytoprotection from TNF-α–induced apoptosis. Application of APC before TNF-α exposure protected endothelial cells from TNF-α–induced apoptosis (Figure 5A). Knockdown of PAR1 using PAR1-specific siRNA reversed the antiapoptotic effect of APC, confirming that APC acts through PAR1 (Figure 5A). In subsequent studies, endothelial cells were exposed to either PM2 or vorapaxar before incubation with APC. Cells were then exposed to TNF-α to determine whether APC-mediated protection from apoptosis was inhibited. Endothelial cells exposed to vorapaxar phenocopied cells in which PAR1 had been knocked down, demonstrating that vorapaxar blocks APC-mediated protection from apoptosis (Figure 5B). In contrast, parmodulin 2 did not block APC-mediated protection from apoptosis (Figure 5B). These results demonstrate that parmodulins interfere with thrombin-induced endothelial stimulation without blocking APC-induced protection from apoptosis.
Vorapaxar, but not parmodulin 2, blocks APC-mediated cytoprotection in endothelial cells. (A) Mock-transfected and PAR1 siRNA–transfected (siPAR1) HUVECs were incubated in the presence of buffer (N/A) or APC before exposure to vehicle or TNF-α. Incubation with APC inhibits apoptosis induced by TNF-α in mock-transfected HUVECs, but not cells in which PAR1 has been knocked down. (B) Endothelial cells were incubated in the presence of vehicle (white), 10 μM parmodulin 2 (black), or 0.3 μM vorapaxar (gray) for 30 minutes. Samples were then exposed to either buffer or APC for 4 hours. Indicated samples were subsequently stimulated with TNF-α and analyzed for apoptosis. Data are presented as means ± SEM (n = 5). *P < .05, **P < .01.
Vorapaxar, but not parmodulin 2, blocks APC-mediated cytoprotection in endothelial cells. (A) Mock-transfected and PAR1 siRNA–transfected (siPAR1) HUVECs were incubated in the presence of buffer (N/A) or APC before exposure to vehicle or TNF-α. Incubation with APC inhibits apoptosis induced by TNF-α in mock-transfected HUVECs, but not cells in which PAR1 has been knocked down. (B) Endothelial cells were incubated in the presence of vehicle (white), 10 μM parmodulin 2 (black), or 0.3 μM vorapaxar (gray) for 30 minutes. Samples were then exposed to either buffer or APC for 4 hours. Indicated samples were subsequently stimulated with TNF-α and analyzed for apoptosis. Data are presented as means ± SEM (n = 5). *P < .05, **P < .01.
Vorapaxar, but not parmodulin 2, induces endothelial cell injury
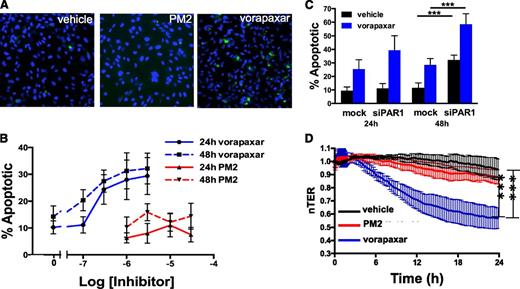
While evaluating the effect of vorapaxar on endothelial cell phenotypes, we observed that longer incubations with vorapaxar induced apoptosis (Figure 6A). A 24-hour exposure to vorapaxar induced endothelial cell apoptosis in a dose-dependent manner, with significant apoptosis occurring at 300 nM (Figure 6B). Increasing the length of exposure increased the sensitivity of endothelial cells to vorapaxar such that after 48 hours, significant apoptosis was observed at 100 nM (Figure 6B). These concentrations are in the range of predicted plasma concentrations achieved with the dosing regimens used in the TRACER and TRA-2P studies.17,18 Similarly, atopaxar, SCH79797, and BMS-200261 all induced apoptosis in endothelial cells (supplemental Figure 5). In contrast, no significant increase in apoptosis was observed when endothelial cells were incubated with parmodulin 2 at concentrations as high as 30 μM (Figure 6A-B) or hirudin (not shown).
Vorapaxar, but not parmodulin 2, induces endothelial dysfunction upon prolonged exposures. (A) HUVECs were exposed to vehicle alone, 10 μM parmodulin 2, or 0.3 μM vorapaxar, as indicated, for 48 hours and subsequently stained for apoptosis using YO-PRO-1. (B) HUVECs were incubated with the indicated concentrations of either vorapaxar (blue) or parmodulin 2 (red) for either 24 hours (solid lines) or 48 hours (dashed lines) and assayed for apoptosis. Data are presented as means ± SEM (n = 5). (C) Mock-transfected (mock) and PAR1 siRNA–transfected (siPAR1) HUVECs were incubated with either vehicle (black) or 0.3 μM vorapaxar (blue) for either 24 or 48 hours. Samples were subsequently assayed for apoptosis. In each condition, addition of vorapaxar led to a significant increase in apoptosis compared with the unexposed sample (P < .001). Knockdown of PAR1 also increased apoptosis. ***P < .001. Data are presented as means ± SEM (n = 5). (D) HMVEC barrier function was continuously monitored by transendothelial resistance for 24 hours after exposure to either 10 μM parmodulin 2 (red) or 0.3 μM vorapaxar (blue). ***P < .001.
Vorapaxar, but not parmodulin 2, induces endothelial dysfunction upon prolonged exposures. (A) HUVECs were exposed to vehicle alone, 10 μM parmodulin 2, or 0.3 μM vorapaxar, as indicated, for 48 hours and subsequently stained for apoptosis using YO-PRO-1. (B) HUVECs were incubated with the indicated concentrations of either vorapaxar (blue) or parmodulin 2 (red) for either 24 hours (solid lines) or 48 hours (dashed lines) and assayed for apoptosis. Data are presented as means ± SEM (n = 5). (C) Mock-transfected (mock) and PAR1 siRNA–transfected (siPAR1) HUVECs were incubated with either vehicle (black) or 0.3 μM vorapaxar (blue) for either 24 or 48 hours. Samples were subsequently assayed for apoptosis. In each condition, addition of vorapaxar led to a significant increase in apoptosis compared with the unexposed sample (P < .001). Knockdown of PAR1 also increased apoptosis. ***P < .001. Data are presented as means ± SEM (n = 5). (D) HMVEC barrier function was continuously monitored by transendothelial resistance for 24 hours after exposure to either 10 μM parmodulin 2 (red) or 0.3 μM vorapaxar (blue). ***P < .001.
PAR1 antagonist–induced endothelial apoptosis could be caused by a biased agonist effect of inhibitors that act at the ligand-binding site. Alternatively, constitutive PAR1 activity could be important for endothelial cell survival33,34 and PAR1 blockade could result in apoptosis. To distinguish between these 2 possibilities, we evaluated the effect of long-term vorapaxar exposure on endothelial cell apoptosis after knockdown of PAR1. Transfection with PAR1-targeted siRNA resulted in >90% knockdown. Incubation of PAR1 siRNA-transfected cells with 300 nM vorapaxar stimulated increased apoptosis compared with incubation with mock-transfected cells at both 24 and 48 hours (Figure 6C). In addition, there was significant apoptosis in the absence of vorapaxar at 48 hours. Endothelial cell apoptosis after PAR1 knockdown and 48-hour exposure to 1 or 3 μM vorapaxar could not be quantified because the cells no longer adhered to the plate. A 24-hour incubation with vorapaxar also induced significant endothelial barrier dysfunction compared with parmodulin 2 (Figure 6D). These observations indicate that long-term exposure to vorapaxar, but not parmodulin 2, elicits endothelial cell injury.
Effect of parmodulin 2 on thrombosis and hemostasis
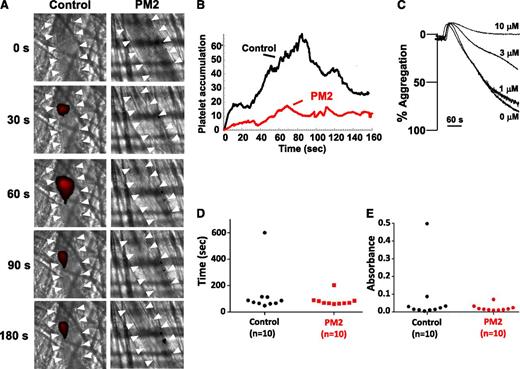
PAR inhibition impairs thrombus formation in both animal models and clinical studies but is associated with bleeding.17-20 We therefore studied parmodulin 2 in assays of both thrombosis and hemostasis. When tested in a murine model in which thrombosis is elicited by laser-induced injury of cremaster arterioles, parmodulin 2 (5 mg/kg) reduced platelet accumulation during thrombus formation by 73% (Figure 7A-B). Because PAR4 is the dominant PAR on mouse platelets, we determined whether parmodulin 2 inhibited aggregation induced through murine PAR4. Previous studies demonstrated that parmodulin 1 interacts with a limited number of GPCRs, including murine PAR4, that have common features at helix 8.30 These features include a predicted ionic interaction between position 1 of helix 8 and the il1 loop, H-bond formation between position 3 of helix 8 and transmembrane domain 7, a hydrophobic interaction between the tyrosine of the NPxxY motif and an aromatic amino acid at position 2 of helix 8, and a palmitoylation site(s) at the C terminus of helix 8.22 Like parmodulin 1, parmodulin 2 blocked AYPGKF-induced aggregation of mouse platelets (Figure 7C), but not AYPGKF-induced aggregation of human platelets (supplemental Figure 6). Thus, parmodulin 2 inhibits activation through murine PAR4, but not human PAR4. For hemostasis assays, parmodulin 2 (5 mg/kg) was infused into mice before tail-tip amputation. Parmodulin 2 failed to affect bleeding times (Figure 7D) or hemoglobin loss (Figure 7E) after transection of the tail tip. These results show that parmodulin 2 inhibits platelet accumulation during thrombus formation but does not affect hemostasis in this model.
Parmodulin 2 inhibits thrombus formation without prolonging bleeding times. (A) Platelet-specific anti-GPIbβ antibody conjugated to Dylight 649 (0.1 µg/g body weight) was infused into mice. Thrombi were induced by laser injury of cremaster arterioles before (n = 62 thrombi in 7 mice) and after (n = 54 thrombi in 7 mice) infusion of parmodulin 2 (5 mg/kg body weight). Thrombus formation was visualized by videomicroscopy for 180 seconds after injury. Representative binarized images of platelets at the injury site before (Control) and after parmodulin 2 infusion (PM2) are shown. White arrowheads indicate the location of the vessel. (B) Median integrated platelet-fluorescence intensity at the injury site in mice before (Control) and after parmodulin 2 infusion (PM2) is plotted over time. (C) Mouse platelet aggregation in response to 65 μM AYPGKF was evaluated after incubation with the indicated concentrations of parmodulin 2. (D) Time to cessation of bleeding and (E) total hemoglobin loss was measured after tail-tip amputation.
Parmodulin 2 inhibits thrombus formation without prolonging bleeding times. (A) Platelet-specific anti-GPIbβ antibody conjugated to Dylight 649 (0.1 µg/g body weight) was infused into mice. Thrombi were induced by laser injury of cremaster arterioles before (n = 62 thrombi in 7 mice) and after (n = 54 thrombi in 7 mice) infusion of parmodulin 2 (5 mg/kg body weight). Thrombus formation was visualized by videomicroscopy for 180 seconds after injury. Representative binarized images of platelets at the injury site before (Control) and after parmodulin 2 infusion (PM2) are shown. White arrowheads indicate the location of the vessel. (B) Median integrated platelet-fluorescence intensity at the injury site in mice before (Control) and after parmodulin 2 infusion (PM2) is plotted over time. (C) Mouse platelet aggregation in response to 65 μM AYPGKF was evaluated after incubation with the indicated concentrations of parmodulin 2. (D) Time to cessation of bleeding and (E) total hemoglobin loss was measured after tail-tip amputation.
Discussion
These studies distinguish between 2 distinct classes of small molecules capable of modulating PAR1 activity. Orthosteric PAR1 inhibitors that induce potent and effective blockade of the binding site of the tethered ligand have been developed,17,18,35,36 and one of these, vorapaxar, has recently been approved for secondary prevention of cardiovascular events. As a class, these inhibitors bind tightly to PAR1. They are inhibitory in the nanomolar range because they must compete with a tethered ligand that has an effective concentration estimated to be 400 μM.1 As a result of the tight binding of orthosteric antagonists, their inhibition is not readily reversible. Blockade at the ligand binding site results in inhibition of all ligand-induced downstream signaling. We now describe a second class of small-molecule PAR1 inhibitors, the parmodulins, which distinguish themselves on the basis of their site of action at the cytosolic face of PAR1 and selectivity at the level of G-protein coupling. As a class, parmodulins are less potent than orthosteric PAR1 antagonists and demonstrate reversible antagonism. Studies in platelets show that they inhibit PAR1-dependent Gαq-mediated elevation of [Ca2+]i flux, but not Gα12/13-mediated signal transduction. The selectivity of parmodulins remains to be fully defined. Our initial profiling and functional assays (supplemental Figure 3), however, indicate that parmodulin 2 is the most selective and parmodulins 3 and 4 are the least selective.25-27 Nonetheless, the fact that parmodulins all demonstrate selectivity at the level of PAR1-mediated signaling transduction defines them as a new class of small-molecule PAR1 modulators that act at an intracellular site on PAR1.
The functional significance of these distinct modes of PAR1 inhibition is best exemplified in the effects of parmodulin 2 and vorapaxar on endothelial cell function (supplemental Figure 7). Both PAR1 antagonists block proinflammatory signaling in endothelial cells (Figure 4). However, although parmodulin 2 spares APC-mediated cytoprotection, vorapaxar blocks this pathway (Figure 5). In addition, incubation with vorapaxar induces endothelial injury upon prolonged exposure (Figure 6). Importantly, endothelial injury was observed at concentrations approximating vorapaxar plasma levels achieved with dosing protocols used in TRACER or TRA-2P, as indicated by earlier pharmacokinetic studies.37,38 The fact that PAR1 knockdown increased susceptibility of endothelial cells to vorapaxar-induced apoptosis suggests that constitutive PAR1 signaling is required for endothelial cell viability. Impaired endothelial growth and vascular formation were observed during development in PAR1−/− mice,33,34 indicating that PAR1 functions in endothelial cell viability in vivo. PAR1 deficiency resulted in 50% embryonic lethality associated with diffuse hemorrhage. Bleeding was reversed with endothelial cell–specific expression of PAR1, raising the possibility that endothelial PAR1 functions in hemostasis.34 The dominant complication of vorapaxar in TRACER and TRA-2P was major bleeding.19,20 Whether exposure to vorapaxar or other orthosteric PAR1 antagonists induces endothelial cell injury that constitutes an additional risk factor for bleeding beyond the antiplatelet effect requires further investigation.
Parmodulins validate the strategy of using small molecules to target the cytosolic face of GPCRs to selectively block one downstream signaling pathway while preserving another. GPCRs demonstrate modularity between the extracellular (ligand-binding) and intracellular (G-protein–binding) regions.39 Association of parmodulin 1 or 2 with PAR1 does not dramatically affect the ligand-binding site, as indicated by their ability to bind PAR1 without significantly modifying the binding of [3H]ha-TRAP. Consistent with this observation, studies using chimeric PARs indicate that interactions at the intracellular face of PAR1 are required for parmodulin activity. Lipidated peptides modeled after the intracellular loops of PAR1 have been developed and have either antagonist or agonist activity at PAR1.40,41 Parmodulin activity, however, is likely more analogous to small-molecule inhibitors directed against the cytoplasmic face of CCR4 and CXCR2. These chemokine inhibitors are thought to act at a binding site within or adjacent to the G-protein–binding pocket.42-44 Mutational studies suggest that different classes of G-proteins associate with different regions on the cytoplasmic face of PAR1.45 Parmodulins could bind at or near the G-protein–binding pocket and selectively compete with Gαq, but not Gα12/13. The ability of antagonists that target an intracellular site to selectively impair downstream G-protein signaling has not previously been studied. Our data show that by targeting the cytosolic face of PAR1, parmodulins inhibit PAR1-induced proinflammatory signaling in endothelial cells without blocking APC-mediated cytoprotection signaling through PAR1. In vivo, these compounds block arteriolar thrombosis without increasing bleeding times after tail clip.
Parmodulins represent a prototype for the identification and characterization of compounds that target the cytosolic face of GPCRs to selectively inhibit coupling to some Gα subunits, but not others. Small molecules targeting the G-protein–binding site will be useful in dissecting the molecular determinants of GPCR–G-protein coupling and developing new approaches to pharmacologically manipulate that coupling. Ultimately, such compounds could be used to inhibit pathways that mediate pathologic signaling while preserving or even stimulating protective signaling pathways.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Shaun R. Coughlin, University of San Francisco Medical Center, for the AP-PAR1 and AP-PAR4 constructs, and Dr. Athan Kuliopulos, Tufts University School of Medicine, for the kind gift of the ΔH8 PAR1 mutant construct. They are grateful to the Psychoactive Drug Screening Program and Dr Bryan L. Roth, University of North Carolina at Chapel Hill, for GPCR profiling.
This work was supported by the National Institutes of Health, National Heart, Lung, and Blood Institute (grants HL87203, HL92125, HL112809, HL07917), National Institute of General Medical Sciences (grant GM55223), the American Heart Association (grant 0840043N), and the Foundation for Women’s Wellness.
Authorship
Contribution: O.A. designed and performed assays with modified PARs, equilibrium binding studies, and [Ca2+]i flux studies; designed figures; and edited manuscript; C.G.P. supervised microscopy and edited manuscript; K.D. performed endothelial cell exocytosis assays; C.C.G. performed TER studies and edited manuscript; J.R.D. performed platelet aggregation studies and RhoA-GTP assays; S.F.G. performed in vivo platelet accumulation studies; M.H. designed chimeric receptors and edited manuscript; C.D. synthesized compounds and edited manuscript; S.M.P. supervised TER studies and edited manuscript; and R.F. initiated the project, led the project team, designed experiments, analyzed results, designed figures, and drafted the manuscript.
Conflict-of-interest disclosure: C.D. and R.F. are named as inventors in a pending patent describing parmodulin 2 (ML161). R.F. is named as an inventor in a patent describing parmodulin 1 (JF5). The remaining authors declare no competing financial interests.
Correspondence: Robert Flaumenhaft, Division of Hemostasis and Thrombosis, Department of Medicine, Beth Israel Deaconess Medical Center, 330 Brookline Ave, Boston, MA 02215; e-mail: rflaumen@bidmc.harvard.edu.

![Figure 1. Identification of 2 classes of PAR1 antagonists. (A) Structure of parmodulins. (B) Structure of orthosteric inhibitors. The effect of vehicle (N/A), 10 μM parmodulin 1 (PM1), 3 μM parmodulin 2 (PM2), 10 μM parmodulin 3 (PM3), 10 μM parmodulin 4 (PM4), 0.3 μM vorapaxar (V), 0.3 μM atopaxar (A), 3 μM SCH79797 (S), or 3 μM BMS-200261 (B) on SFLLRN-induced [Ca2+]i was evaluated in COS-7 transfected with (C) PAR1 or (D) PAR1/PAR4CT, in which the cytoplasmic tail of PAR1 (white) is replaced with that of PAR4 (gray). Increase in [Ca2+]i flux after stimulation with 5 μM SFLLRN was evaluated. Data are presented as means ± standard error of the mean (SEM) (n = 6).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/12/10.1182_blood-2014-09-599910/4/m_1976f1.jpeg?Expires=1769831427&Signature=RTXihByaEeQqS~r9nf5rqApkGVfxoRlIrG3KYj5tVFXTq3SKBQ8Q2Ohjl3-qGSc~5VI32sZjsa5q5CyMcAQ0BIUvO8skqiC~v6oZWyb-x7pLcTsx8QgYDzahQCfuFIgrPI-nJgYZ7KlWJFiA-ylvNU51DaGfcEcaZ2iCHvM0F2lbtUeQzuLIia9KTD4PNM6iyzQYPzTMJGMA2gVak3ki-exB7untXaW5aKOqeu2hjzHR5mVpvDZfLPAHdrYadbWKRFzvfyOxGAAx9kaYYjoStEMQd6Z4gBp83IoSopIU2wnVm3QAtrOFu-c7SU2Tuzked8Epdw0njDHqBGXzKLUIcw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 2. Parmodulins inhibit signaling mediated through platelet Gαq, but not Gα13. Platelets were incubated in the presence (black line) or absence (dark gray line) of (A) 10 μM parmodulin 1 (PM1), 3 μM parmodulin 2 (PM2), 10 μM parmodulin 3 (PM3), or 10 μM parmodulin 4 (PM4); or (B) 0.3 μM vorapaxar, 0.3 μM atopaxar, 1 μM SCH79797, or 3 μM BMS-200261, and subsequently stimulated with 5 μM SFLLRN. Reversibility was assessed after washing platelets incubated with PAR1 antagonists and exposing them to 5 μM SFLLRN (light gray line). (C) Platelets were incubated with parmodulins or orthosteric PAR1 agonists as described in (A) and (B), respectively, and evaluated for [Ca2+]i flux after incubation with 5 μM SFLLRN. Data are presented as means ± SEM (n = 4). (D) Platelets were incubated with the indicated PAR1 antagonists at the concentrations described in the previous legends and subsequently exposed to vehicle (–) or 10 μM SFLLRN (+). Activation of RhoA after exposure to SFLLRN was evaluated.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/12/10.1182_blood-2014-09-599910/4/m_1976f2.jpeg?Expires=1769831427&Signature=k4b0idknN4IO~AeeyB3ZpObHrDxFsQwLkHPxRTc~VLO-Auf3CowI4h3VOBzHKb3Evg2TT2CGfU2jV6gsK9lqXUgjGEGQzbZiJQUALHVuSnrNCzmFm-uMmrxGaQZI4nwpEf8J6NUmYUZc4p6JoCzMqxySw~U5J~corskso9SBjNSWOXdiltBGGAN5Bp2UKe-B~WEFxQCmK5Dj7tNCXJQcKRrHkm~vH4Ki~edAlf~ytWHyJxkmA6ssnab-B~HZIlN4EC1uDSxwlnA5r5V3tllNGtcn-aDUCQEM4RZVq6~gVNY89I4xKn0NZThxZfVYbzLj0mEPrp0T7L~mZS4fYerdMA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. Parmodulins bind outside the ligand-binding pocket and act at the intracellular face of PAR1. (A) Equilibrium radioligand-binding studies were performed as described in “Methods.” The effect of vehicle (closed triangles), parmodulin 1 (open diamonds), parmodulin 2 (closed squares), and SCH79797 (open circles) on bound (B) and free (F) [3H]TRAP after incubation with platelet membranes was evaluated. The effect of vehicle (N/A), 10 μM parmodulin 1 (PM1), 3 μM parmodulin 2 (PM2), and 1 μM SCH79797 (S) on agonist-induced [Ca2+]i flux was evaluated in COS-7 transfected with (B) wild-type (WT) PAR1, (C) WT PAR4, (D) the ΔH8 PAR1 mutant, (E) a chimeric PAR1 in which il2 was replaced with il2 of PAR4, and (F) a chimeric PAR1 in which il3 was replaced with il3 of PAR4. [Ca2+]i flux was evaluated after stimulation with either 5 μM SFLLRN (B,D-F) or 300 μM AYPGKF (C). Data are presented as means ± SEM (n = 3-6). Results were analyzed using a Bonferroni multiple comparison test as described in supplemental Table 1 (*P < .05).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/12/10.1182_blood-2014-09-599910/4/m_1976f3.jpeg?Expires=1769831427&Signature=2y5qQ8-1rwRMPV5oY0k0ATfk2CCS6VMJibYie5towMuZkwv2ep17mZ-aqqEVjDimVYFZx3QjkDFHpqX-Ddr3RR0tV7~1UQ-0N55ef7k~JUQEENG5V-F3Lu5cACaCGEoQmVZ8jjVQ6m~OBLuWt7sirHGQDFr8EufHJnrV5vEjO7hGRclArMRljKgomO4lJsSsuC~LI0cRMwxChNdp8-hsIEefrpke19qhljyyBr1~~HQpoy8FsniVA0Hi2t70e4LUjPRnqzkuVZT2ryoKAAvRKgk-qhVSXBLkSWIYTPQYnGJEZ-Cx4cErEm6CjUfInoYikCnd6YIonUArZkMFoO5ynA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. Parmodulin 2 and vorapaxar inhibit proinflammatory signaling in endothelial cells. HUVECs were incubated with the indicated concentrations of (A) parmodulins or (B) orthosteric inhibitors for 30 minutes before exposure to SFLLRN. Increase in [Ca2+]i flux after stimulation with 5 μM SFLLRN was evaluated and compared with vehicle controls that were not exposed to PAR1 antagonists. Data are presented as means ± SEM (n = 3-6). (C) HUVECs were incubated with either 3 μM parmodulin 2 (PM2) or 0.3 μM vorapaxar before exposure to vehicle (black) or SFLLRN (red). Release of von Willebrand factor (VWF) into supernatants was quantified by ELISA. Data are presented as means ± SEM (n = 4). ***P < .001. (D) Immunofluorescence microscopy of HUVECs incubated with vehicle, parmodulin 2, or vorapaxar, exposed to buffer or 1 U/ml thrombin, and subsequently stained with PE-phalloidin and DAPI. (E) Representative tracings of real-time monitoring of transendothelial electric resistance (TER) of HMVECs incubated with vehicle, 10 μM parmodulin 2, or 0.3 μM vorapaxar and subsequently exposed to SFLLRN (arrow). (F) Quantification of inhibition of SFLLRN- and thrombin-induced barrier dysfunction in HMVECs by parmodulin 2 and vorapaxar. *P < .05, **P < .01, ***P < .001.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/12/10.1182_blood-2014-09-599910/4/m_1976f4.jpeg?Expires=1769831427&Signature=qI0s6gZKCRPhbq2YeV1mFqK1Sex~Q-SRNdyNJ9EvRDCIE29Ot~jZSxLXpHjDY9cuaBXF1OtS3ZO0QXMio5im5P4p~ma~yeztY9ygYt6LnXgjJSU0iwYOStMz~c1sdgSPudAGgUJble0LkpBOKsscSzTZSvMae31SqHcZjLvoyl0ewCHSKhd1QhYu-hVEW768KTUqEIkCqvShy~sC8WJcjsoJIQY~jZxBtyT5vXXLk0xKDo4UwMAiACgpQVuPRnkdTBcxW41~DaH6S4Zbha9NiRD9X9w8P0XCdmzHFE6S4E5LgQ-EPjAmR3lrLwV~2eLw5qwClm7ahHYHBYPpMzRs4w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)