Key Points
Different driver mutations have distinct effects on phenotype of myelodysplastic syndromes (MDS) and myelodysplastic/myeloproliferative neoplasms (MDS/MPN).
Accounting for driver mutations may allow a classification of these disorders that is considerably relevant for clinical decision-making.
Abstract
Our knowledge of the genetic basis of myelodysplastic syndromes (MDS) and myelodysplastic/myeloproliferative neoplasms (MDS/MPN) has considerably improved. To define genotype/phenotype relationships of clinical relevance, we studied 308 patients with MDS, MDS/MPN, or acute myeloid leukemia evolving from MDS. Unsupervised statistical analysis, including the World Health Organization classification criteria and somatic mutations, showed that MDS associated with SF3B1-mutation (51 of 245 patients, 20.8%) is a distinct nosologic entity irrespective of current morphologic classification criteria. Conversely, MDS with ring sideroblasts with nonmutated SF3B1 segregated in different clusters with other MDS subtypes. Mutations of genes involved in DNA methylation, splicing factors other than SF3B1, and genes of the RAS pathway and cohesin complex were independently associated with multilineage dysplasia and identified a distinct subset (51 of 245 patients, 20.8%). No recurrent mutation pattern correlated with unilineage dysplasia without ring sideroblasts. Irrespective of driver somatic mutations, a threshold of 5% bone marrow blasts retained a significant discriminant value for identifying cases with clonal evolution. Comutation of TET2 and SRSF2 was highly predictive of a myeloid neoplasm characterized by myelodysplasia and monocytosis, including but not limited to, chronic myelomonocytic leukemia. These results serve as a proof of concept that a molecular classification of myeloid neoplasms is feasible.
Introduction
The World Health Organization (WHO) classification currently provides the best diagnostic approach to myeloid neoplasms, which include myelodysplastic syndromes (MDS), myelodysplastic/myeloproliferative neoplasms (MDS/MPN), and acute myeloid leukemia (AML).1,2 The presence of dysplasia in one or more of myeloid cell lines is one of the major criteria for the diagnosis of MDS; however, dysplasia is not specific, and its morphologic criteria are poorly reproducible.3,4
In a group of myeloid disorders classified on the basis of morphologic criteria, identifying specific associations between genotype and disease phenotypes is essential to defining disease entities according to their distinctive genetic profiles.5 This genotype-phenotype relationship in MDS is illustrated by the 5q- syndrome, described as a distinct clinical entity by Van den Berghe et al in 1974.6 In 2001, the WHO classification recognized MDS with isolated del(5q) as a distinct category, representing the first subtype of MDS defined by a genetic abnormality.7 The molecular basis of this MDS subtype has been then identified as the haploinsufficiency of genes that map in the common deleted region.8-10
A major step forward in genotype-phenotype relationship has been the identification of somatically acquired mutations of SF3B1 in MDS patients with ring sideroblasts (RS).11-13 The analysis of a subgroup of patients in whom a quantitative enumeration of RS was performed irrespective of the WHO category, showed that SF3B1 mutation status had a positive predictive value for disease phenotype with RS of 98%.12,14
We previously characterized the genomic landscape of MDS.15 In the present study, based on this comprehensive mutation analysis in a large and clinically well characterized cohort of MDS patients, we sought to determine if significant associations between genotype and disease phenotype existed and adopted unsupervised hierarchical clustering analyses to identify genetically defined MDS subtypes.
Patients and methods
Patients and clinical procedures
These investigations were approved by the Ethics Committee of the Fondazione IRCCS Policlinico San Matteo, Pavia, Italy, and other local Institutional Review Boards. The procedures followed were in accordance with the Declaration of Helsinki, as revised in 2000, and samples were obtained with informed consent of the subjects.
We studied 308 patients with myeloid neoplasms, 245 with MDS, 34 with MDS/MPN, and 29 with AML evolving from MDS (AML-MDS). Diagnostic procedures were performed according to the recommendations of the European LeukemiaNet.2 In order to classify patients, peripheral blood and bone marrow (BM) specimens were analyzed by two independent cytologists who were blinded to clinical data, as previously reported.16,17 The diagnostic criteria of the WHO classification of tumors of hematopoietic and lymphoid tissues were adopted.1,18 Quantitative enumeration of myeloblasts, RS and monocytes, and their precursors was performed using recently established consensus criteria.19,20
For clinical and hematologic features of patients included in the study, see supplemental Table 1 on the Blood Web site. Patients were studied at diagnosis or during follow-up before any disease-modifying treatment (ie, allogeneic stem cell transplantation, intensive chemotherapy, or hypomethylating agents). These patients were included in our recent study of targeted gene sequencing in myelodysplasia.15
Sample collection and cell separation
Mononuclear cells were separated from BM samples by standard density gradient centrifugation, and granulocytes were isolated from peripheral blood as previously described.21 Genomic DNA was obtained from BM mononuclear cells or peripheral blood granulocytes by following standard protocols for human tissue.
Targeted gene sequencing
A panel of 111 genes was selected on the basis of prior implication in the pathogenesis of myeloid disease by recurrent somatic mutation, recurrent mutation or aberrations in common cancers, or candidate gene mapping within regions of common copy number alterations (supplemental Table 2). Genomic DNA samples underwent whole genome amplification. Bar-coded sequencing libraries were prepared and target enrichment for 111 coded genes was performed before samples were pooled (n = 96) and sequenced using 2 lanes of Illumina HiSeq (Illumina Inc., San Diego, CA), as previously reported.15
JAK2 and CALR mutation analysis
Janus kinase 2 (JAK2) (V617F) mutation was analyzed using a quantitative real-time polymerase chain reaction-based allelic discrimination assay in order to obtain a precise quantification of mutant allele burden.21 Mutations of calreticulin (CALR), not included in the original panel of 111 genes were analyzed by sequencing, as previously described.22
Statistical analysis
Numerical variables were summarized by median and range; categorical variables were described with count and relative frequency (%) of subjects in each category. Comparison of numerical variables between groups was carried out using a nonparametric approach (Mann-Whitney U test or Kruskal Wallis analysis of variance). Comparison of the distribution of categorical variables in different groups was performed with either the Fisher’s exact test (when computationally feasible) or the χ2 test (larger tables). Variant allele fraction estimates were used to evaluate clonal and subclonal variant relationships within each sample, as previously reported.15
Unsupervised hierarchical clustering analyses were performed in order to identify homogeneous groups of patients with respect to the variables of interest. Multinomial and logistic multivariable regressions were performed for predicting disease phenotype based on gene mutation status. Survival analyses were performed with the Kaplan-Meier method. Overall survival (OS) was defined as the time (in months) between the date of diagnosis, and the date of death (for cases) or last follow-up (for censored patients). Statistical comparison of survival curves was carried out with the log-rank test. Leukemia-free survival was defined as the time (in months) between the date of diagnosis, and the date of leukemic transformation (for cases) or last follow-up (for censored patients). Multivariate survival analyses were performed by means of Cox proportional hazards regression. All analyses accounted for left censoring of the observations at the time of mutation assessment. Additional information is provided in the supplemental Methods. Analyses were performed using the Stata SE 12.1 (StataCorp LP, College Station, TX; http://www.stata.com) software.
Results
Spectrum of mutation in MDS and MDS/MPN
High-confidence oncogenic mutations were identified in 43 genes, with a median number of 2 mutations per patient (range, 0 to 9). A significantly lower number of mutations per patient was found in MDS, compared with MDS/MPN or AML-MDS (P = .003) (supplemental Figure 1A).
A significantly higher prevalence of mutations in splicing factors were observed in MDS (122 of 245, 50%) and MDS/MPN (23 of 34, 74%) compared with AML-MDS (9 of 29, 31%) (P = .003), whereas mutations in DNA methylators were more frequent in MDS/MPN (17 of 34, 50%) and AML-MDS (14 of 29, 48%) than in MDS (71 of 245, 29%) (P = .01) (supplemental Table 3 and supplemental Figure 1B).
A significantly higher prevalence of mutations in SF3B1 was observed in MDS and MDS/MPN compared with AML-MDS (P = .026), whereas TET2 mutations were more frequent in MDS/MPN than in MDS or AML-MDS (P = .01). NMP1 (P < .001), WT1 (P = .004), and IDH1/2 (P = .005) were more frequent in AML-MDS compared with MDS or MDS/MPN.
Correlations between genotype and disease phenotype in MDS with RS
RS were identified at variable percentages (range, 1% to 97%) in 126 of 245 patients (51%). Of these, 62 were classified within WHO categories identified by this morphologic feature (sideroblastic categories, refractory anemia with RS [RARS] or refractory cytopenia with multilineage dysplasia [RCMD]-RS), whereas 64 were assigned to different WHO categories as follows: 6 refractory anemia (RA), 5 MDS with isolated del(5q), 18 RCMD, 16 refractory anemia with excess blasts type 1 (RAEB-1), and 19 RAEB-2. A significantly higher prevalence of SF3B1 mutation was found in patients with RARS (31 of 34, 91.2%) or RCMD-RS (16 of 28, 57.1%) compared with nonsideroblastic categories (P < .001).
We studied the relationships between mutant genes and RS in patients wild-type for SF3B1, and found that mutations of SRSF2 were significantly associated with the presence of BM RS in variable proportion (range, 1% to 68%) (P = .04). These cases were classified as 2 RA or RARS, 9 RCMD or RCMD-RS, and 6 RAEB-1 or RAEB-2. The mutation patterns in SRSF2-positive MDS patients with or without RS is shown in supplemental Figure 2.
In order to identify homogeneous groups of patients within MDS with RS, we performed unsupervised hierarchical clustering analyses including current classification features according to WHO criteria (ie, unilineage or multilineage dysplasia [MD], RS lower or ≥15%), and gene mutations associated with RS (ie, SF3B1 and SRSF2). Clustering analysis identified 2 groups of patients: one cluster included patients carrying the SF3B1 mutation, irrespective of current WHO classification criteria, whereas the second cluster included patients carrying the SRSF2 mutation as well as those negative for both SF3B1 and SRSF2 mutations. With an extended clustering analysis to the whole population of patients with MDS without excess blasts including current classification features according to WHO criteria and SF3B1 mutational status, two main clusters were identified: one cluster included patients with the SF3B1 mutation and RS (31 RARS, 16 RCMD-RS, 3 RA, and 1 RCMD according to WHO criteria), whereas the second cluster included patients negative for SF3B1 mutation either without or with RS (25 RA, 3 RARS, 66 RCMD, and 12 RCMD-RS according to WHO criteria).
Patients classified in the first cluster (MDS associated with SF3B1 mutation) showed a significantly better OS compared with those included in the second cluster (P = .018) (supplemental Figure 3). Within MDS associated with SF3B1 mutation, no significant differences were observed between patients with unilineage or MD in clinical and hematologic parameters, OS, and risk of leukemic evolution (P = .64 and P = .96, respectively) (supplemental Figure 4). In addition, no significantly different patterns of co-occurring or mutually exclusive mutations were found between SF3B1-mutated patients with unilineage or MD.
Among MDS patients with the SF3B1 mutation, 80% showed a clonal SF3B1 mutation, whereas 5% of cases had statistical evidence for subclonal SF3B1 mutation. In the remaining 15% of patients, phylogenetic relationships among mutations could not be determined unambiguously. No difference in disease phenotype was found between cases in which the SF3B1 mutation was clonal or subclonal.
Genetic determinants of MD and excess of BM blasts in MDS
We then focused on nonsideroblastic categories of MDS (RA, RCMD, RAEB-1, and RAEB-2) with the aim of identifying genetic determinants of unilineage or MD and excess of BM blasts. We found that mutations in the following genes were significantly associated with disease phenotype with MD or excess blasts: DNA methylation (DNMT3A, TET2, IDH1, and IDH2); splicing factors other than SF3B1 (SRSF2, U2AF1, and ZRSR2); RAS pathway (KRAS, NRAS, CBL, and NF1); cohesin complex (STAG2 and RAD21); and RUNX1 (P values ranging from .007 to .033). Positive predictive values of these mutations for MD or advanced disease, defined as either MD or excess blasts, ranged from 83% to 92.9% and from 93.8% to 100%, respectively.
We then performed multivariable logistic regression analyses to identify genotypic variables that were independently associated with disease phenotype with MD or excess blasts. We first performed a logistic regression model including mutation in DNA methylation, splicing factors other than SF3B1, RAS pathway, cohesion complex, and RUNX1 as predictive variables, and unilineage dysplasia (baseline) or MD as outcome. We found that mutations in DNA methylation (Odds ratio [OR]: 9.734; 95% CI, 9.560-9.881; P < .001), splicing factors (OR: 4.213; 95% CI, 4.178-4.248; P < .001), RAS pathway (OR: 1.945; 95% CI, 1.668-2.268; P < .001), and cohesin complex (OR: 1.440; 95% CI, 1.178-1.760; P < .001) independently predicted disease phenotype with MD.
Next, we performed a logistic regression model including mutation in DNA methylation, splicing factors other than SF3B1, RAS pathway, cohesion complex, and RUNX1 as predictive variables and MD (baseline) or excess blasts as outcome. We found that mutation in RUNX1 (OR: 4.065; 95% CI, 3.974-4.158; P < .001), cohesin complex (OR: 2.050; 95% CI, 2.002-2.100; P < .001), RAS pathway (OR: 1.384; 95% CI, 1.368-1.401; P < .001), and splicing factors (OR: 1.140; 95% CI, 1.116-1.166, P < .001) were independently associated with disease phenotype with excess blasts, whereas mutations in DNA methylation were independently associated with MD (OR: .566; 95% CI, .557-.575; P < .001). No significant association was found between mutation pattern and RAEB subcategories (ie, RAEB-1 and RAEB-2).
Based on these results, we performed unsupervised hierarchical clustering analyses including current classification features according to WHO criteria and gene mutations, in order to identify homogeneous groups of patients with respect to the variables of interest.
First, we focused on MDS with no excess blasts, and performed unsupervised hierarchical clustering analyses including current WHO classification criteria (ie, unilineage or MD, RS lower or ≥15%), and gene mutations independently associated with MD (MD-associated mutations: DNA methylation, splicing factors other than SF3B1, RAS pathway, and cohesin complex). Patients with isolated del(5q) or SF3B1 mutation were excluded from this analysis. This analysis identified 2 main clusters: one cluster included all patients with MD-associated mutations classified as RCMD or SF3B1-negative RCMD-RS according to WHO criteria, as well as the 4 patients with unilineage dysplasia carrying mutation(s) in these genes. The second cluster comprised all the nonmutated patients irrespective of the presence of unilineage or MD (MDS not otherwise specified [NOS]). When SF3B1 mutation status was included in the analysis, these two clusters were replicated and a third cluster represented by patients with the SF3B1 mutation was identified. Finally, when the analysis was extended to include MDS with excess blasts entering all the mutations associated with advanced disease phenotype (DNA methylation, splicing factors other than SF3B1, RAS pathway, cohesin complex, and RUNX1), patients with RAEB clustered in a single group irrespective of their genotype.
Among MDS patients carrying MD-associated mutation(s), in 16% of patients relationships among mutations could not be determined unambiguously, whereas 80% of patients showed a clonal mutation. No significant difference in disease phenotype was found between cases in which these driver mutations were clonal or subclonal.
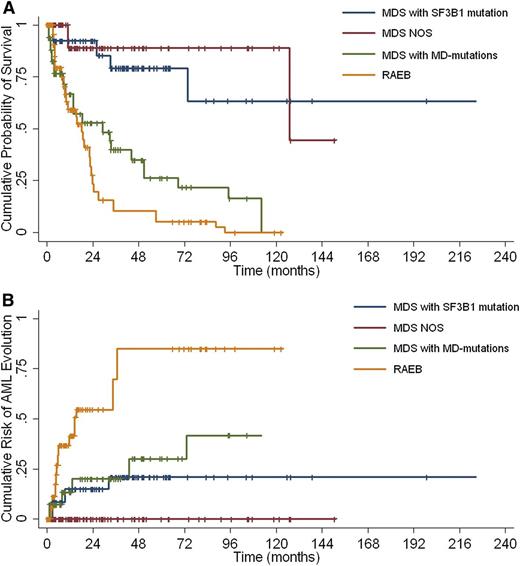
Clinical and hematologic characteristics, and mutation patterns of MDS patients without excess blasts classified according to genotype-based clusters (MDS associated with SF3B1 mutation; MDS with MD-associated mutations; and MDS NOS) are reported in Table 1 and Figure 1, respectively. When comparing the outcome of these 3 categories, as well as of the fourth cluster including MDS with excess blasts, we found that these 4 categories had significantly different OS (P < .001) and a risk of AML progression (P < .001) (Figure 2).
Clinical and hematologic features of patients with MDS without excess blasts classified according to the unsupervised clustering analysis including WHO classification criteria and mutation pattern
| Clinical variables . | MDS associated with SF3B1 mutation . | MDS NOS . | MDS with MD-associated mutations* . |
|---|---|---|---|
| No. of patients | 51 | 55 | 51 |
| Age (years) | 69 (46-88) | 67 (29-88) | 71 (45-88) |
| Sex (male/female) | 24/27 | 27/28 | 32/19 |
| WHO category | |||
| RA | 3 | 22 | 3 |
| RARS | 31 | 2 | 1 |
| RCMD | 1 | 26 | 40 |
| RCMD-RS | 16 | 5 | 7 |
| Hemoglobin (g/dL) | 9.4 (6.9-12.8) | 9.5 (6.5-13.9) | 9.2 (6.1-12.9) |
| WBC count (×109/L) | 5.1 (1.4-9.3) | 3.9 (1.18-10.6) | 3.4 (0.6-11.1) |
| ANC† (×109/L) | 2.7 (0.8-7.5) | 2.0 (0.1-7.6) | 1.5 (0.1-7.8) |
| Platelets (×109/L) | 247 (16-536) | 98 (5-434) | 111 (7-315) |
| BM blasts (%) | 1 (0-4) | 2 (0-4) | 3 (1-4) |
| RS (%) | 65 (2-95) | 0 (0-77) | 0 (0-62) |
| Karyotype (IPSS-R) | |||
| Very good | 2 | 2 | — |
| Good | 38 | 39 | 37 |
| Intermediate | 5 | 6 | 6 |
| High | 1 | 4 | 3 |
| Very high | — | 1 | — |
| Failed | 5 | 3 | 5 |
| No. of mutations | 2 (1-9) | 1 (0-5) | 3 (1-7) |
| Clinical variables . | MDS associated with SF3B1 mutation . | MDS NOS . | MDS with MD-associated mutations* . |
|---|---|---|---|
| No. of patients | 51 | 55 | 51 |
| Age (years) | 69 (46-88) | 67 (29-88) | 71 (45-88) |
| Sex (male/female) | 24/27 | 27/28 | 32/19 |
| WHO category | |||
| RA | 3 | 22 | 3 |
| RARS | 31 | 2 | 1 |
| RCMD | 1 | 26 | 40 |
| RCMD-RS | 16 | 5 | 7 |
| Hemoglobin (g/dL) | 9.4 (6.9-12.8) | 9.5 (6.5-13.9) | 9.2 (6.1-12.9) |
| WBC count (×109/L) | 5.1 (1.4-9.3) | 3.9 (1.18-10.6) | 3.4 (0.6-11.1) |
| ANC† (×109/L) | 2.7 (0.8-7.5) | 2.0 (0.1-7.6) | 1.5 (0.1-7.8) |
| Platelets (×109/L) | 247 (16-536) | 98 (5-434) | 111 (7-315) |
| BM blasts (%) | 1 (0-4) | 2 (0-4) | 3 (1-4) |
| RS (%) | 65 (2-95) | 0 (0-77) | 0 (0-62) |
| Karyotype (IPSS-R) | |||
| Very good | 2 | 2 | — |
| Good | 38 | 39 | 37 |
| Intermediate | 5 | 6 | 6 |
| High | 1 | 4 | 3 |
| Very high | — | 1 | — |
| Failed | 5 | 3 | 5 |
| No. of mutations | 2 (1-9) | 1 (0-5) | 3 (1-7) |
IPSS-R, revised international prognostic scoring system; WBC, white blood cell.
MD-associated mutations: DNA methylation, splicing factors other than SF3B1, RAS pathway, and cohesin complex.
ANC indicates absolute neutrophil count.
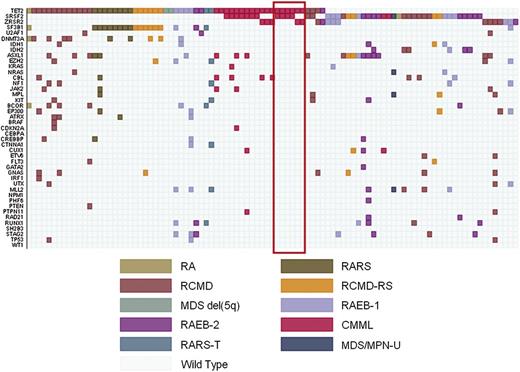
Representation of unsupervised hierarchical clustering analyses including somatic mutations and current classification features according to WHO criteria within MDS without excess blasts. Diagnosis of each sample is shown by indicated colors. These analyses identified 3 main clusters: patients carrying the SF3B1 mutation, irrespective of current WHO classification criteria (MDS associated with SF3B1 mutation) (31 patients classified as RARS, 16 as RCMD-RS, 3 as RA, and 1 as RCMD according to WHO criteria) (left); patients with MD-associated mutations (mutations in genes involved in DNA methylation, splicing factors other than SF3B1, RAS pathway, and cohesin complex) classified as RCMD or SF3B1-negative RCMD-RS according to WHO criteria, as well as the 4 patients with unilineage dysplasia carrying mutation(s) in these genes (MDS with MD-associated mutations) (right); and all the patients with different mutation patterns (MDS NOS) irrespective of the presence of unilineage or MD (middle).
Representation of unsupervised hierarchical clustering analyses including somatic mutations and current classification features according to WHO criteria within MDS without excess blasts. Diagnosis of each sample is shown by indicated colors. These analyses identified 3 main clusters: patients carrying the SF3B1 mutation, irrespective of current WHO classification criteria (MDS associated with SF3B1 mutation) (31 patients classified as RARS, 16 as RCMD-RS, 3 as RA, and 1 as RCMD according to WHO criteria) (left); patients with MD-associated mutations (mutations in genes involved in DNA methylation, splicing factors other than SF3B1, RAS pathway, and cohesin complex) classified as RCMD or SF3B1-negative RCMD-RS according to WHO criteria, as well as the 4 patients with unilineage dysplasia carrying mutation(s) in these genes (MDS with MD-associated mutations) (right); and all the patients with different mutation patterns (MDS NOS) irrespective of the presence of unilineage or MD (middle).
Survival and risk of leukemic evolution of patients with MDS classified according to the clusters resulting from the unsupervised analysis including WHO classification criteria and mutation patterns. (A) Overall survival and (B) risk of AML evolution of MDS patients clustered as follows: MDS associated with SF3B1 mutation; MDS with MD–associated mutations; MDS NOS; and MDS with excess blasts (RAEB).
Survival and risk of leukemic evolution of patients with MDS classified according to the clusters resulting from the unsupervised analysis including WHO classification criteria and mutation patterns. (A) Overall survival and (B) risk of AML evolution of MDS patients clustered as follows: MDS associated with SF3B1 mutation; MDS with MD–associated mutations; MDS NOS; and MDS with excess blasts (RAEB).
Somatic mutations with prognostic value in genotype-based MDS categories
We tested the prognostic value of somatic mutations in patients with MDS. As a first step, we performed uni- and multivariable Cox regression analyses including age, sex, peripheral blood cytopenias, BM blasts, karyotype, and gene mutations on the whole cohort of MDS patients. Mutations in the following genes showed an independent effect on OS in multivariable analysis: TP53 (hazard ratio [HR]: 6.00; P = .001), U2AF1 (HR: 3.85; P = .009), RUNX1 (HR: 3.33; P = .005), ASXL1 (HR: 2.08; P = .03), EZH2 (HR: 2.32; P = .04), and SF3B1 (HR: 0.32; P = .01).
We then performed multivariable analyses on MDS categories identified by the unsupervised hierarchical clustering analyses (MDS associated with SF3B1 mutation; MDS with MD-associated mutations; MDS NOS; and MDS with excess blasts). In MDS associated with SF3B1 mutation, we found that the only gene that showed a negative prognostic value was RUNX1 (HR: 15.94; P = .017). Among MDS with MD-associated mutations and MDS NOS, the following genes retained an independent negative prognostic value: RUNX1 (HR: 10.80; P = .005), U2AF1 (HR: 7.32; P = .003), and ASXL1 (HR: 4.76; P = .006). Finally, when the analysis was extended to include MDS with excess blasts: U2AF1 (HR: 3.97; P = .008), ASXL1 (HR: 2.40; P = .009), and TP53 (HR: 5.89; P = .002) showed a significant independent prognostic value.
Genetic determinants of monocytosis and thrombocytosis in myeloid neoplasms with myelodysplasia
We focused on myeloid neoplasms with dysplasia with the aim of identifying mutation patterns associated with myeloproliferative features, including monocytosis and thrombocytosis.
Among patients with monocytosis (20 patients with diagnosis of chronic myelomonocytic leukemia [CMML] according to WHO criteria), 9 patients carried a mutation in SRSF2 (9 of 20, 45%); of these, 7 patients (77.8%) had comutation in TET2. When analyzing the whole cohort of myeloid neoplasms with dysplasia, we found that the association of SRSF2 and TET2 mutations was highly specific for CMML disease phenotype (specificity, 97.6%). Interestingly, of the 6 double-mutated patients with a diagnosis of MDS according to WHO criteria, 2 had relative monocytosis at the time of mutation analysis and 2 developed an overt CMML during follow-up. Three of the 5 TET2-positive and SRSF2-negative CMML patients carried a mutation in ZRSR2. In addition, among 4 patients with comutation of TET2 and ZRSR2 and a diagnosis of MDS, 2 had relative monocytosis and 1 developed overt CMML during follow-up. When accounting for these cases, the co-occurrence of TET2 and SRSF2 or ZRSR2 mutations showed a specificity for myelomonocytic phenotype of 98.4%. A multivariable logistic regression having disease phenotype with monocytosis as outcome and including mutations in SRSF2, ZRSR2, and TET2, showed that the association between TET2 and SRSF2, or ZRSR2, was highly predictive of this disease phenotype (OR: 62.32; P = .001). Mutation patterns of TET2, SRSF2, and ZRSR2-positive cases are shown in Figure 3.
Relationship between mutation pattern and disease phenotype in TET2, SRSF2, and ZRSR2-mutated myeloid neoplasms with myelodysplasia. The red rectangle identifies patients with monocytosis nonfulfilling criteria for classification of chronic myelomonocytic leukemia at the time of mutation analysis.
Relationship between mutation pattern and disease phenotype in TET2, SRSF2, and ZRSR2-mutated myeloid neoplasms with myelodysplasia. The red rectangle identifies patients with monocytosis nonfulfilling criteria for classification of chronic myelomonocytic leukemia at the time of mutation analysis.
Patients with concurrent mutations of TET2 and SRSF2 or ZRSR2, showed significantly higher hemoglobin levels (12.1 vs 9.6 g/dL; P = .003) and higher monocyte count (2.95 vs 2.05 × 109/L; P = .017) compared with those without comutation. Mutant allele burdens in patients with TET2 and SRSF2 or ZRSR2 mutations, are reported in supplemental Figure 5.
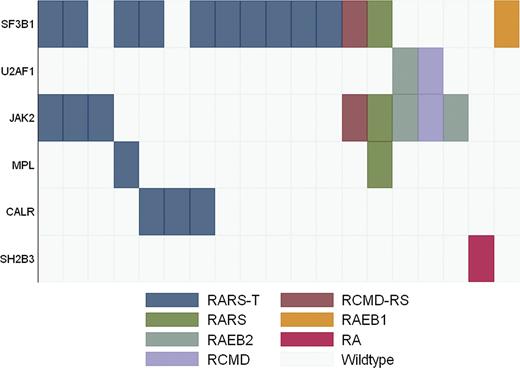
We then focused on myeloid neoplasms with dysplasia and thrombocytosis. A total of 22 of 308 patients in our cohort showed a platelet count ≥450 × 109/L at the time of mutation analysis. In 6 cases, mild thrombocytosis was observed in patients with typical RARS or RCMD-RS carrying the SF3B1 mutation (2 cases) or with del(5q) or t(3;3) (4 cases). Thirteen of the remaining 16 patients (12 classified as RARS-T and 4 as RA, RCMD, or RAEB) showed mutation in splicing factors (81.2%): 11 in SF3B1 (68.7%) (10 RARS-T and 1 RAEB-1), and 2 in U2AF1 (12.5%) (1 RCMD and 1 RAEB-2). Ten out of 16 patients (62.5%) showed mutations in JAK2(V617F) (5 cases), MPL (1 case), SH2B3 (1 case), or CALR (3 cases); 7 had a typical RARS-T phenotype, whereas the remaining 3 showed a proportion of RS lower than 15%.
Three additional patients (1 with RARS, 1 with RCMD-RS, and 1 with RAEB-2) carried the JAK2(V617F) mutation associated with megakaryocyte proliferation. One patient with a diagnosis of RAEB-2 at the time of mutation assessment had a previous history of RARS-T, followed by a progressive increase in blast and decrease in platelet counts, whereas the remaining 2 patients (1 with RARS and 1 with RCMD-RS) had SF3B1 mutation and a platelet count slightly lower than 450 × 109/L, but then progressively increased leading to re-classification as RARS-T. Mutation patterns of patients with RS and megakaryocyte proliferation are shown in Figure 4.
Mutation pattern in MDS and MDS/MPN with thrombocytosis. Diagnosis of each sample according to WHO criteria is shown by indicated colors.
Mutation pattern in MDS and MDS/MPN with thrombocytosis. Diagnosis of each sample according to WHO criteria is shown by indicated colors.
Discussion
The diagnosis and classification of myeloid neoplasms with myelodysplasia are hampered by the poor specificity of dysplastic changes and the scarce reproducibility of morphologic analysis of dysplasia. Identifying specific associations between mutation pattern and disease phenotype is essential to recognizing genetically defined disease entities and to differentiate between these neoplasms and nonclonal conditions. The purpose of this study was to define reliable genotype/phenotype relationships in MDS and MDS/MPN, and the findings obtained serve as a proof of concept that a molecular classification of myeloid neoplasms is feasible.
We first focused on WHO categories defined by the presence of RS (RARS and RCMD-RS). In agreement with previous reports, a significantly higher prevalence of SF3B1 mutation was found in patients classified in these categories compared with WHO subtypes not defined by this morphologic feature.11,12,23 In this study, more than 90% of patients with RARS were found to carry a SF3B1 mutation, whereas this proportion was lower in patients with MD. In addition, we found a significant association between SRSF2 and RS. At variance with SF3B1-positive cases, more than 90% of these SRSF2-mutated cases were classified as RCMD or RAEB according to WHO criteria. This supports previous observations suggesting that depletion of SRSF2 can lead to genomic instability.24
Unsupervised hierarchical clustering analyses showed that SF3B1 mutation is a major clustering criterion per se, and able to identify a distinct subset of MDS patients with homogeneous genotypic and phenotypic features and favorable prognosis, irrespective of current classification criteria. In fact, in this group, neither the threshold of 15% or more RS nor the presence of unilineage or MD were able to recognize separate subsets. In particular, cases with MD, according to current WHO morphologic criteria, had only mild dysplasia in myeloid or megakyocytic lineage (data not shown), and showed peripheral cytopenias and outcome not significantly different from those with unilineage dysplasia. Conversely, MDS with RS negative for the SF3B1 mutation, mainly classified as RCMD, segregated in a different cluster with other MDS subtypes showing a significantly worse prognosis. Taken together, these data identify SF3B1 mutation as a classification criterion, and suggest that MDS associated with SF3B1 mutation should be recognized as a separate subtype within myelodysplastic syndromes/neoplasms.
We then focused on MDS categories without RS with the aim of identifying genetic determinants of unilineage or MD and excess BM blasts. We found that mutations in genes implicated in DNA methylation independently predicted disease phenotype with MD. In addition, mutations of splicing factors other than SF3B1, and those of the RAS pathway and cohesin complex were independently associated with either MD and excess blasts. Among MDS categories without excess blasts, unsupervised clustering analysis suggested that these mutations were able to discriminate a homogeneous group of patients, invariably characterized by MD and with significantly worse prognosis compared with cases with different mutation patterns. Interestingly, mutations in splicing factors, mainly SRSF2 and U2AF1, were recently found to identify a homogeneous group of patients with advanced disease phenotype overriding the boundaries of MDS and AML.25
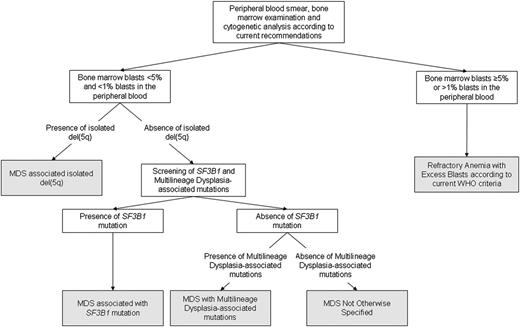
Unsupervised analysis showed that, irrespective of the underlying driver mutation pattern, a threshold of 5% BM blasts retains a significant discriminant value for identifying cases with clonal evolution. This suggests that disease progression may result in a genotypic heterogeneity that would prevent the identification of recurrent mutation patterns overcoming current classification criteria. An algorithm outlining the classification process based on morphologic and genetic criteria is shown in Figure 5.
Algorithm illustrating the classification process based on morphologic and genetic criteria identified by the unsupervised clustering analyses. According to these analyses, the threshold of 5% BM blasts retains a significant discriminant value, irrespective of the underlying driver mutation pattern. In MDS with no excess blasts, the presence of isolated del(5q), SF3B1 mutation or multilineage dysplasia-associated mutations recognize genetically-defined disease subtypes.
Algorithm illustrating the classification process based on morphologic and genetic criteria identified by the unsupervised clustering analyses. According to these analyses, the threshold of 5% BM blasts retains a significant discriminant value, irrespective of the underlying driver mutation pattern. In MDS with no excess blasts, the presence of isolated del(5q), SF3B1 mutation or multilineage dysplasia-associated mutations recognize genetically-defined disease subtypes.
We failed to identify mutation patterns specifically associated with unilineage erythroid dysplasia. Several genes were found to be mutated in this category; however, none of these genes were specifically associated with this phenotypic feature. Undetected mutations, as well as additional variables such as differences in hematopoietic progenitor primarily involved by mutation or BM microenvironment abnormalities, might contribute to sustain this as yet unexplained, phenotypic heterogeneity.26,27
We performed multivariable Cox regression analyses in order to identify gene mutations with additive prognostic value to genotype-based MDS categories. Not surprisingly, a fraction of mutations that were found to be associated with specific disease phenotypes in this study also showed an independent prognostic value, such as SF3B1, U2AF1, and RUNX1. However, in some cases, such as TET2 mutations, disease phenotype with multilineage involvement may be related to the specific effect of mutation on hematopoiesis, without significant propensity to disease progression.28,29 In addition, other mutations were found to have an independent prognostic value without being associated with specific phenotypic features, such as TP53 or ASXL1.30-32 When the analysis was focused on the genotype-based categories identified in this study, we found that within MDS associated with SF3B1 mutation, the only additional mutant gene with negative prognostic significance was RUNX1. Conversely, in SF3B1-negative subgroups, TP53 and ASXL1 had an independent negative prognostic impact on survival.
Finally, we focused on myeloid neoplasms with myelodysplasia with the aim of identifying mutation patterns associated with coexisting myeloproliferative features, including monocytosis and thrombocytosis. Interestingly, we found that mutations in SF3B1 and SRSF2 were associated with a high rate of evolution of the MDS clone with occurrence of myeloproliferative features.
A high prevalence of mutations in SRSF2 was previously reported in patients with CMML,26 and cooperation between SRSF2 and TET2 mutations in this disorder has been suggested.13,26 Our data show that the co-occurrence of TET2 and SRSF2 or ZRSR2 mutations, is highly specific for disease phenotype characterized by the coexistence of dysplastic and proliferative features with monocytosis.
As reported in previous studies, most of the patients with thrombocytosis associated with abnormal megakaryocyte proliferation carried mutations in SF3B1 in association with mutations of JAK2, MPL, or CALR.12,21,33-35 In addition, we found a single patient with a mutation in SH2B3, which was previously reported in patients with typical MPN.36 Taken together, these data confirm that the co-occurrence of mutations in SF3B1 and in JAK2, MPL, or CALR is strongly associated with a myelodysplastic/myeloproliferative phenotype characterized by thrombocytosis.
Although a validation of these results in independent cohorts of patients is warranted, our observations may represent a basis for a molecular classification of myeloid neoplasms with myelodysplasia.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by grants from the Associazione Italiana per la Ricerca sul Cancro, Fondo per gli Investimenti della Ricerca di Base (project no. RBAP11CZLK), and Ministero dell’Istruzione, dell’Università e della Ricerca (MIUR, PRIN 2010-2011) (M.C.); and from Fondazione Berlucchi, Fondazione Veronesi, Fondazione Cariplo, and Regione Lombardia, Milan, Italy (M.G.D.P.).
Authorship
Contribution: L.M. designed the study, performed statistical analysis, and wrote the manuscript; E.P. and P.J.C. analyzed sequencing data and performed bioinformatic analysis; I.A., C.E., M.G.D.P., M.U., E.B., and M.C.D.V. collected clinical data; E.T., R.I., and E.B. performed blinded revision of diagnostic specimens; A.G., D.P., A.B., F.B., L.C., and M.F. analyzed sequencing data; C.P. performed statistical analysis; and M.C. designed the study, reviewed data analysis, and wrote the manuscript. All authors reviewed the manuscript during its preparation.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Luca Malcovati, Department of Hematology Oncology, Fondazione IRCCS Policlinico San Matteo and Department of Molecular Medicine, University of Pavia, 27100 Pavia, Italy; e-mail: luca.malcovati@unipv.it.