Key Points
MiR-125b induces tumorigenesis in myeloid cells by repressing the expression of IRF4 at the mRNA and protein level.
MiR-125b promotes oncogenesis in B cells that involves selection of cells that acquire genetic deletion of the gene encoding IRF4.
Abstract
The oncomir microRNA-125b (miR-125b) is upregulated in a variety of human neoplastic blood disorders and constitutive upregulation of miR-125b in mice can promote myeloid and B-cell leukemia. We found that miR-125b promotes myeloid and B-cell neoplasm by inducing tumorigenesis in hematopoietic progenitor cells. Our study demonstrates that miR-125b induces myeloid leukemia by enhancing myeloid progenitor output from stem cells as well as inducing immortality, self-renewal, and tumorigenesis in myeloid progenitors. Through functional and genetic analyses, we demonstrated that miR-125b induces myeloid and B-cell leukemia by inhibiting interferon regulatory factor 4 (IRF4) but through distinct mechanisms; it induces myeloid leukemia through repressing IRF4 at the messenger RNA (mRNA) level without altering the genomic DNA and induces B-cell leukemia via genetic deletion of the gene encoding IRF4.
Introduction
MicroRNAs have been found to be dysregulated in several types of human and mouse cancers, including carcinomas and leukemias. As happens with protein-coding oncogenes, noncoding oncomirs can provoke cancers by dysregulating developmental, signaling, and cell survival pathways in different cell types. For the majority of oncomirs, it is not clear how a single microRNA can induce cancer development in various cell types. Potentially, an oncomir can suppress the same target(s) in different cell types to promote tumorigenesis, or it can inhibit distinct cell-specific targets to induce cancer development.
The oncomir microRNA-125b (miR-125b) is upregulated in a myriad of neoplastic blood disorders, including acute myeloid leukemia (AML) and B-cell precursor acute lymphoblastic leukemia (BCP-ALL).1-3 Importantly, we and other researchers showed that enforced constitutive overexpression of miR-125b in mice induces myeloid, B-cell, and T-cell leukemia,4-7 indicating that miR-125b can provoke the oncogenic state in a range of hematopoietic cells. Interestingly, we found that miR-125b overexpression initially impairs the development of B cells whereas others found that it induces B-cell leukemia.5,6 This suggests that miR-125b might initially repress the development of B cells but that these cells might acquire secondary mutational or epigenetic events that transform them into cancer cells. To date, the mechanism by which miR-125b induces tumorigenesis in different hematopoietic lineages is unknown.
Previously, we found that downregulating the expression of the direct miR-125b target interferon regulatory factor 4 (IRF4) was sufficient to recapitulate the activated phenotype observed upon overexpressing miR-125b in bone marrow–derived macrophages.8 Relevant to leukemia, the expression of IRF4 is downregulated in a range of hematopoietic cancer cell lines9 as well as in human patients with AML, chronic myeloid leukemia (CML), and acute lymphoblastic leukemia (ALL).10,11 Also, deletion of IRF4 in mice exacerbates the development of myeloid leukemia and leads to development of B-cell leukemia.12-14 However, whether repression of IRF4 plays a functional role in miR-125b–induced myeloid and B-cell leukemia remains to be tested.
In this study, we have investigated the cellular and molecular mechanisms by which miR-125b induces the development of myeloid and B-cell leukemia. We found that miR-125b induces cancer development by initiating tumorigenesis in myeloid and B precursor cells. Our data also indicate that in both cases miR-125b induces myeloid and B-cell leukemia by inhibiting IRF4 expression. Whereas miR-125b induces tumorigenesis in myeloid cells by repressing the expression of IRF4 at the messenger RNA (mRNA) and protein level, it promotes oncogenesis in B cells by provoking genetic deletion of IRF4. Thus, miR-125b represents a novel paradigm by which an oncomir induces cancer development in multiple cell lineages by modulating the same signaling pathway but via distinct mechanisms.
Methods
DNA constructs
pMG, pMSCV-IRES-GFP (MIG), pMG-miR-125b, and pMiR-report IRF4 3′ untranslated region (3′UTR) vectors have been described.5,8 pMIG-miR-125b coexpresses green fluorescent protein (GFP) and miR-125b. pMIG-IRF4 coexpresses GFP and IRF4. pHcRed-miR125b and pmCherry-miR125b coexpress miR-125b and HcRed or mCherry, respectively (supplemental Table 1 [see supplemental Data available at the Blood Web site] for cloning primers).
Infection of BMCs, bone marrow reconstitution, and in vitro cell proliferation assays
To generate MG and MG-125b mice, lethally irradiated C57bl/6 recipient mice were injected with virally transduced bone marrow cells (BMCs). Briefly, donor C57bl/6 BMCs were transduced with miR-125b overexpressing vector (pMG-miR-125b or pMIG-miR-125b) through 2 to 4 rounds of spin infection, which achieved 25- and 186-fold higher miR-125b overexpression (supplemental Figure 7). The expression levels are within range of the level of miR-125b overexpression observed in human patients with leukemia or myelodysplastic syndromes, which range from several to 262-fold above normal.1,6,15 For the in vitro proliferation assays, BMCs and sorted Lin−cKit+Sca1− myeloid progenitors (MPs) were cultured and passaged in 50 ng/mL stem cell factor (SCF), and sorted Lin−cKit+Sca1+ hematopoietic stem and progenitor cells (HSPCs) were cultured in media containing 50 ng/mL SCF, 50 ng/mL interleukin 6 (IL6), and 25 ng/mL IL3. All animal studies were approved by the institutional animal care and use committee (IACUC).
Transplantation of miR-125b–induced cancer cells and fluorescent-activated cell sorting
Splenic cells, BMCs, sorted GFP+Lin−cKit+Sca1− cells, and sorted GFP+CD19+ cells were harvested from MG-125b mice when they developed leukemia and transplanted into sublethally irradiated C57bl/6 recipients. For transplantation of common myeloid progenitors, 3500 Lin−cKit+Sca1− cells (Lin = ILR7a, Thy1, Cd11b, Cd11c, B220, Ter119, CD3e, NK1.1, GR-1) were sorted from MG-125b mice when they were moribund. Cells were sorted using the FACSAria (BD Biosciences) or iCyte (Sony Biotech) instrument. Flow cytometric antibodies were purchased from eBioscience or Biolegend.
Single-cell analysis and cell differentiation assay
MiR-125b–overexpressing Lin−cKit+Sca1− MPs were passaged 6 times in 50 ng/mL SCF media. Single cells were plated. One month later, an aliquot of the expanded cells were assayed for granulocytes formation and some cells were cultured in 20 ng/mL macrophage colony-stimulating factor (MCSF) or J558L–granulocyte MSCF (GMCSF) conditioned media to differentiate into macrophages and dendritic cells, respectively.
In vitro proliferation assay with rescued expression of IRF4
For in vitro proliferation assays, BMCs were first infected with pHcRed-miR-125b retroviruses that express miR-125b and HcRed. Subsequently, cells were infected with pMIG or pMIG-IRF4 retroviruses. HcRed+GFP+ cells were sorted and cultured in 50 ng/mL SCF.
Competitive repopulation of miR-125b–overexpressing cells with rescued IRF4 expression
Donor BMCs were infected with mCherry+ miR-125b–overexpressing virus (pmCherry-miR-125b vector). The cells were then infected with GFP+ virus (pMIG vector) or GFP+ IRF4–overexpressing virus (pMIG-IRF4 vector) and transplanted into lethally irradiated C57bl/6 mice.
Transplantation of sorted miR-125b–overexpressing cells with restored IRF4 expression
BMCs were infected with GFP+ miR-125b–overexpressing retroviruses (pMG-miR-125b vector) or coinfected with HcRed+ miR-125b–overexpressing (pHcRed-miR-125b vector) and GFP+ IRF4–overexpressing retroviruses (pMIG-IRF4 vector). GFP+ cells or GFP+HcRed+ cells were sorted from the former and latter group of cells, respectively. The sorted BMCs were transplanted into sublethally irradiated C57bl/6 mice.
Exome sequencing and comparative genomic hybridization microarray
See the supplemental Methods for detailed experimental setup and data analysis. The microarray GEO accession number is GSE58900.
Results
MiR-125b–overexpressing leukemic cells are serially transplantable
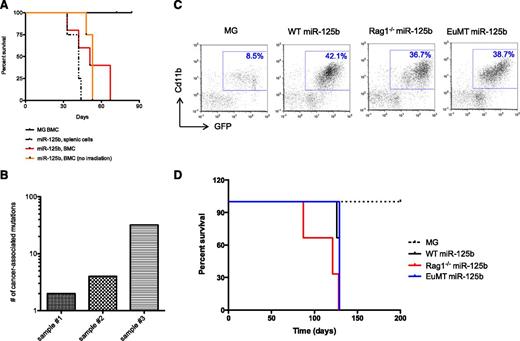
To examine the ability of miR-125b–induced myeloid cancer cells to serially propagate, we overexpressed miR-125b through bone marrow reconstitution experiments using the murine stem cell virus (MSCV)–GFP+ (MG)–based retroviral system in mice (hereon denoted as “MG-125b”).5 When the MG-125b mice developed frank myeloid leukemia, cells from the spleen or bone marrow were transferred into sublethally irradiated secondary recipients. These recipients were moribund or died within 70 days (Figure 1A) and developed leukemia indicated by increased white blood cell count (supplemental Figure 1A) and splenomegaly (supplemental Figure 1B). Recipient mice that received cells from control “MG” mice did not develop cancer and remained healthy through the duration of the study (Figure 1A). Remarkably, transplantation of miR-125b–induced cancer cells from secondary recipients into tertiary nonirradiated immunocompetent recipients also induced myeloid leukemia in the mice (Figure 1A and supplemental Figure 1C-D). Collectively, our data indicates that the myeloid cancer induced by miR-125b overexpression is serially transplantable.
Features of miR-125b-induced myeloid cancer cells. (A) Leukemic cells from MG-125b are serially transplantable. Cells from MG and MG-125b mice were harvested 3 to 6 months after bone marrow reconstitution. 200K GFP+ splenic cells (black dotted line) or 400 to 1000K GFP+ BMCs (red line) from MG-125b mice were transferred into secondary sublethally irradiated C57bl/6 recipients. Two million GFP+ BMCs from the secondary mice were injected into tertiary nonirradiated C57bl/6 recipients (orange line indicated as “no irradiation”). Death point of the recipients (at least 4 mice per group) were recorded when found dead or moribund. (B) Myeloid cancer cells induced by miR-125b overexpression were harvested and subjected to exome deep-sequencing analysis. Every exon was sequenced an average of 50 times. The sequencing data sets were filtered on a 0.1% false-positive discovery rate and cancer-associated mutations were obtained by excluding those that were identified in normal noncancerous mice. Plot shows the number of cancer-associated exonic mutations identified in each of the 3 independent experiments. (C) Donor BMCs from C57bl/6, Rag1−/−, and EμMT (B6.129S2-Ighmtm1Cgn/J) mice were transduced with MG-125b retroviruses, which encode for miR-125b and GFP expression. As control, donor BMCs from C57bl/6 animals were transduced with empty MG retroviruses, which encode for GFP. BMCs were transplanted into lethally irradiated C57bl/6 recipients, and the blood of the mice was subjected to flow cytometric analyses 3 months after reconstitution. (D) The survival curve of the mice described in panel C is shown (3 mice per group). Mice were considered dead when they became moribund or found dead.
Features of miR-125b-induced myeloid cancer cells. (A) Leukemic cells from MG-125b are serially transplantable. Cells from MG and MG-125b mice were harvested 3 to 6 months after bone marrow reconstitution. 200K GFP+ splenic cells (black dotted line) or 400 to 1000K GFP+ BMCs (red line) from MG-125b mice were transferred into secondary sublethally irradiated C57bl/6 recipients. Two million GFP+ BMCs from the secondary mice were injected into tertiary nonirradiated C57bl/6 recipients (orange line indicated as “no irradiation”). Death point of the recipients (at least 4 mice per group) were recorded when found dead or moribund. (B) Myeloid cancer cells induced by miR-125b overexpression were harvested and subjected to exome deep-sequencing analysis. Every exon was sequenced an average of 50 times. The sequencing data sets were filtered on a 0.1% false-positive discovery rate and cancer-associated mutations were obtained by excluding those that were identified in normal noncancerous mice. Plot shows the number of cancer-associated exonic mutations identified in each of the 3 independent experiments. (C) Donor BMCs from C57bl/6, Rag1−/−, and EμMT (B6.129S2-Ighmtm1Cgn/J) mice were transduced with MG-125b retroviruses, which encode for miR-125b and GFP expression. As control, donor BMCs from C57bl/6 animals were transduced with empty MG retroviruses, which encode for GFP. BMCs were transplanted into lethally irradiated C57bl/6 recipients, and the blood of the mice was subjected to flow cytometric analyses 3 months after reconstitution. (D) The survival curve of the mice described in panel C is shown (3 mice per group). Mice were considered dead when they became moribund or found dead.
Genetic analyses of miR-125b myeloid cancer
Using deep-sequencing technologies, it has been determined that cancer cells can acquire few to thousands of secondary mutations in their exomes.16-20 Thus, we sorted GFP+Cd11b+ myeloid cancer cells induced by miR-125b overexpression and subjected them to exome deep-sequencing analysis. The first sample (sample 1) was harvested from a mouse 2 months after bone marrow reconstitution, which we validated as cancerous cells as demonstrated by the ability of these cells to be transplanted into a nonirradiated recipient mouse (data not shown). The second sample (sample 2) was harvested from a miR-125b–overexpressing mouse 3 months after reconstitution; this mouse developed frank myeloid leukemia as characterized by moribund feature, exacerbated myeloid cell count in the blood (27× higher than normal), and splenomegaly (data not shown). The third sample (sample 3) was harvested from miR-125b–induced myeloid leukemic cells that were serially transplanted into recipient mice to ensure that we were analyzing cancerous cells in our deep-sequencing analysis. The deep-sequencing data sets were filtered based on a 0.1% false discovery rate, and cancer-specific mutations were obtained by subtracting those that were also identified in normal healthy mice. From the 3 independent experiments, we identified 2, 4, and 32 mutations from samples 1, 2, and 3, respectively (Figure 1B and supplemental Table 2). The variations in the number of mutations from the different samples might be due to the different time at which they were harvested after bone marrow reconstitution. We have not ruled out the possibility that a small fraction of the myeloid cancer cells might have random mutations, which is outside of the deep-sequencing detection limit. However, our data suggest that the majority of miR-125b–driven myeloid cancer cell populations in the samples tested have relatively few nucleotide mutations in the coding regions and do not have widespread genomic instability.
Overexpression of miR-125b in primary BMCs in vitro recapitulates myeloid malignant phenotype
Previously, we showed that induction of myeloid leukemia in mice deleted of the gene for a different microRNA, miR-146a, was in part dependent on extrinsic factors released by miR-146a−/− lymphocytes.21 Thus, we investigated whether miR-125b–overexpressing lymphocytes contribute to myeloid cancer development in MG-125b mice. BMCs harvested from B-cell–deficient EμMT22 or lymphocyte-deficient Rag1−/−23 donor mice were transduced with miR-125b encoding retroviruses and transplanted into normal C57bl/6 recipient mice. These recipient mice developed myeloid leukemia (Figure 1C) at a similar time scale and rate (100%) compared with those transplanted with miR-125b–overexpressing wild-type (WT) BMCs (Figure 1D), indicating that the development of myeloid leukemia provoked by miR-125b does not require miR-125b–overexpressing lymphocytes.
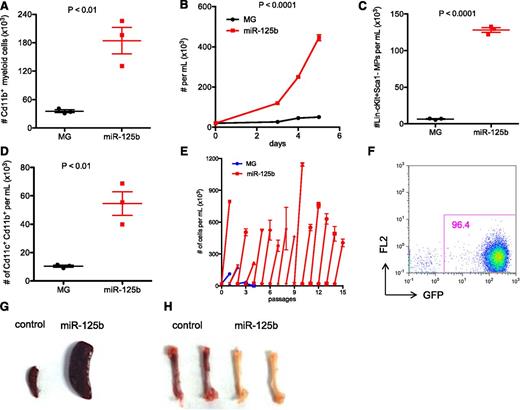
Next, we tested whether overexpressing miR-125b in BMCs in vitro could induce cellular hyperproliferation and recapitulate the myeloproliferative phenotype observed in mice. We overexpressed miR-125b in BMCs and cultured them in cytokine-supplemented media (SCF + IL3 + IL6, SCF + IL-3, or SCF + IL-6) that supports growth and differentiation of HSCs, myeloid progenitors, and mature myeloid cells. Indeed, overexpression of miR-125b in vitro elevated the growth rate of BMCs (supplemental Figure 2A-C) and induced hyperexpansion of myeloid cells (Figure 2A). MiR-125b–overexpressing BMCs cultured in SCF alone, which supports growth of hematopoietic stem and progenitor cells, also grew faster (Figure 2B), suggesting that miR-125b induces hyperproliferation in part due to its effect in stem/progenitor cells. Indeed, growth of BMCs with ectopic miR-125b expression generated higher number of Lin−cKit+Sca1− cells (Figure 2C). In normal counterparts, Lin−cKit+Sca1− cells represent myeloid progenitors/MPs. Consistently with elevated number of myeloid progenitors, miR-125b–overexpressing BMCs generated more dendritic cells when subjected to a methylcellulose-based colony-forming assay (supplemental Figure 2D) or differentiated into dendritic cells in liquid culture (Figure 2D). MiR-125b–overexpressing cells also formed more colonies in methylcellulose assay when cultured in present of SCF + IL3 + IL6 (supplemental Figure 2E) although the cells did not serially replate (data not shown). Thus, our in vitro assay recapitulated the phenotype observed with overexpressing miR-125b in mice with higher numbers of myeloid progenitors and differentiated Cd11b+ myeloid cells.5
Overexpressing miR-125b in BMCs induces myeloproliferative disorder in vitro. (A) HSPC-enriched BMCs, which were obtained by injecting C57bl/6 mice with 5-fluorouracil, were transduced with MG control or MG-125b retroviruses. Equal numbers of BMCs were cultured in 50 ng/mL SCF, 50 ng/mL IL-6, and 25 ng/mL IL-3. After 5 days, the total number of myeloid cells (CD11b+) cells was determined by flow cytometry. (B) MG control or miR-125b–overexpressing HSPC-enriched cells were expanded at same starting density in 50 ng/mL SCF. Cells were counted by flow cytometry. (C) Equal numbers of HSPC-enriched cells transduced with MG or miR-125b overexpression cassette were expanded in 50 ng/mL SCF. The density of Lin−Sca1−cKit+ cells after 4 days was determined by flow cytometry. (D) Equal numbers of MG control or miR-125b–overexpressing BMCs were cultured in 20 ng/mL GMCSF to induce differentiation into dendritic cells. The number of dendritic cells was determined by flow cytometry 6 days after culture. (E) MG or miR-125b–transduced BMCs were cultured in 50 ng/mL SCF. When the miR-125b–overexpressing cells were confluent between 4 and 8 days after plating, the cells were reseeded at a starting density of 20K per mL. The x-axis represents the passage number, and the y-axis represents the cell density. Panels A through E are representative of at least 2 independent experiments. (F) MiR-125b–overexpressing BMCs were cultured and expanded in vitro. One million GFP+ cells were injected into sublethally irradiated C57bl/6 mice. Two months posttransplantation, recipient blood was subjected to flow cytometric analysis to determine engraftment of GFP+ cells. The pictures of (G) spleen and (H) femur are representative examples of these organs taken from moribund miR-125b–transduced mice (right panel) and age-matched C57bl/6 controls (left panel). Representative of 6 mice.
Overexpressing miR-125b in BMCs induces myeloproliferative disorder in vitro. (A) HSPC-enriched BMCs, which were obtained by injecting C57bl/6 mice with 5-fluorouracil, were transduced with MG control or MG-125b retroviruses. Equal numbers of BMCs were cultured in 50 ng/mL SCF, 50 ng/mL IL-6, and 25 ng/mL IL-3. After 5 days, the total number of myeloid cells (CD11b+) cells was determined by flow cytometry. (B) MG control or miR-125b–overexpressing HSPC-enriched cells were expanded at same starting density in 50 ng/mL SCF. Cells were counted by flow cytometry. (C) Equal numbers of HSPC-enriched cells transduced with MG or miR-125b overexpression cassette were expanded in 50 ng/mL SCF. The density of Lin−Sca1−cKit+ cells after 4 days was determined by flow cytometry. (D) Equal numbers of MG control or miR-125b–overexpressing BMCs were cultured in 20 ng/mL GMCSF to induce differentiation into dendritic cells. The number of dendritic cells was determined by flow cytometry 6 days after culture. (E) MG or miR-125b–transduced BMCs were cultured in 50 ng/mL SCF. When the miR-125b–overexpressing cells were confluent between 4 and 8 days after plating, the cells were reseeded at a starting density of 20K per mL. The x-axis represents the passage number, and the y-axis represents the cell density. Panels A through E are representative of at least 2 independent experiments. (F) MiR-125b–overexpressing BMCs were cultured and expanded in vitro. One million GFP+ cells were injected into sublethally irradiated C57bl/6 mice. Two months posttransplantation, recipient blood was subjected to flow cytometric analysis to determine engraftment of GFP+ cells. The pictures of (G) spleen and (H) femur are representative examples of these organs taken from moribund miR-125b–transduced mice (right panel) and age-matched C57bl/6 controls (left panel). Representative of 6 mice.
Moreover, we investigated whether miR-125b overexpression is also sufficient to transform and immortalize BMCs in vitro. Indeed, whereas control cells died after several passages, miR-125b–overexpressing cells continued to grow indefinitely (Figure 2E). When we injected miR-125b–overexpressing BMCs that had been cultured in vitro into sublethally irradiated recipient mice, the cells dominated the blood of these mice and produced myeloid leukemia (Figure 2F) marked by splenomegaly (Figure 2G) and bone marrow pallor (Figure 2H). These data suggest that the cultured miR-125b–overexpressing BMCs were transformed in vitro into cancer cells capable of inducing malignancy in vivo.
Effect of overexpressing miR-125b in HSPCs
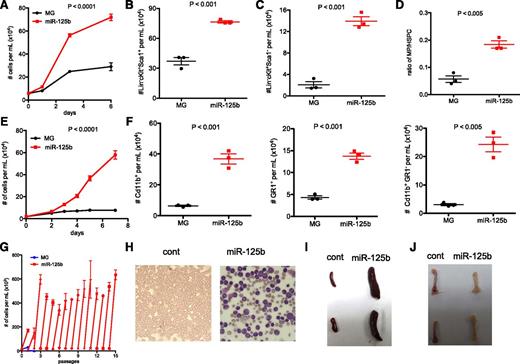
Next, we tested whether miR-125b overexpression elevates the myeloid progenitor pool by enhancing the hematopoietic output of cells upstream of MPs, such as HSPCs. We cultured sorted MG and miR-125b Lin−cKit+Sca1+ (LKS+) cells (supplemental Figure 3A) and found that those with miR-125b overexpression generated higher number of cells overall (Figure 3A) with a moderate increase in absolute LKS+ cell numbers (Figure 3B) after 6 days of growth. When we instead looked at the Lin−cKit+Sca1− (LKS−) population, the cellular output was dramatically higher in the miR-125b–overexpressing samples (Figure 3C) with exaggerated ratio of LKS− to LKS+ cells (Figure 3D). Overall, our data suggests that miR-125b functions in HSPCs and increases the output of downstream MPs. Next, we tested whether miR-125b also functions intrinsically in MPs to accelerate the proliferation of these cells. We cultured LKS− cells sorted from BMCs transduced with MG or miR-125b–encoding retroviruses. We found that the cells with ectopic miR-125b expression grew much faster (Figure 3E) and generated more myeloid cells (Cd11b+) of both granulocytic (GR1+) and nongranulocytic lineages (Cd11b+GR1−) (Figure 3F). In addition, the miR-125b–overexpressing cells were also more resistant to cell death as indicated by propidium iodide staining (supplemental Figure 3B). Thus, miR-125b induces hyperproliferation and resistance to cell death of MPs in vitro.
Effect of overexpressing miR-125b in HSPCs and MPs. (A) Equal numbers of MG or MG-125b–transduced Lin−cKit+Sca1+ HSPCs were cultured, and the cell density was determined using flow cytometry. Two-way analysis of variance was used to obtain P values. The number of (B) Lin−cKit+Sca1+ HSPCs and (C) Lin−cKit+Sca1− MPs was determined by flow cytometry 6 days after culture. (D) The ratio of MPs to HSPCs was calculated and plotted. Two independent experiments were performed. (E) Equal number of MG or MG-125b–transduced Lin−cKit+Sca1− MPs were cultured. The cell density was determined according to the indicated time. Representative of two independent experiments. (F) The density of myeloids (Cd11b+), granulocytes (GR1+), or nongranulocytic myeloids (CD11b+GR1−) cells after growing miR-125b–overexpressing MPs was determined by flow cytometry 10 days later. Two independent experiments performed. (G) Sorted MG or miR-125b–transduced Lin−cKit+ Sca1− MPs were cultured and passaged similarly as described in the Figure 2E legend. (H) Sublethally irradiated C57bl/6 recipients were injected with common myeloid progenitors sorted from MG-125b mice. Wright stain of the blood was performed when the recipients were moribund. (I) The spleen and (J) femur were harvested and imaged when the mice were moribund and sacrificed. Representative of 4 mice. Control mice represent recipient mice injected with BMCs from MG mice.
Effect of overexpressing miR-125b in HSPCs and MPs. (A) Equal numbers of MG or MG-125b–transduced Lin−cKit+Sca1+ HSPCs were cultured, and the cell density was determined using flow cytometry. Two-way analysis of variance was used to obtain P values. The number of (B) Lin−cKit+Sca1+ HSPCs and (C) Lin−cKit+Sca1− MPs was determined by flow cytometry 6 days after culture. (D) The ratio of MPs to HSPCs was calculated and plotted. Two independent experiments were performed. (E) Equal number of MG or MG-125b–transduced Lin−cKit+Sca1− MPs were cultured. The cell density was determined according to the indicated time. Representative of two independent experiments. (F) The density of myeloids (Cd11b+), granulocytes (GR1+), or nongranulocytic myeloids (CD11b+GR1−) cells after growing miR-125b–overexpressing MPs was determined by flow cytometry 10 days later. Two independent experiments performed. (G) Sorted MG or miR-125b–transduced Lin−cKit+ Sca1− MPs were cultured and passaged similarly as described in the Figure 2E legend. (H) Sublethally irradiated C57bl/6 recipients were injected with common myeloid progenitors sorted from MG-125b mice. Wright stain of the blood was performed when the recipients were moribund. (I) The spleen and (J) femur were harvested and imaged when the mice were moribund and sacrificed. Representative of 4 mice. Control mice represent recipient mice injected with BMCs from MG mice.
Moreover, we found that whereas control LKS− cells died after several passages, miR-125b–overexpressing cells continued to grow indefinitely, suggesting that miR-125b MPs were immortalized (Figure 3G). We also tested whether cultured miR-125b–overexpressing LKS− retain their ability to differentiate and whether a single cell can repopulate the myeloid repertoire. After passaging miR-125b–overexpressing MPs 6 times, single cells were plated into individual wells. Of the 96 single cells plated, 11 clones expanded beyond 1 month and were able differentiate into GR1+ granulocytes, F4/80+ macrophages, and Cd11c+ dendritic cells (supplemental Figure 3C). Collectively, this data indicate that miR-125b overexpression induces self-renewal of MPs in vitro.
To test whether miR-125b overexpression induces tumorigenesis in myeloid progenitors, we transplanted sorted MPs from MG-125b mice into sublethally irradiated C57bl/6 recipients. Indeed, the recipient mice were moribund within 70 days and developed frank myeloid leukemia characterized by large excess of myeloid and myeloblast cells in the peripheral blood (Figure 3H), splenomegaly (Figure 3I) and bone marrow pallor (Figure 3J), and dissemination of cancer cells into nonlymphoid tissues (liver, lung, kidney) (data not shown). In further support that miR-125b induces myeloid leukemia through progenitor cells, injection of terminally differentiated miR-125b–overexpressing dendritic cells into mice did not induce leukemia (supplemental Figure 3D-F), indicating the requirement of progenitor state for miR-125b to initiate tumorigenesis.
IRF4 expression rescues miR-125b–induced myeloid leukemia
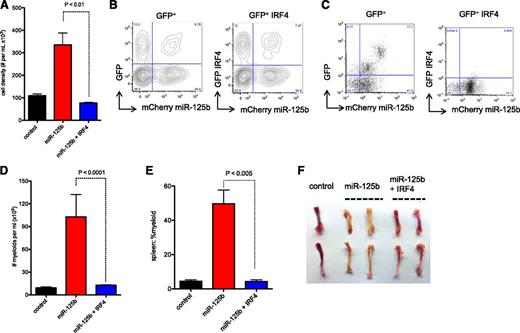
Next, we investigated signaling pathways regulated by miR-125b to induce cancer development. Previously, we and other laboratories showed that miR-125b directly inhibits the expression of the transcription factor IRF4.8,24,25 We first confirmed that miR-125b inhibits IRF4 expression in primary mouse sorted LKS+ and LKS− cells (supplemental Figure 4A-B) and represses a luciferase reporter linked to the 3′UTR of IRF4 (supplemental Figure 4C). Next, we tested and found that reintroduction of IRF4 without its 3′UTR (supplemental Figure 4D-E) into miR-125b–overexpressing BMCs was sufficient to reverse the hyperproliferative phenotype induced by miR-125b (Figure 4A) and prevented miR-125b–mediated myeloid expansion in vitro (supplemental Figure 4F).
Rescued expression of IRF4 inhibits miR-125b–induced myeloid leukemia. (A) IRF4 inhibits miR-125b–induced hyperproliferation in vitro. Control cells correspond to MG-infected BMCs. BMCs were transduced with retroviruses that encode miR-125b and HcRed. These cells were then transduced with MG or MIG-IRF4 retroviruses, which coexpresses GFP and IRF4. HcRed+GFP+ cells were sorted, and 20K cells per mL of cells were plated. The cell number was determined 3 days later. Representative of 3 experiments. (B) BMCs were transduced with retroviruses that encode miR-125b and mCherry. These cells were then infected with MG or MIG-IRF4 viruses. Shown are flow cytometric plots of these cells and infection efficiency before transplantation into mice. Plots show cells overexpressing miR-125b in the x-axis (mCherry+) and either GFP+ or GFP+ IRF4-overexpressing cells in the y-axis. (C) BMCs described in panel B were transplanted into recipient C57bl/6 mice. One month after transplantation, the peripheral blood of the recipient mice was analyzed by flow cytometry. The left and right panels represent the blood of recipient mice transplanted with BMCs coinfected with miR125b-mCherry with GFP vector and miR125b-mCherry with IRF4-GFP vector, respectively. Representative of 4 mice. (D) Recipient mice were transplanted with donor BMCs that overexpress miR-125b alone or along with restored IRF4 expression mice. The peripheral blood of the recipient mice (>6 mice per group) was analyzed by flow cytometry. Plot displays the Cd11b+ myeloid cell count 3 months after transplantation of donor cells. Control mice represent normal healthy C57bl/6 mice. P value obtained through Mann-Whitney t test. (E) The percent splenic Cd11b+ myeloid was determined using flow cytometry ∼3 months after cell transplantation. (F) Representative images of the femur and tibia of the corresponding recipient mice are shown.
Rescued expression of IRF4 inhibits miR-125b–induced myeloid leukemia. (A) IRF4 inhibits miR-125b–induced hyperproliferation in vitro. Control cells correspond to MG-infected BMCs. BMCs were transduced with retroviruses that encode miR-125b and HcRed. These cells were then transduced with MG or MIG-IRF4 retroviruses, which coexpresses GFP and IRF4. HcRed+GFP+ cells were sorted, and 20K cells per mL of cells were plated. The cell number was determined 3 days later. Representative of 3 experiments. (B) BMCs were transduced with retroviruses that encode miR-125b and mCherry. These cells were then infected with MG or MIG-IRF4 viruses. Shown are flow cytometric plots of these cells and infection efficiency before transplantation into mice. Plots show cells overexpressing miR-125b in the x-axis (mCherry+) and either GFP+ or GFP+ IRF4-overexpressing cells in the y-axis. (C) BMCs described in panel B were transplanted into recipient C57bl/6 mice. One month after transplantation, the peripheral blood of the recipient mice was analyzed by flow cytometry. The left and right panels represent the blood of recipient mice transplanted with BMCs coinfected with miR125b-mCherry with GFP vector and miR125b-mCherry with IRF4-GFP vector, respectively. Representative of 4 mice. (D) Recipient mice were transplanted with donor BMCs that overexpress miR-125b alone or along with restored IRF4 expression mice. The peripheral blood of the recipient mice (>6 mice per group) was analyzed by flow cytometry. Plot displays the Cd11b+ myeloid cell count 3 months after transplantation of donor cells. Control mice represent normal healthy C57bl/6 mice. P value obtained through Mann-Whitney t test. (E) The percent splenic Cd11b+ myeloid was determined using flow cytometry ∼3 months after cell transplantation. (F) Representative images of the femur and tibia of the corresponding recipient mice are shown.
Moreover, we tested the effect of uncontrolled IRF4 expression on miR-125b–overexpressing mice. To this end, we retrovirally transduced BMCs with a mCherry+ miR-125b–overexpressing vector and then coinfected these cells with retroviruses expressing both GFP and IRF4 (lacking its UTR) or with GFP controls (Figure 4B). These unsorted retrovirally transduced BMCs were then transplanted into C57Bl/6 recipient animals. After reconstitution, we found that mice transplanted with cells coinfected with IRF4-GFP and miR125b-mCherry vectors were deficient of cells expressing both miR-125b and IRF4 (mCherry+GFP+ population) (Figure 4C, supplemental Figure 4G). They did, however, have a prominent population of mCherry+GFP− cells (Figure 4C), demonstrating that the cells that persisted in these mice were those expressing only miR-125b but not ones coexpressing IRF4. In contrast, recipient mice transplanted with cells transduced with 125b-mCherry and GFP control vector contained prominent amounts of mCherry+GFP+ cells (Figure 4C, supplemental Figure 4G). Collectively, our data indicate that hematopoietic cells that overexpress miR-125b alone robustly outcompete those with uncontrolled expression of IRF4, suggesting that IRF4 inhibits the enhanced proliferative capacity induced by miR-125b in vivo.
Finally, we examined whether rescued expression of IRF4 could inhibit myeloid leukemia provoked by miR-125b overexpression in mice. To this end, we transplanted into recipient mice GFP+ BMCs that overexpress miR-125b alone or along with restored IRF4 expression. Both populations of BMCs engrafted in the recipients as demonstrated by GFP positivity in the blood of these mice. Whereas the recipient mice transplanted with the former group of cells exhibited severe myeloid hyperplasia after 3 months, the animals that received cells with restored IRF4 expression had normal myeloid counts (Figure 4D). As expected, the mice transplanted with miR-125b–overexpressing cells had exaggerated development of myeloid in their spleens (Figure 4E) accompanied by bone marrow pallor (Figure 4F), which are pathognomonic features of miR-125b–mediated myeloid leukemia, while those with restored IRF4 expression had normal splenic myeloipoiesis and displayed normal reddish bone marrow (Figure 4E-F). Collectively, our data indicate that suppression of IRF4 is important for miR-125b to induce myeloid leukemia and restoration of IRF4 can inhibit disease development.
MiR-125b overexpression induces B-cell cancer harboring IRF4 deletion
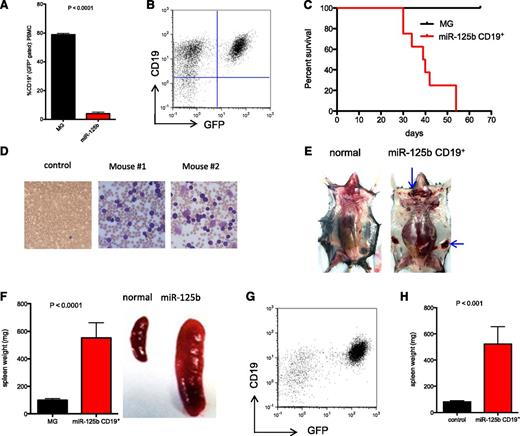
We confirmed our previous finding that MG-125b mice have impaired B-cell development (Figure 5A).5 Curiously, others have shown that B-cell–restricted overexpression of miR-125b induces lymphoblastic leukemia in mice.6 We posited that our experimental mice might be dying from complications associated with myeloid leukemia before manifesting features of B-cell leukemia. To examine whether miR-125b promotes B-cell leukemia in our experimental setting, we sorted the few GFP+CD19+ B cells from MG-125b mice when they developed myeloid leukemia and transplanted these B cells into C57bl/6 recipients. The cells were able to engraft in the secondary recipients (Figure 5B) and proved lethal within 2 months, generating a fulminant CD19+ cancer (Figure 5C). Moreover, the resultant GFP+CD19+ cancer cells were serially transplantable into sublethally irradiated or nonirradiated recipient mice for up to 11 generations (supplemental Figure 5A), inducing B-cell leukemia (Figure 5D) and in some cases B-cell lymphoma (Figure 5E) in the recipients. The development of B-cell leukemia in these mice was also marked by splenomegaly (Figure 5F) and dissemination of GFP+CD19+ into nonhematopoietic organs, including the lung, liver, and kidney (supplemental Figure 5B). Thus, miR-125b overexpression promotes tumorigenesis in B cells in addition to myeloid cells.
MiR-125b induces tumorigenesis in pre-B cells. (A) MG and MG-125b were bled 12 weeks after bone marrow reconstitution. The figure represents the percent of B cells (CD19+) within the GFP+ gated population in the peripheral blood of these mice (at least 3 mice per group). P value was calculated by Student t test. (B) GFP+CD19+ B cells were sorted from MG-125b mice 4 to 6 months after bone marrow reconstitution. CD19+ B cells (35-52K) were transplanted into sublethally irradiated mice. The figure shows the flow cytometric analysis of the secondary recipient mice (bone marrow) 6 weeks after transplantation. Representative of 8 mice. (C) The figure shows the survival of curve of the secondary recipient mice transplanted with GFP+CD19+ cells from MG-125b mice (8 mice) or total BMCs from MG mice (6 mice). (D) Wright stain was performed from the blood of recipient mice injected with miR-125b–overexpressing CD19+ cells. The dark purple cells represent leukocytes. The smaller cells with central pallor are red blood cells. (E) Some recipients of miR-125b–overexpressing CD19+ cells develop lymphomas. Lymphomas shown at the superficial cervical (top blue arrow) and inguinal lymph node sites (side blue arrow). The normal mouse shown is a healthy C57bl/6 mouse. (F) Left panel, The spleen weight of the recipient mice transplanted with GFP+CD19+ cells (denoted as “miR-125b CD19+”) from MG-125b mice or total BMCs from MG animals were obtained 4 to 6 months after transplantation. Six mice per group. Right panel, Representative images of spleens harvested from normal C57bl/6 control and miR-125b CD19 transplanted mice 6 months posttransplant. (G) MiR-125b induces pre-B-cell cancer. Sorted GFP+CD19+ cells (10K) harvested from mice reconstituted with miR-125b–overexpressing EμMT BMCs were injected into sublethally irradiated secondary C57bl/6 recipients. Bone marrow of the secondary recipient harvested when the mice became moribund, and figure shows flow cytometric plot of the leukocyte population of the bone marrow. (H) Spleen weights of the secondary recipients described in panel H are shown (4 mice). The controls signify spleen weight from secondary recipients injected with cells from MG reconstituted mice (8 mice). P value was calculated by Student t test.
MiR-125b induces tumorigenesis in pre-B cells. (A) MG and MG-125b were bled 12 weeks after bone marrow reconstitution. The figure represents the percent of B cells (CD19+) within the GFP+ gated population in the peripheral blood of these mice (at least 3 mice per group). P value was calculated by Student t test. (B) GFP+CD19+ B cells were sorted from MG-125b mice 4 to 6 months after bone marrow reconstitution. CD19+ B cells (35-52K) were transplanted into sublethally irradiated mice. The figure shows the flow cytometric analysis of the secondary recipient mice (bone marrow) 6 weeks after transplantation. Representative of 8 mice. (C) The figure shows the survival of curve of the secondary recipient mice transplanted with GFP+CD19+ cells from MG-125b mice (8 mice) or total BMCs from MG mice (6 mice). (D) Wright stain was performed from the blood of recipient mice injected with miR-125b–overexpressing CD19+ cells. The dark purple cells represent leukocytes. The smaller cells with central pallor are red blood cells. (E) Some recipients of miR-125b–overexpressing CD19+ cells develop lymphomas. Lymphomas shown at the superficial cervical (top blue arrow) and inguinal lymph node sites (side blue arrow). The normal mouse shown is a healthy C57bl/6 mouse. (F) Left panel, The spleen weight of the recipient mice transplanted with GFP+CD19+ cells (denoted as “miR-125b CD19+”) from MG-125b mice or total BMCs from MG animals were obtained 4 to 6 months after transplantation. Six mice per group. Right panel, Representative images of spleens harvested from normal C57bl/6 control and miR-125b CD19 transplanted mice 6 months posttransplant. (G) MiR-125b induces pre-B-cell cancer. Sorted GFP+CD19+ cells (10K) harvested from mice reconstituted with miR-125b–overexpressing EμMT BMCs were injected into sublethally irradiated secondary C57bl/6 recipients. Bone marrow of the secondary recipient harvested when the mice became moribund, and figure shows flow cytometric plot of the leukocyte population of the bone marrow. (H) Spleen weights of the secondary recipients described in panel H are shown (4 mice). The controls signify spleen weight from secondary recipients injected with cells from MG reconstituted mice (8 mice). P value was calculated by Student t test.
Next, we examined the stage of B-cell development at which cells are sensitive to miR-125b–induced transformation; specifically, we investigated whether B-cell maturation is required for miR-125b to induce transformation. To this end, we overexpressed GFP+ miR-125b in donor BMCs from EμMT mice, which do not develop mature B cells but produce CD19+ pre-B cells.22 C57Bl/6 mice were reconstituted with these cells and 3 months after transplantation, sorted GFP+ CD19+ cells were injected into sublethally irradiated mice. These cells became leukemic in the recipient mice, as indicated by domination of GFP+ CD19+ cells in the bone marrow (Figure 5G) and induction of splenomegaly (Figure 5H). Thus, miR-125b can initiate tumorigenesis at or earlier than the pre-B-cell stage of development. Potentially, miR-125b can directly transform pre-B cells, or miR-125b can initiate tumorigenesis at earlier stages of cell development with complete transformation occurring at the B-cell progenitor stage. Nevertheless, our experiment indicates that B-cell maturation is not required for miR-125b to induce lymphomagenesis.
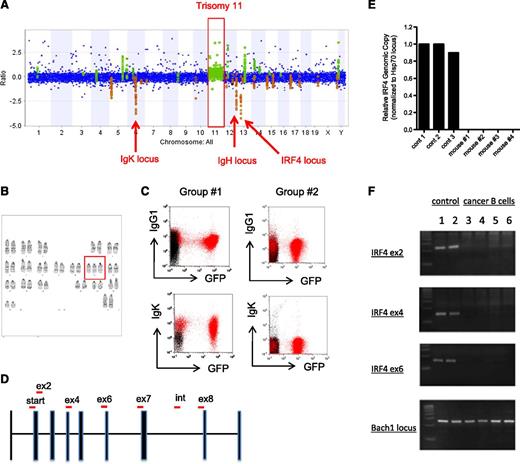
In our experiments, miR-125b ectopic overexpression initially repressed B-cell genesis (Figure 5A) but some cells of this lineage became cancerous later (Figure 5B-F), leading to the possibility that induction of B-cell leukemia may result from the accumulation of secondary genetic mutations involved in cellular transformation. To test this possibility, we performed comparative genomic hybridization microarray experiments (aCGH) of miR-125b B cancer cells to identify genomic alterations that might have occurred. In two independent experiments, we found that the B cancer cells induced by overexpression of miR-125b had an extra copy of chromosome 11 (Figure 6A and supplemental Figure 6A). This characteristic was specific to miR-125b–induced B cancer cells and was not found in the myeloid cancer cells (supplemental Figure 6A). G-band karyotyping analysis confirmed that the miR-125b–induced B cancer cells indeed harbor trisomy 11 (Figure 6B). No obvious inversions or translocations were identified through karyotyping. Further, the aCGH experiments indicated that the IgK (chromosome 6) and IgH (chromosome 12) loci had suffered deletions in the cancerous B cells harvested from 2 separate groups of mice (Figure 6A and supplemental Figure 6B), suggesting that these loci had undergone VDJ rearrangement. Thus, we performed flow cytometric analyses to characterize these cancer B cells further and found that they were CD43+CD24+IgM−cKit− (data not shown) with 1 group of cells being IgK+IgG1+ (group 1) and another being predominantly IgK−IgG1− (Figure 6C), indicating the former group of cells have productively rearranged their immunoglobulin loci whereas the latter did so nonproductively.
Deletion of IRF4 in miR-125b cells. (A) Analysis of genetic mutations in cancer B cells through comparative genomic hybridization microarray (aCGH). Genomic DNA harvested from sorted miR-125b–overexpressing GFP+CD19+ cancer B cells was subjected to aCGH analysis. Genomic DNA from normal C57bl/6 mice of the same gender (female) was used as controls. The figure represents the relative amount of genetic content of the cancer sample vs the C57bl/6 control (plotted as ratio in y-axis). A genomic region was considered different between the samples if 3 consecutive probes exhibited different signal intensities in the microarray. Regions highlighted in green and orange indicate genomic areas in which the cancer cells have higher and lower DNA content, respectively. The other aCGH analysis is displayed in supplemental Figure 6A. (B) G-band karyotyping of miR-125b–induced cancer B cells. Trisomy 11 is highlighted in red box. (C) Flow cytometric analysis of cancer B cells. Sorted GFP+CD19+ cells isolated from independent groups of MG-125b mice were transplanted into recipient mice. Upon cancer development, the bone marrow of these mice (highlight in red) was analyzed by flow cytometry. The plot shows the samples within the CD19+ gated population. The sample overlaid in black represents total BMCs harvested from healthy control C57bl/6 mice. Group 1 and group 2 corresponds to the aCGH samples displayed in panel A and supplemental Figure 6A, respectively. (D) IRF4 locus and genotyping primer sequences. IRF4 locus is shown with exons represented as solid bars. “Start,” “ex,” and “int” signifies translation start site, exon, and intron, respectively. The red bars represent the PCR product amplified by the primers used for genotyping in panel E-F and supplemental Figure 6E. (E) Quantitative PCR analysis of IRF4 locus. The relative amount of genomic DNA from miR-125b–induced cancer B cells were quantified by quantitative PCR and normalized to control Hsp70 locus. The control samples are genomic DNA harvested from normal C57bl/6 mice. The PCR amplifies a region spanning the translational start site (denoted as “start” in panel C). (F) Genomic DNA from miR-125b–induced cancer B cells was subjected to genotyping analysis. Exon2 (ex2), exon4 (ex4), and exon6 (ex6) of IRF4 were assessed. Bach1 locus was used as positive control. Samples 3-4 and 5-6 correspond to miR-125b–induced cancer B cells harvested from mice originating from group 1 and group 2 described in Figure 5G, respectively. The agarose gel images of the PCRs are displayed.
Deletion of IRF4 in miR-125b cells. (A) Analysis of genetic mutations in cancer B cells through comparative genomic hybridization microarray (aCGH). Genomic DNA harvested from sorted miR-125b–overexpressing GFP+CD19+ cancer B cells was subjected to aCGH analysis. Genomic DNA from normal C57bl/6 mice of the same gender (female) was used as controls. The figure represents the relative amount of genetic content of the cancer sample vs the C57bl/6 control (plotted as ratio in y-axis). A genomic region was considered different between the samples if 3 consecutive probes exhibited different signal intensities in the microarray. Regions highlighted in green and orange indicate genomic areas in which the cancer cells have higher and lower DNA content, respectively. The other aCGH analysis is displayed in supplemental Figure 6A. (B) G-band karyotyping of miR-125b–induced cancer B cells. Trisomy 11 is highlighted in red box. (C) Flow cytometric analysis of cancer B cells. Sorted GFP+CD19+ cells isolated from independent groups of MG-125b mice were transplanted into recipient mice. Upon cancer development, the bone marrow of these mice (highlight in red) was analyzed by flow cytometry. The plot shows the samples within the CD19+ gated population. The sample overlaid in black represents total BMCs harvested from healthy control C57bl/6 mice. Group 1 and group 2 corresponds to the aCGH samples displayed in panel A and supplemental Figure 6A, respectively. (D) IRF4 locus and genotyping primer sequences. IRF4 locus is shown with exons represented as solid bars. “Start,” “ex,” and “int” signifies translation start site, exon, and intron, respectively. The red bars represent the PCR product amplified by the primers used for genotyping in panel E-F and supplemental Figure 6E. (E) Quantitative PCR analysis of IRF4 locus. The relative amount of genomic DNA from miR-125b–induced cancer B cells were quantified by quantitative PCR and normalized to control Hsp70 locus. The control samples are genomic DNA harvested from normal C57bl/6 mice. The PCR amplifies a region spanning the translational start site (denoted as “start” in panel C). (F) Genomic DNA from miR-125b–induced cancer B cells was subjected to genotyping analysis. Exon2 (ex2), exon4 (ex4), and exon6 (ex6) of IRF4 were assessed. Bach1 locus was used as positive control. Samples 3-4 and 5-6 correspond to miR-125b–induced cancer B cells harvested from mice originating from group 1 and group 2 described in Figure 5G, respectively. The agarose gel images of the PCRs are displayed.
Strikingly, in both of our independent aCGH experiments, we found that cancer B cells induced by miR-125b overexpression harbored genetic deficiency at the tumor suppressor IRF4 locus (Figure 6A and supplemental Figure 6A). The IRF4 genetic deletion was specific to the miR-125b–induced B cancer cells and was not observed in miR-125b–induced myeloid cancer cells (data not shown). Quantitative PCR analysis confirmed that the translational start site of both copies of IRF4 was deleted in the B-cell cancer samples (Figure 6D-E), and genotyping analyses revealed that almost the entire IRF4 coding region (exon2-exon7, amino acids 1-366 of 450 amino acids full-length protein) was deleted (Figure 6D-F and supplemental Figure 6C). Interestingly, the population of cancer B cells was found to be heterogeneous with some expressing CD47 whereas others did not (supplemental Figure 6D); also, individual cancer B-cell samples exhibited different IRF4 genetic breaks (supplemental Figure 6E), indicating that although there were different cancer clones, IRF4 deletion is ubiquitous. The strong selective pressure for genetically eliminating both copies of IRF4 suggests a functional role of IRF4 in miR-125b–induced B-cell cancer. Indeed, multiple studies have recently demonstrated that deletion of IRF4 in mice leads to the development of B-cell leukemia.12-14,26-29
Discussion
We set out to determine the mechanisms by which miR-125b induces development of leukemia. Relating to myeloid leukemia, we found that: (1) myeloid cancer cells induced by miR-125b exhibit relatively few mutational alterations; (2) miR-125b is able to promote tumorigenesis in vitro in a cell intrinsic manner; (3) miR-125b causes myeloproliferation by enhancing output of MPs from HSPC as well as promoting hyperproliferation of MPs; (4) constitutive overexpression of miR-125b promotes self-renewal of MPs in vitro and induces the transformed state in these cells; (5) miR-125b represses IRF4 expression to induce myeloid leukemia. In relation to miR-125b–induced B-cell leukemia we found that: (1) overexpression of miR-125b initially represses B-cell development but constitutive long-term upregulated expression provokes a highly aggressive serially transplantable B-cell leukemia; (2) miR-125b can promote B-cell leukemia by inducing tumorigenesis in cells at or earlier than the pre-B-cell stage; (3) The miR-125b–induced B cancer cells are capable of immunoglobulin recombination; and (4) leukemic B cells induced by miR-125b acquire common genetic deletion of the tumor suppressor IRF4.
Our deep-sequencing analysis indicates that miR-125b–driven myeloid cancer cells acquire relatively few nucleotide mutations in their exomes, suggesting that the majority of these cells have not acquired permanent common genetic mutations. Thus, we speculate that reducing miR-125b expression might produce a regression of the myeloid tumors. In support of this notion, it was recently shown that survival of myeloid leukemic cells induced by miR-125a requires continual overexpression of this microRNA and removal of miR-125a after cancer development was accompanied by regression of the leukemia.30 Furthermore, we established that miR-125b functions within both HSPCs and MPs to drive a myeloproliferative disorder in vitro. Indeed, transplanting miR-125b–overexpressing MPs could induce leukemia in recipient mice. These data along with the fact that miR-125b–overexpressing MPs could grow indefinitely and also retain their ability to generate the entire myeloid lineage from a single cell suggests that miR-125b induces an abnormal self-renewal program in MPs in vitro.
Our data suggests that in both cases miR-125b induces myeloid and B-cell leukemia by modulating IRF4 but through distinct mechanisms. Whereas miR-125b induces tumorigenesis in myeloid cells by repressing the expression of IRF4 at the mRNA and protein levels without an affect on its genomic content, it promotes oncogenesis in B cells by a process involving genetic deletion of IRF4. Thus, miR-125b represents a novel paradigm by which an oncomir might induce cancer development in multiple cell lineages by modulating the same signaling pathway but involving distinct mechanisms. In one case, the oncomir works directly to yield tumor cells; in the other, it blocks differentiation but a presumably spontaneous deletion then appears to induce the oncogenic state. It is likely that the miR-125b–induced block of B-cell differentiation is then overcome by the effect of the IRF4 deletion, leading to uncontrolled growth of the cells. It has been shown that IRF4−/− but not IRF4+/− mice develop B-cell cancer,14 indicating that complete ablation of IRF4 is sufficient but reduced expression of IRF4 is insufficient to drive tumorigenesis in B cells. Thus, we speculate that although miR-125b overexpression represses IRF4 expression at the mRNA level, the magnitude is not sufficient to promote tumorigenesis in B cells and thus require selection of cells that acquire genetic deletion of IRF4. In addition to IRF4 deletion, other genes may play a role in this process as indicated by the occurrence of trisomy 11.
It has also been shown that human patients with B-cell leukemia have elevated expression of miR-125b.1,2,31 There are also reports of patients that acquire genetic mutations at the IRF4 locus.32-35 The increased expression of miR-125b could be caused by a chromosomal translocation that joins the miR-125b locus to, for instance, the immunoglobulin heavy chain gene regulatory elements1,6,31 or it could be epigenetic. Because of our finding that overexpression of miR-125b associates with genetic aberration at the IRF4 gene, we speculate that miR-125b upregulation in human cancer patients might result in a selection of IRF4 genetic mutations and consequently induce lymphoblastic leukemia. In support of this idea, it has already been established that IRF4 deletion in mice leads to the development of B-cell leukemia, and overexpression of IRF4 inhibits the development of B-cell leukemia in mice using cancer models induced by c-Myc and BCR/ABL.26,28 Of note, our finding that the IRF4 tumor suppressor gene is deleted in miR-125b–induced B cancer cells suggests that persistent upregulation of miR-125b might no longer be required for maintaining oncogenicity of these cancer cells. Although additional experiments will be required to test this point, we speculate that inhibiting miR-125b will not be a suitable therapy for treating B-cell leukemia invoked by this microRNA because permanent genetic lesions have occurred.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all members of the Baltimore laboratory and funding sources.
This work was supported by National Institutes of Health (National Cancer Institute) 1F32 CA139883-01A1 and (National Heart, Lung, and Blood Institute) K99HL118754 (A.Y.-L.S.), the Paul and Daisy Soros Fellowship and the National Science Foundation Graduate Research Fellowship (A.A.C.), and National Institutes of Health (National Institute of Allergy and Infectious Diseases) 1RO1AI079243 and 1R01AI093531 (D.B.).
Authorship
Contribution: A.Y.-L.S., A.A.C., R.S., A.M., D.C., C.X., J.T.K., E.L.L., Y.G.F., S.J., C.K., and P.R. performed the experiments and analyzed the data; A.Y.-L.S. and D.B. conceived the study and wrote the manuscript; A.Y.-L.S., A.A.C., R.S., A.M., D.C., C.X., J.T.K., E.L.L., and S.J. provided crucial reagents; and all authors provided input for the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: David Baltimore, Department of Biology, California Institute of Technology, Pasadena, CA 91125; e-mail: baltimo@caltech.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal