Key Points
ALK-negative ALCLs have chromosomal rearrangements of DUSP22 or TP63 in 30% and 8% of cases, respectively.
DUSP22-rearranged cases have favorable outcomes similar to ALK-positive ALCLs, whereas other genetic subtypes have inferior outcomes.
Abstract
Anaplastic lymphoma kinase (ALK)-negative anaplastic large cell lymphoma (ALCL) is a CD30-positive T-cell non-Hodgkin lymphoma that morphologically resembles ALK-positive ALCL but lacks chromosomal rearrangements of the ALK gene. The genetic and clinical heterogeneity of ALK-negative ALCL has not been delineated. We performed immunohistochemistry and fluorescence in situ hybridization on 73 ALK-negative ALCLs and 32 ALK-positive ALCLs and evaluated the associations among pathology, genetics, and clinical outcome. Chromosomal rearrangements of DUSP22 and TP63 were identified in 30% and 8% of ALK-negative ALCLs, respectively. These rearrangements were mutually exclusive and were absent in ALK-positive ALCLs. Five-year overall survival rates were 85% for ALK-positive ALCLs, 90% for DUSP22-rearranged ALCLs, 17% for TP63-rearranged ALCLs, and 42% for cases lacking all 3 genetic markers (P < .0001). Hazard ratios for death in these 4 groups after adjusting for International Prognostic Index and age were 1.0 (reference group), 0.58, 8.63, and 4.16, respectively (P = 7.10 × 10−5). These results were similar when restricted to patients receiving anthracycline-based chemotherapy, as well as to patients not receiving stem cell transplantation. Thus, ALK-negative ALCL is a genetically heterogeneous disease with widely disparate outcomes following standard therapy. DUSP22 and TP63 rearrangements may serve as predictive biomarkers to help guide patient management.
Introduction
Anaplastic large cell lymphomas (ALCLs) are malignancies of mature T lymphocytes that express the lymphocyte activation marker CD30.1 About one-half express anaplastic lymphoma kinase (ALK) fusion proteins resulting from chromosomal rearrangements involving the ALK gene on 2p23, and the pathogenic role of these fusion proteins has been studied extensively.2 In contrast, ALCLs lacking ALK rearrangements are poorly understood, with ALK-negative ALCL accepted as a provisional entity in the World Health Organization (WHO) classification system only in 2008.3 Criteria to distinguish ALK-negative ALCLs from other CD30-positive peripheral T-cell lymphomas (PTCLs) remain imprecise, a difficulty compounded by lack of genetic biomarkers for ALK-negative ALCL. Finally, although patients with ALK-positive ALCLs typically have 5-year overall survival (OS) rates of ≥70%, patients with ALK-negative ALCL have 5-year OS rates consistently <50%.4-7
To better understand ALK-negative ALCL, our group used next-generation sequencing and complementary approaches to study the genetics of this disease. We identified 2 recurrent rearrangements seen almost exclusively in ALK-negative ALCL.8,9 One involved rearrangement of the TP53 homolog, TP63, on 3q28. These rearrangements encoded p63 fusion proteins with structural homology to oncogenic ΔNp63 and were associated with poor outcomes.9 The other rearrangement involved the DUSP22-IRF4 locus on 6p25.3 (DUSP22 rearrangement), which was associated with markedly decreased expression of DUSP22.8 The prognosis of patients with DUSP22-rearranged systemic ALCL is unknown. Because there is a critical gap in the understanding of ALK-negative ALCL in general, we undertook the present multi-institution study to determine whether combined biomarker testing for TP63 and DUSP22 rearrangements would facilitate diagnosis and risk stratification of this disease.
Materials and methods
Patient samples and clinical data
Studies were approved by the respective Institutional Review Boards at the participating institutions. Studies were conducted in accordance with the Declaration of Helsinki. Patients were identified based on diagnosis of systemic ALCL or other PTCL expressing CD30 in ≥80% of cells. Clinical data included age, gender, anatomic site, skin involvement, International Prognostic Index (IPI) score, treatment, time to progression, date/status at last follow-up, and immunophenotype (CD2, CD3, CD4, CD5, CD7, CD8, CD30, p63, T-cell-restricted intracellular antigen [TIA]-1, granzyme B, epithelial membrane antigen [EMA], clusterin, ALK, T-cell receptor βF1, and T-cell receptor γ/δ). The threshold for immunopositivity was ≥20% of tumor cells (CD30 and p63, ≥80%). When immunophenotype was unknown, immunohistochemistry was performed using previously published methods and antibodies.10,11 A subset of samples also was stained for perforin. Fluorescence in situ hybridization (FISH) was performed using breakapart probes for the DUSP22-IRF4 and TP63 loci and a dual-fusion probe for TBL1XR1/TP63 fusion [inv(3)(q26q28)], as previously described.8,9
Blinded, 2-tier pathology review
Cases were evaluated by a panel of 3 expert hematopathologists (ESJ, JWS, and SHS). Cases first were assessed by each panel member independently using hematoxylin and eosin and CD30 stains only, without knowledge of the clinical, immunophenotypic, or molecular/genetic data (tier 1 review). Cases were scored for the presence or absence of a classic ALCL pattern, based on a composite assessment of presence of hallmark and/or multinucleated/wreath-like cells; sheet-like growth pattern; sinusoidal involvement; and homogenous CD30 staining with a predominantly membranous and Golgi-zone distribution.1,3,12 Cases then were rereviewed with knowledge of immunophenotypic findings and clinical presentation but without molecular/genetic data and classified by WHO criteria (tier 2 review). Cases in which a diagnosis of ALCL was supported by ≥2 of the 3 reviewers on tier 2 review were considered for further analysis.
Statistics and survival analysis
OS was defined as the time from diagnosis to death due to any cause or date of last known follow-up in patients still alive. Progression-free survival (PFS) was defined as the time from diagnosis to the date of progression, initiation of second-line treatment, or death due to any cause. Patients in remission after initial therapy were censored at date of last known disease status. Association of genetic subgroups with OS and PFS were assessed using Kaplan-Meier curves and Cox proportional hazards models. Differences in patient characteristics, tumor phenotype, and other clinical factors between genetic subgroups were assessed using χ2 tests and Wilcoxon rank-sum tests as appropriate.
Results
Patients and panel diagnoses
Of 128 cases evaluated, 105 were eligible for further analysis (73 ALK-negative ALCLs and 32 ALK-positive ALCLs). Demographic and clinical characteristics are shown in Table 1. Year of diagnosis ranged from 1982 to 2012 (median, 2002; interquartile range, 1996-2008). Median follow-up time from diagnosis for patients still alive was 78 months (range, 0.3-354 months). The remaining cases were excluded because they were not diagnosed as ALCL (n = 10); a primary cutaneous T-cell neoplasm could not be completely ruled out from the clinical history (n = 5); or the genetic status could not be determined by FISH (n = 8). Of the 10 cases not classified as ALCL, there were 5 PTCLs, not otherwise specified (NOS); 2 enteropathy-associated T-cell lymphomas; and 3 cases where a majority consensus was not reached.
Clinical characteristics of 105 patients with ALCL
| Characteristic . | ALCL stratified by ALK status, N (%) . | ALK-negative ALCL stratified by genetics, N (%) . | All ALCL, N (%) . | |||
|---|---|---|---|---|---|---|
| ALK positive (N = 32) . | ALK negative (N = 73) . | DUSP22 (N = 22) . | TP63 (N = 6) . | −/−/− (N = 45) . | Total (N = 105) . | |
| Age | ||||||
| Median | 27.5 | 58 | 53.5 | 48 | 62 | 51 |
| Range | 6-77 | 22-94 | 36-76 | 30-73 | 22-94 | 6-94 |
| Gender | ||||||
| Female | 11 (34) | 24 (33) | 7 (32) | 3 (50) | 14 (31) | 35 (33) |
| Male | 21 (66) | 49 (67) | 15 (68) | 3 (50) | 31 (69) | 70 (67) |
| Ann Arbor stage | ||||||
| I | 7 (26) | 4 (11) | 0 (0) | 0 (0) | 4 (17) | 11 (17) |
| II | 6 (22) | 5 (14) | 1 (14) | 1 (20) | 3 (12) | 11 (17) |
| III | 0 (0) | 7 (19) | 2 (28) | 1 (20) | 4 (17) | 7 (11) |
| IV | 14 (52) | 20 (56) | 4 (57) | 3 (60) | 13 (54) | 34 (54) |
| Missing | 5 | 37 | 15 | 1 | 21 | 42 |
| IPI | ||||||
| 0-1 | 20 (69) | 21 (40) | 6 (43) | 2 (33) | 13 (41) | 41 (50) |
| 2 | 6 (21) | 15 (29) | 2 (14) | 3 (50) | 10 (31) | 21 (26) |
| 3 | 3 (10) | 9 (17) | 3 (21) | 1 (17) | 5 (16) | 12 (15) |
| 4-5 | 0 (0) | 7 (14) | 3 (21) | 0 (0) | 4 (12) | 7 (9) |
| Missing | 3 | 21 | 8 | 0 | 13 | 24 |
| Extranodal involvement at diagnosis | ||||||
| Yes | 18 (56) | 30 (44) | 7 (33) | 2 (33) | 21 (49) | 48 (48) |
| No | 14 (44) | 38 (56) | 14 (67) | 4 (67) | 20 (51) | 52 (52) |
| Missing | 0 | 5 | 1 | 0 | 4 | 5 |
| Skin involvement during disease course | ||||||
| Yes | 4 (13) | 14 (22) | 5 (28) | 2 (33) | 7 (17) | 18 (19) |
| No | 26 (87) | 50 (78) | 13 (72) | 4 (67) | 33 (83) | 76 (81) |
| Missing | 2 | 9 | 4 | 0 | 5 | 11 |
| Initial treatment | ||||||
| CHOP/CHOP-like | 22 (76) | 51 (78) | 18 (90) | 5 (83) | 28 (72) | 73 (78) |
| RT only | 1 (3) | 5 (8) | 0 (0) | 0 (0) | 5 (13) | 6 (6) |
| Other | 6 (21) | 5 (8) | 1 (5) | 0 (0) | 4 (10) | 11 (12) |
| None | 0 (0) | 4 (6) | 1 (5) | 1 (17) | 2 (5) | 4 (4) |
| Missing/unknown | 3 | 8 | 2 | 0 | 6 | 11 |
| Stem cell transplant | ||||||
| No | 22 (79) | 55 (86) | 15 (79) | 5 (83) | 35 (90) | 77 (84) |
| Yes | 6 (21) | 9 (14) | 4 (21) | 1 (17) | 4 (10) | 15 (16) |
| Consolidative | 1 (4) | 2 (3) | 1 (5) | 0 (0) | 1 (3) | 3 (3) |
| Salvage | 4 (14) | 4 (6) | 2 (11) | 1 (17) | 1 (3) | 8 (9) |
| Unknown timing | 1 (4) | 3 (5) | 1 (5) | 0 (0) | 2 (5) | 4 (4) |
| Missing | 4 | 9 | 3 | 0 | 6 | 13 |
| Characteristic . | ALCL stratified by ALK status, N (%) . | ALK-negative ALCL stratified by genetics, N (%) . | All ALCL, N (%) . | |||
|---|---|---|---|---|---|---|
| ALK positive (N = 32) . | ALK negative (N = 73) . | DUSP22 (N = 22) . | TP63 (N = 6) . | −/−/− (N = 45) . | Total (N = 105) . | |
| Age | ||||||
| Median | 27.5 | 58 | 53.5 | 48 | 62 | 51 |
| Range | 6-77 | 22-94 | 36-76 | 30-73 | 22-94 | 6-94 |
| Gender | ||||||
| Female | 11 (34) | 24 (33) | 7 (32) | 3 (50) | 14 (31) | 35 (33) |
| Male | 21 (66) | 49 (67) | 15 (68) | 3 (50) | 31 (69) | 70 (67) |
| Ann Arbor stage | ||||||
| I | 7 (26) | 4 (11) | 0 (0) | 0 (0) | 4 (17) | 11 (17) |
| II | 6 (22) | 5 (14) | 1 (14) | 1 (20) | 3 (12) | 11 (17) |
| III | 0 (0) | 7 (19) | 2 (28) | 1 (20) | 4 (17) | 7 (11) |
| IV | 14 (52) | 20 (56) | 4 (57) | 3 (60) | 13 (54) | 34 (54) |
| Missing | 5 | 37 | 15 | 1 | 21 | 42 |
| IPI | ||||||
| 0-1 | 20 (69) | 21 (40) | 6 (43) | 2 (33) | 13 (41) | 41 (50) |
| 2 | 6 (21) | 15 (29) | 2 (14) | 3 (50) | 10 (31) | 21 (26) |
| 3 | 3 (10) | 9 (17) | 3 (21) | 1 (17) | 5 (16) | 12 (15) |
| 4-5 | 0 (0) | 7 (14) | 3 (21) | 0 (0) | 4 (12) | 7 (9) |
| Missing | 3 | 21 | 8 | 0 | 13 | 24 |
| Extranodal involvement at diagnosis | ||||||
| Yes | 18 (56) | 30 (44) | 7 (33) | 2 (33) | 21 (49) | 48 (48) |
| No | 14 (44) | 38 (56) | 14 (67) | 4 (67) | 20 (51) | 52 (52) |
| Missing | 0 | 5 | 1 | 0 | 4 | 5 |
| Skin involvement during disease course | ||||||
| Yes | 4 (13) | 14 (22) | 5 (28) | 2 (33) | 7 (17) | 18 (19) |
| No | 26 (87) | 50 (78) | 13 (72) | 4 (67) | 33 (83) | 76 (81) |
| Missing | 2 | 9 | 4 | 0 | 5 | 11 |
| Initial treatment | ||||||
| CHOP/CHOP-like | 22 (76) | 51 (78) | 18 (90) | 5 (83) | 28 (72) | 73 (78) |
| RT only | 1 (3) | 5 (8) | 0 (0) | 0 (0) | 5 (13) | 6 (6) |
| Other | 6 (21) | 5 (8) | 1 (5) | 0 (0) | 4 (10) | 11 (12) |
| None | 0 (0) | 4 (6) | 1 (5) | 1 (17) | 2 (5) | 4 (4) |
| Missing/unknown | 3 | 8 | 2 | 0 | 6 | 11 |
| Stem cell transplant | ||||||
| No | 22 (79) | 55 (86) | 15 (79) | 5 (83) | 35 (90) | 77 (84) |
| Yes | 6 (21) | 9 (14) | 4 (21) | 1 (17) | 4 (10) | 15 (16) |
| Consolidative | 1 (4) | 2 (3) | 1 (5) | 0 (0) | 1 (3) | 3 (3) |
| Salvage | 4 (14) | 4 (6) | 2 (11) | 1 (17) | 1 (3) | 8 (9) |
| Unknown timing | 1 (4) | 3 (5) | 1 (5) | 0 (0) | 2 (5) | 4 (4) |
| Missing | 4 | 9 | 3 | 0 | 6 | 13 |
RT, radiotherapy; −/−/−, triple-negative ALCL.
Genetics of ALK-negative ALCL
Of the 73 ALK-negative ALCLs, 22 (30%) had DUSP22 rearrangements and 6 (8%) had TP63 rearrangements; these 2 events were mutually exclusive and were not seen in any ALK-positive ALCL. Forty-five cases lacked both rearrangements and ALK expression and are referred to as triple-negative ALCLs. Clinical data on patients with genetic subtypes of ALK-negative ALCL are shown in Table 1. Of the 10 cases excluded because they were not diagnosed as ALCL, no DUSP22 or TP63 rearrangement was detected in 8 cases, and the genetic status could not be determined in 2.
Outcomes of ALK-negative ALCL
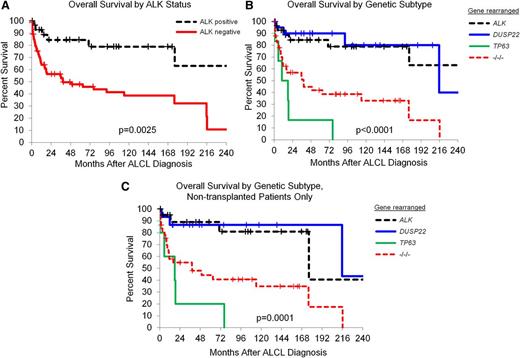
Consistent with published data, patients with ALK-negative ALCL had OS rates inferior to those with ALK-positive ALCL (5-year OS: 52%, 95% confidence interval [CI]: 41-67% vs 5-year OS: 85%, 95% CI: 72-100%, respectively; log rank, P = .0025; Figure 1A). OS rates among patients with ALK-negative ALCL varied widely when stratified by genetics (Figure 1B). Patients with DUSP22-rearranged ALCL had OS rates similar to those with ALK-positive ALCL (5-year OS: 90%, 95% CI: 78-100%; hazard ratio [HR] = 0.95, 95% CI: 0.27-3.36, P = .95; Table 2); OS was somewhat better after adjusting for age and IPI (HR = 0.58, 95% CI: 0.12-2.79, P = .50). Patients with TP63-rearranged ALCL had OS rates significantly worse than those with ALK-positive ALCL (5-year OS: 17%, 95% CI: 3-100%; HR = 11.01, 95% CI: 3.42-35.50, P < .0001; age- and IPI-adjusted HR = 8.63, 95% CI: 2.08-35.77, P = .003). Patients with triple-negative ALCL had intermediate OS rates similar to those with ALK-negative ALCL as a whole and significantly worse than those ALK-positive ALCL (5-year OS: 42%, 95% CI: 26-59%; HR = 5.20, 95% CI: 2.10-12.86, P = .0004; age- and IPI-adjusted HR = 4.16, 95% CI: 1.32-13.09, P = .015).
Outcomes in patients with ALCL based on genetic subtype. (A) OS rates in patients with ALCL, stratified by ALK status only (ALK positive, N = 29; ALK negative, N = 67). (B) OS rates in patients with ALCL, stratified by rearrangements of ALK (N = 29), DUSP22 (N = 21), and TP63 (N = 6). −/−/−, triple-negative cases lacking all 3 rearrangements (N = 40). (C) OS rates in patients with ALCL who did not undergo transplantation, stratified by rearrangements of ALK (N = 21), DUSP22 (N = 15), and TP63 (N = 5). −/−/−, N = 34.
Outcomes in patients with ALCL based on genetic subtype. (A) OS rates in patients with ALCL, stratified by ALK status only (ALK positive, N = 29; ALK negative, N = 67). (B) OS rates in patients with ALCL, stratified by rearrangements of ALK (N = 29), DUSP22 (N = 21), and TP63 (N = 6). −/−/−, triple-negative cases lacking all 3 rearrangements (N = 40). (C) OS rates in patients with ALCL who did not undergo transplantation, stratified by rearrangements of ALK (N = 21), DUSP22 (N = 15), and TP63 (N = 5). −/−/−, N = 34.
Cox regression model of OS and PFS survival based on ALCL genetics
| Analysis . | Gene rearranged . | N* . | No. of events . | Unadjusted . | Adjusted for age and IPI . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HR . | 95% CI . | P value . | Genetic P value . | HR . | 95% CI . | P value . | Genetic P value . | ||||
| All patients, OS | ALK | 29 | 6 | Reference | 1.09E-05 | Reference | 7.10E-05 | ||||
| DUSP22 | 21 | 4 | 0.95 | 0.27-3.36 | .9500 | 0.58 | 0.12-2.79 | .5000 | |||
| TP63 | 6 | 6 | 11.01 | 3.42-35.50 | <.0001 | 8.63 | 2.08-35.77 | .0030 | |||
| −/−/− | 40 | 26 | 5.20 | 2.10-12.86 | .0004 | 4.16 | 1.32-13.09 | .0150 | |||
| All patients, PFS | ALK | 27 | 11 | Reference | 0.0020 | Reference | .01390 | ||||
| DUSP22 | 10 | 7 | 1.38 | 0.51-3.74 | .5300 | 0.76 | 0.23-2.44 | .6400 | |||
| TP63 | 6 | 6 | 5.02 | 1.82-13.82 | .0018 | 3.48 | 1.13-10.69 | .0300 | |||
| −/−/− | 30 | 27 | 3.01 | 1.48-6.14 | .0025 | 2.32 | 1.05-5.13 | .0380 | |||
| Anthracycline-treated, OS | ALK | 21 | 4 | Reference | 1.02E-05 | Reference | .00026 | ||||
| DUSP22 | 18 | 2 | 0.52 | 0.10-2.86 | .4500 | 0.41 | 0.07-2.55 | .3400 | |||
| TP63 | 5 | 5 | 11.17 | 2.79-44.77 | .0007 | 9.53 | 2.04-44.49 | .0041 | |||
| −/−/− | 27 | 17 | 5.51 | 1.74-17.43 | .0037 | 4.19 | 1.20-14.64 | .0250 | |||
| Nontransplanted, OS | ALK | 21 | 4 | Reference | 4.96E-05 | Reference | .00065 | ||||
| DUSP22 | 15 | 3 | 0.95 | 0.21-4.27 | .9500 | 0.51 | 0.08-3.14 | .4700 | |||
| TP63 | 5 | 5 | 9.90 | 2.61-37.58 | .0008 | 7.72 | 1.72-34.53 | .0075 | |||
| −/−/− | 34 | 23 | 5.02 | 1.73-14.58 | .0030 | 3.27 | 1.00-10.70 | .0500 | |||
| Analysis . | Gene rearranged . | N* . | No. of events . | Unadjusted . | Adjusted for age and IPI . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HR . | 95% CI . | P value . | Genetic P value . | HR . | 95% CI . | P value . | Genetic P value . | ||||
| All patients, OS | ALK | 29 | 6 | Reference | 1.09E-05 | Reference | 7.10E-05 | ||||
| DUSP22 | 21 | 4 | 0.95 | 0.27-3.36 | .9500 | 0.58 | 0.12-2.79 | .5000 | |||
| TP63 | 6 | 6 | 11.01 | 3.42-35.50 | <.0001 | 8.63 | 2.08-35.77 | .0030 | |||
| −/−/− | 40 | 26 | 5.20 | 2.10-12.86 | .0004 | 4.16 | 1.32-13.09 | .0150 | |||
| All patients, PFS | ALK | 27 | 11 | Reference | 0.0020 | Reference | .01390 | ||||
| DUSP22 | 10 | 7 | 1.38 | 0.51-3.74 | .5300 | 0.76 | 0.23-2.44 | .6400 | |||
| TP63 | 6 | 6 | 5.02 | 1.82-13.82 | .0018 | 3.48 | 1.13-10.69 | .0300 | |||
| −/−/− | 30 | 27 | 3.01 | 1.48-6.14 | .0025 | 2.32 | 1.05-5.13 | .0380 | |||
| Anthracycline-treated, OS | ALK | 21 | 4 | Reference | 1.02E-05 | Reference | .00026 | ||||
| DUSP22 | 18 | 2 | 0.52 | 0.10-2.86 | .4500 | 0.41 | 0.07-2.55 | .3400 | |||
| TP63 | 5 | 5 | 11.17 | 2.79-44.77 | .0007 | 9.53 | 2.04-44.49 | .0041 | |||
| −/−/− | 27 | 17 | 5.51 | 1.74-17.43 | .0037 | 4.19 | 1.20-14.64 | .0250 | |||
| Nontransplanted, OS | ALK | 21 | 4 | Reference | 4.96E-05 | Reference | .00065 | ||||
| DUSP22 | 15 | 3 | 0.95 | 0.21-4.27 | .9500 | 0.51 | 0.08-3.14 | .4700 | |||
| TP63 | 5 | 5 | 9.90 | 2.61-37.58 | .0008 | 7.72 | 1.72-34.53 | .0075 | |||
| −/−/− | 34 | 23 | 5.02 | 1.73-14.58 | .0030 | 3.27 | 1.00-10.70 | .0500 | |||
−/−/−, triple-negative ALCL.
The numbers of patients with OS and PFS data are different from the total number of patients in Table 1 because some patients did not have follow-up data available (missing OS data: 3 ALK, 1 DUSP22, 5 −/−/−; missing PFS data: 5 ALK, 12 DUSP22, 15 −/−/−).
In a sensitivity analysis, differences between outcomes in genetic groups were similar when restricted to patients receiving anthracycline-based chemotherapy and patients not receiving stem cell transplantation (SCT) (Table 2). Notably, the favorable outcome of DUSP22-rearranged ALCL could not be attributed to the use of SCT in these patients, as similar data were obtained when only nontransplanted cases were considered (5-year OS: 87%, 95% CI: 71-100%; Figure 1C).
Pathology of ALK-negative ALCL
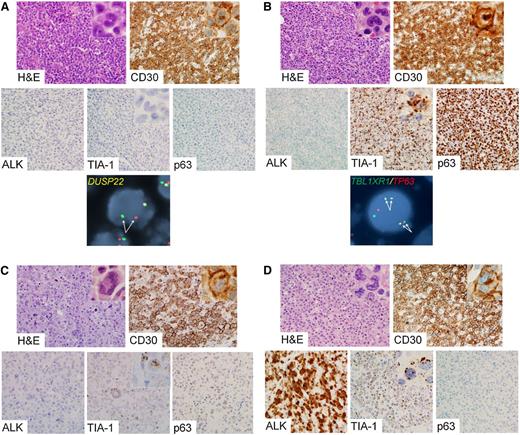
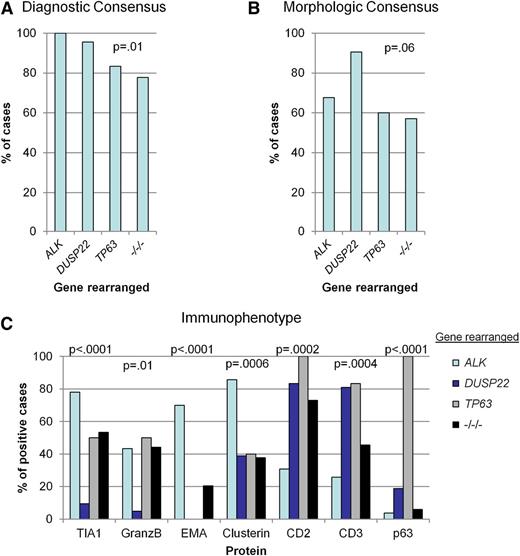
Representative examples of genetic subtypes of ALCL are shown in Figure 2. Although the diagnosis of ALCL required consensus by ≥2 of the 3 expert reviewers, the degree to which this consensus was unanimous (3 of 3 reviewers) varied by genetic subtype. As expected, all ALK-positive ALCLs were diagnosed unanimously. Among ALK-negative ALCLs, there was unanimous consensus in 96% of DUSP22-rearranged cases, 83% of TP63-rearranged cases, and 78% of triple-negative cases (P = .01; Figure 3A; Table 3).
Representative cases of genetic subtypes of ALCL. (A) ALK-negative ALCL with DUSP22 rearrangement. The tumor cells are positive for CD30 and are negative for ALK, TIA-1, and p63. FISH using a breakapart probe to the DUSP22-IRF4 locus on 6p25.3 shows abnormal separation of the red and green signals on 1 allele (pair of arrows). (B) ALK-negative ALCL with TP63 rearrangement. The tumor cells are positive for CD30, TIA-1, and p63 and are negative for ALK. FISH using a dual-fusion probe to TBL1XR1 on 3q26 and TP63 on 3q28 demonstrates 2 pairs of abnormal fusion signals, indicating 2 copies of inv(3)(q26q28) (TBL1XR1/TP63 fusion; 2 pairs of arrows). (C) ALK-negative ALCL lacking DUSP22 and TP63 rearrangements (triple-negative ALCL). The tumor cells are positive for CD30 and TIA-1 and are negative for ALK and p63. (D) ALK-positive ALCL. The tumor cells are positive for CD30, ALK, and TIA-1 and are negative for p63. Photomicrographs were taken using an Olympus DP71 camera, Olympus BX51 microscope, and Olympus DP Manager image acquisition software at an original magnification of ×400 (insets, ×1000). Original magnification of FISH images, ×600.
Representative cases of genetic subtypes of ALCL. (A) ALK-negative ALCL with DUSP22 rearrangement. The tumor cells are positive for CD30 and are negative for ALK, TIA-1, and p63. FISH using a breakapart probe to the DUSP22-IRF4 locus on 6p25.3 shows abnormal separation of the red and green signals on 1 allele (pair of arrows). (B) ALK-negative ALCL with TP63 rearrangement. The tumor cells are positive for CD30, TIA-1, and p63 and are negative for ALK. FISH using a dual-fusion probe to TBL1XR1 on 3q26 and TP63 on 3q28 demonstrates 2 pairs of abnormal fusion signals, indicating 2 copies of inv(3)(q26q28) (TBL1XR1/TP63 fusion; 2 pairs of arrows). (C) ALK-negative ALCL lacking DUSP22 and TP63 rearrangements (triple-negative ALCL). The tumor cells are positive for CD30 and TIA-1 and are negative for ALK and p63. (D) ALK-positive ALCL. The tumor cells are positive for CD30, ALK, and TIA-1 and are negative for p63. Photomicrographs were taken using an Olympus DP71 camera, Olympus BX51 microscope, and Olympus DP Manager image acquisition software at an original magnification of ×400 (insets, ×1000). Original magnification of FISH images, ×600.
Comparative pathologic features of genetic subtypes of ALCL. (A) Percentage of cases with unanimous consensus on a diagnosis of ALCL, based on morphology, phenotype, and clinical data. (B) Percentage of cases with unanimous consensus on the presence of classic histologic features of ALCL, as assessed without knowledge of phenotype (except CD30 expression) or clinical data. (C) Immunophenotypic markers that differed significantly among genetic subtypes. As a defining feature of ALK-positive ALCL, immunohistochemistry for ALK is not included here.
Comparative pathologic features of genetic subtypes of ALCL. (A) Percentage of cases with unanimous consensus on a diagnosis of ALCL, based on morphology, phenotype, and clinical data. (B) Percentage of cases with unanimous consensus on the presence of classic histologic features of ALCL, as assessed without knowledge of phenotype (except CD30 expression) or clinical data. (C) Immunophenotypic markers that differed significantly among genetic subtypes. As a defining feature of ALK-positive ALCL, immunohistochemistry for ALK is not included here.
Pathologic data stratified by ALCL genetics
| Variable . | Gene rearranged, N (%) . | Total (N = 105) . | P value* . | |||
|---|---|---|---|---|---|---|
| ALK (N = 32) . | DUSP22 (N = 22) . | TP63 (N = 6) . | −/−/− (N = 45) . | |||
| Diagnosis agreement | .0148 | |||||
| 2 reviewers | 0 (0) | 1 (5) | 1 (17) | 10 (22) | 12 (11) | |
| 3 reviewers | 32 (100) | 21 (95) | 5 (83) | 35 (78) | 93 (89) | |
| Classic histology | .06421 | |||||
| <3 reviewers | 10 (32) | 2 (10) | 2 (40) | 18 (43) | 32 (32) | |
| 3 reviewers | 21 (67) | 19 (90) | 3 (60) | 24 (57) | 67 (68) | |
| TIA1 | <.0001 | |||||
| Negative | 7 (22) | 19 (90) | 3 (50) | 21 (47) | 50 (48) | |
| Positive | 25 (78) | 2 (10) | 3 (50) | 24 (53) | 54 (52) | |
| Granzyme B | .0100 | |||||
| Negative | 17 (57) | 20 (95) | 3 (50) | 24 (56) | 64 (64) | |
| Positive | 13 (43) | 1 (5) | 3 (50) | 19 (44) | 36 (36) | |
| EMA | <.0001 | |||||
| Negative | 9 (30) | 20 (100) | 5 (100) | 31 (80) | 65 (69) | |
| Positive | 21 (70) | 0 (0) | 0 (0) | 8 (20) | 29 (31) | |
| Clusterin | .0006 | |||||
| Negative | 4 (14) | 11 (61) | 3 (60) | 23 (62) | 41 (47) | |
| Positive | 24 (86) | 7 (39) | 2 (40) | 14 (38) | 47 (53) | |
| TCR βF1 | .1272 | |||||
| Negative | 12 (75) | 7 (50) | 1 (20) | 20 (65) | 40 (61) | |
| Positive | 4 (25) | 7 (50) | 4 (80) | 11 (35) | 26 (39) | |
| TCR γ-δ | .7963 | |||||
| Negative | 13 (100) | 9 (100) | 3 (100) | 24 (96) | 49 (98) | |
| Positive | 0 (0) | 0 (0) | 0 (0) | 1 (4) | 1 (2) | |
| CD2 | .0002 | |||||
| Negative | 18 (69) | 3 (17) | 0 (0) | 10 (27) | 31 (36) | |
| Positive | 8 (31) | 15 (83) | 5 (100) | 27 (73) | 55 (64) | |
| CD3 | .0004 | |||||
| Negative | 23 (74) | 4 (19) | 1 (17) | 24 (55) | 52 (51) | |
| Positive | 8 (26) | 17 (81) | 5 (83) | 20 (45) | 50 (49) | |
| CD4 | .9922 | |||||
| Negative | 10 (45) | 8 (47) | 2 (40) | 16 (47) | 36 (46) | |
| Positive | 12 (55) | 9 (53) | 3 (60) | 18 (53) | 42 (54) | |
| CD5 | .3770 | |||||
| Negative | 13 (65) | 15 (88) | 3 (60) | 26 (72) | 57 (73) | |
| Positive | 7 (35) | 2 (12) | 2 (40) | 10 (28) | 21 (27) | |
| CD8 | .4206 | |||||
| Negative | 20 (100) | 15 (88) | 5 (100) | 30 (94) | 70 (95) | |
| Positive | 0 (0) | 2 (12) | 0 (0) | 2 (6) | 4 (5) | |
| p63 | <.0001 | |||||
| Negative | 26 (96) | 13 (81) | 0 (0) | 32 (94) | 71 (85) | |
| Positive | 1 (4) | 3 (19) | 6 (100) | 2 (6) | 12 (15) | |
| Variable . | Gene rearranged, N (%) . | Total (N = 105) . | P value* . | |||
|---|---|---|---|---|---|---|
| ALK (N = 32) . | DUSP22 (N = 22) . | TP63 (N = 6) . | −/−/− (N = 45) . | |||
| Diagnosis agreement | .0148 | |||||
| 2 reviewers | 0 (0) | 1 (5) | 1 (17) | 10 (22) | 12 (11) | |
| 3 reviewers | 32 (100) | 21 (95) | 5 (83) | 35 (78) | 93 (89) | |
| Classic histology | .06421 | |||||
| <3 reviewers | 10 (32) | 2 (10) | 2 (40) | 18 (43) | 32 (32) | |
| 3 reviewers | 21 (67) | 19 (90) | 3 (60) | 24 (57) | 67 (68) | |
| TIA1 | <.0001 | |||||
| Negative | 7 (22) | 19 (90) | 3 (50) | 21 (47) | 50 (48) | |
| Positive | 25 (78) | 2 (10) | 3 (50) | 24 (53) | 54 (52) | |
| Granzyme B | .0100 | |||||
| Negative | 17 (57) | 20 (95) | 3 (50) | 24 (56) | 64 (64) | |
| Positive | 13 (43) | 1 (5) | 3 (50) | 19 (44) | 36 (36) | |
| EMA | <.0001 | |||||
| Negative | 9 (30) | 20 (100) | 5 (100) | 31 (80) | 65 (69) | |
| Positive | 21 (70) | 0 (0) | 0 (0) | 8 (20) | 29 (31) | |
| Clusterin | .0006 | |||||
| Negative | 4 (14) | 11 (61) | 3 (60) | 23 (62) | 41 (47) | |
| Positive | 24 (86) | 7 (39) | 2 (40) | 14 (38) | 47 (53) | |
| TCR βF1 | .1272 | |||||
| Negative | 12 (75) | 7 (50) | 1 (20) | 20 (65) | 40 (61) | |
| Positive | 4 (25) | 7 (50) | 4 (80) | 11 (35) | 26 (39) | |
| TCR γ-δ | .7963 | |||||
| Negative | 13 (100) | 9 (100) | 3 (100) | 24 (96) | 49 (98) | |
| Positive | 0 (0) | 0 (0) | 0 (0) | 1 (4) | 1 (2) | |
| CD2 | .0002 | |||||
| Negative | 18 (69) | 3 (17) | 0 (0) | 10 (27) | 31 (36) | |
| Positive | 8 (31) | 15 (83) | 5 (100) | 27 (73) | 55 (64) | |
| CD3 | .0004 | |||||
| Negative | 23 (74) | 4 (19) | 1 (17) | 24 (55) | 52 (51) | |
| Positive | 8 (26) | 17 (81) | 5 (83) | 20 (45) | 50 (49) | |
| CD4 | .9922 | |||||
| Negative | 10 (45) | 8 (47) | 2 (40) | 16 (47) | 36 (46) | |
| Positive | 12 (55) | 9 (53) | 3 (60) | 18 (53) | 42 (54) | |
| CD5 | .3770 | |||||
| Negative | 13 (65) | 15 (88) | 3 (60) | 26 (72) | 57 (73) | |
| Positive | 7 (35) | 2 (12) | 2 (40) | 10 (28) | 21 (27) | |
| CD8 | .4206 | |||||
| Negative | 20 (100) | 15 (88) | 5 (100) | 30 (94) | 70 (95) | |
| Positive | 0 (0) | 2 (12) | 0 (0) | 2 (6) | 4 (5) | |
| p63 | <.0001 | |||||
| Negative | 26 (96) | 13 (81) | 0 (0) | 32 (94) | 71 (85) | |
| Positive | 1 (4) | 3 (19) | 6 (100) | 2 (6) | 12 (15) | |
−/−/−, triple-negative ALCL.
χ2.
Although both morphology and immunophenotype are important in diagnosing ALK-negative ALCL, the relative contribution of each toward the diagnosis has not been established.3 To address this, reviewers first rendered a tier 1 opinion on whether each case had classic morphologic features of ALCL, without knowledge of the immunophenotype or ALK status. Unanimous consensus on the presence of classic morphology was observed more frequently in DUSP22-rearranged ALCLs (91%) than in TP63-rearranged ALCLs (60%), triple-negative ALCLs (57%), or ALK-positive ALCLs (68%; P = .06; Figure 3B; Table 3). In keeping with the high degree of consensus for DUSP22-rearranged ALCL, these cases generally showed sheets of hallmark cells as described for the common pattern of ALK-positive ALCL,1 although in some cases, the cells were slightly smaller than those seen in other genetic subtypes (Figure 2A). The cytologic features of TP63-rearranged ALCLs were somewhat more variable, but characteristic hallmark cells could always be identified with careful observation (Figure 2B). Large, pleomorphic, and/or multinucleated tumor cells, which have been observed more frequently in ALK-negative ALCL than in ALK-positive ALCL (Figure 2C),5,13 tended to be absent in DUSP22- and TP63-rearranged cases. The lower frequency of classic morphologic findings in ALK-positive ALCL was expected, because use of ALK expression as a diagnostic criterion has led to inclusion of cases with a broad spectrum of morphologic variation (Figure 2D).1,12
In contrast to other ALCLs, the cytotoxic markers TIA-1 and granzyme B generally were absent in DUSP22-rearranged ALCL (present in 10% and 5% of cases, respectively; P < .0001 and P = .01, respectively; Figures 2A and 3C; Table 3). ALK-negative ALCLs negative for TIA-1 and granzyme B had OS rates more favorable than those positive for these markers but not as favorable as the group of patients identified by presence of DUSP22 rearrangements (supplemental Figure 1A-B, available on the Blood Web site). In a multivariate model, the prognostic significance of DUSP22 rearrangements was independent of cytotoxic marker expression (P = .02). Because perforin represents an additional cytotoxic marker that can be expressed in ALK-negative ALCL,7 we examined expression of this protein in 10 cases with DUSP22 rearrangements that were negative for both TIA-1 and granzyme B. All 10 cases were also negative for perforin. EMA and clusterin were expressed less frequently in ALK-negative ALCL than in ALK-positive ALCL but did not distinguish between genetic subtypes of ALK-negative ALCL. Conversely, CD2 and CD3 were expressed more frequently in all genetic subtypes of ALK-negative ALCL than in ALK-positive ALCL. p63 was expressed in all cases with TP63 rearrangements; however, p63 also was expressed in some nonrearranged cases, and p63 protein expression was not significantly associated with OS in ALK-negative ALCL (supplemental Figure 1C).
Discussion
This is the first study to demonstrate unequivocal genetic and clinical heterogeneity among systemic ALK-negative ALCLs. Most strikingly, patients with DUSP22-rearranged ALCL had excellent OS rates, similar to patients with ALK-positive ALCL. Conversely, patients with TP63-rearranged ALCL had dismal outcomes and nearly always failed standard therapy. Among all ALCLs, rearrangements of DUSP22, TP63, and ALK were mutually exclusive. A fourth group of triple-negative ALCLs had intermediate OS rates similar to those previously reported for ALK-negative ALCL as a whole. These findings have critical implications for the diagnostic evaluation, classification, and therapeutic management of ALCL.
All cases with either DUSP22 or TP63 rearrangements evaluated in this study were classified as ALK-negative ALCL using current WHO criteria by a panel of expert reviewers blinded to the genetic data. Thus, current diagnostic standards did not distinguish among genetic subtypes of ALK-negative ALCL. Despite pathologic features similar to other ALCLs, DUSP22-rearranged ALCLs were associated with significantly better outcomes than TP63-rearranged or triple-negative ALCLs. Patients with DUSP22-rearranged ALCL had slightly better OS than those with ALK-positive ALCL when adjusted for the older age of patients with DUSP22-rearranged ALCL, but this trend was not statistically significant.
Optimal upfront therapy for ALCL has not been defined.14 Most patients receive anthracycline-based multiagent chemotherapy, typically cyclophosphamide, doxorubicin, vincristine, and prednisone (CHOP). Although alternative upfront regimens have been investigated, none has been definitively superior to CHOP. The CD30-targeted immunoconjugate brentuximab vedotin has clinical efficacy in relapsed/refractory ALCL, and its utility combined with anthracycline-based chemotherapy in the frontline setting is being assessed in ongoing trials.15 In some centers, patients with ALK-negative ALCL who respond to CHOP are offered high-dose chemotherapy (eg, bis-chloroethylnitrosourea, etoposide, cytarabine, and melphalan [BEAM]) followed by SCT.1,7,16 Because of the favorable results with CHOP alone, SCT is not recommended for patients with ALK-positive ALCL in most clinical settings.
We considered that the comparison of OS rates in DUSP22-rearranged ALCLs and ALK-positive ALCLs might be biased by more aggressive treatment in the former group. However, there was no significant difference between groups with regard to initial treatment with an anthracycline-containing regimen or subsequent SCT. In fact, when only nontransplanted patients were considered, the outcomes in DUSP22-rearranged ALCL and ALK-positive ALCL remained nearly identical to each other. Based on these data, if the lack of need for SCT in most patients with ALK-positive ALCL is accepted, it would be difficult to justify the risks and cost of routine SCT in DUSP22-rearranged ALCL.
Although cases with DUSP22 and TP63 rearrangements were classified as ALK-negative ALCL based on current WHO criteria, some pathologic differences among the genetic subsets of ALCL were observed. Notably, DUSP22-rearranged ALCLs had classic histologic features of ALCL more consistently than other genetic subtypes and nearly always lacked cytotoxic marker expression. In keeping with previously reported data, all 3 genetic subtypes of ALK-negative ALCL shared phenotypic features that were different than ALK-positive ALCLs, namely less common expression of EMA and clusterin and more common expression of the pan-T cell antigens CD2 and CD3.7
DUSP22 rearrangements are associated with markedly decreased expression of DUSP22, which encodes a dual specificity phosphatase that regulates mitogen-activated protein kinase signaling.8,17 These rearrangements are not associated with altered expression of the neighboring IRF4 gene and are distinct from rare IRF4/TRA rearrangements in CD30-negative PTCL, NOS.8,18 Importantly, DUSP22 rearrangements also can be seen in primary cutaneous ALCL and lymphomatoid papulosis, which are both cutaneous T-cell neoplasms with excellent prognosis.18-21 The cases in the present study, however, all presented with systemic disease in the absence of cutaneous involvement. Interestingly, although ALK-positive ALCL has been considered a systemic disease, a number of cases limited to the skin recently have been reported.22,23 Thus, DUSP22 rearrangements and ALK rearrangements both may define distinct groups of ALCLs that can occur in either systemic or cutaneous forms and are associated with better prognosis than most other PTCL subtypes.
In contrast to other patient subgroups in this study, patients with TP63-rearranged ALCLs nearly uniformly failed standard therapy and had very poor OS. Most did not have sufficient response to initial therapy to be candidates for SCT. Thus, identification of TP63 rearrangements is critical to help clinicians and patients make informed decisions based on the very poor prognosis of these tumors. It is not clear whether these patients might benefit from alternative upfront approaches. TP63 rearrangements lead to fusion proteins homologous to oncogenic ΔNp63 isoforms; as the latter is a candidate therapeutic target, investigation into strategies to target p63 fusion proteins is merited.9,24
One argument for distinguishing ALK-negative ALCL from PTCL, NOS has been the superior OS of the former (49% vs 32%, respectively, at 5 years), which is even more pronounced when considering only CD30-positive PTCL, NOS (5-year OS, 19%).7,25 The latter OS rate is similar to that of the TP63-rearranged ALCLs in the current series; however, the TP63-rearranged cases met morphologic and phenotypic criteria for ALK-negative ALCL. The finding that consensus on both histology and final diagnosis for TP63-rearranged ALCLs and triple-negative ALCLs was weaker than for DUSP22-rearranged cases or ALK-positive ALCLs suggests the need for improved WHO criteria to distinguish these cases from CD30-positive PTCL, NOS.
Based on the above considerations, we recommend that all ALK-negative ALCLs undergo FISH testing for rearrangements involving DUSP22 or TP63. If a TP63 rearrangement is present, we would advocate that the pathology report indicate that ALK-negative ALCLs with TP63 rearrangements are associated with poor prognosis. If a TP63 rearrangement is absent and a DUSP22 rearrangement is present, we would recommend the designation ALK-negative ALCL with DUSP22 rearrangement. Immunohistochemistry for p63 and cytotoxic markers is not specific for genetic subtype and should not replace FISH based on currently available data. Absence of TIA-1 or granzyme B expression had prognostic significance; however, the OS of patients identified by this analysis was not as favorable as the OS of patients identified by DUSP22 rearrangements. Although expression of p63 protein did not have prognostic significance, immunohistochemistry for p63 might help identify cases in which TP63 FISH would be most informative. It should be noted that this study was retrospective, had a small number of patients in each genetic subgroup (especially TP63), and was heterogeneous with regard to treatment. Thus, validation of our results will be important and may help to refine our recommendations further.
Triple-negative cases remain genetically uncharacterized, and it is unclear whether triple-negative ALCL encompasses further biological or clinical heterogeneity or whether it represents a homogeneous group. Interestingly, Boi et al recently found that losses at 6q21 and/or 17p, encompassing the PRDM1 and TP53 genes, respectively, were associated with poor OS in ALK-negative ALCL.26 Combining these and other newly discovered genetic biomarkers with the findings described here may lead to even more robust risk stratification for patients with this disease.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Center for Individualized Medicine and the Department of Laboratory Medicine and Pathology, Mayo Clinic (Rochester, MN) and National Cancer Institute awards R01 CA177734 (to A.L.F.) and P50 CA97274 (University of Iowa/Mayo Clinic Lymphoma Specialized Program of Research Excellence [SPORE]). A.L.F. is a Damon Runyon Clinical Investigator supported by the Damon Runyon Cancer Research Foundation (CI-48-09).
Authorship
Contribution: E.R.P.C. analyzed data and wrote the manuscript; E.S.J., J.W.S., and S.H.S. performed pathology review and provided patient samples and clinical data; R.P.K., R.A.K., and M.E.L. performed and interpreted fluorescence in situ hybridization studies; J.S.S., E.D.H., S.K., L.J., S.E.G., S.O., A.N., K.L.G., K.M.R., W.H.W., W.R.M., J.R.C., T.M.H., S.M.A., and A.D. provided patient samples and/or clinical data; G.V. analyzed data; C.A. performed statistical analysis; M.J.M. performed statistical analysis and wrote the manuscript; and A.L.F. designed the study, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for E.R.P.C. is University of Washington, Seattle, WA.
The current affiliation for A.D. is Memorial Sloan-Kettering Cancer Center, New York, NY.
Correspondence: Andrew L. Feldman, Department of Laboratory Medicine and Pathology, Hilton Building, Room 8-00F, Mayo Clinic, 200 First St SW, Rochester, MN 55905; e-mail: feldman.andrew@mayo.edu.