Key Points
We identify gain-of-function mutations involving IL2RG, JAK1/3, and STAT5B as well as deleterious mutations affecting EZH2, FBXW10, and CHEK2 in T-PLL.
Pharmacologic targeting of primary T-PLL cells with the STAT5 inhibitor pimozide leads to apoptosis.
Abstract
The comprehensive genetic alterations underlying the pathogenesis of T-cell prolymphocytic leukemia (T-PLL) are unknown. To address this, we performed whole-genome sequencing (WGS), whole-exome sequencing (WES), high-resolution copy-number analysis, and Sanger resequencing of a large cohort of T-PLL. WGS and WES identified novel mutations in recurrently altered genes not previously implicated in T-PLL including EZH2, FBXW10, and CHEK2. Strikingly, WGS and/or WES showed largely mutually exclusive mutations affecting IL2RG, JAK1, JAK3, or STAT5B in 38 of 50 T-PLL genomes (76.0%). Notably, gain-of-function IL2RG mutations are novel and have not been reported in any form of cancer. Further, high-frequency mutations in STAT5B have not been previously reported in T-PLL. Functionally, IL2RG-JAK1-JAK3-STAT5B mutations led to signal transducer and activator of transcription 5 (STAT5) hyperactivation, transformed Ba/F3 cells resulting in cytokine-independent growth, and/or enhanced colony formation in Jurkat T cells. Importantly, primary T-PLL cells exhibited constitutive activation of STAT5, and targeted pharmacologic inhibition of STAT5 with pimozide induced apoptosis in primary T-PLL cells. These results for the first time provide a portrait of the mutational landscape of T-PLL and implicate deregulation of DNA repair and epigenetic modulators as well as high-frequency mutational activation of the IL2RG-JAK1-JAK3-STAT5B axis in the pathogenesis of T-PLL. These findings offer opportunities for novel targeted therapies in this aggressive leukemia.
Introduction
T-cell prolymphocytic leukemia (T-PLL) is an aggressive neoplasm of mature T-lymphocytes characterized by a rapid clinical course and resistance to conventional chemotherapy.1,2 Patients typically present with elevated white blood cell counts, hepatosplenomegaly, generalized lymphadenopathy, anemia, and/or thrombocytopenia as abnormal T cells infiltrate the peripheral blood, bone marrow, lymph nodes, spleen, and occasionally pleural fluid and skin. Previously, few therapeutic options existed for T-PLL patients. However, the recent use of alemtuzumab-based chemotherapy has allowed many patients to achieve complete remissions. Unfortunately, these remissions are typically short-lived, and treatment results in significant therapy-related immune suppression. The prognosis for patients diagnosed with T-PLL remains poor.3
Rearrangements between the TCL1A/B locus on chromosome 144,5 or its homolog MTCP1 on chromosome X and the T-cell receptor locus on chromosome 143,6,7 are characteristic of T-PLL. Additionally, the ATM gene located at chromosome 11q23 is frequently deleted or mutated,8-10 and trisomies of the long arm of chromosome 8 are often observed.3 However, the comprehensive mutational spectrum underlying the pathogenesis of T-PLL is unknown. Moreover, murine models have shown that the highly recurrent rearrangements affecting the TCL1A/B or MTCP1 loci are insufficient to recapitulate the aggressive oncogenic properties of human T-PLL.11,12 The protracted latency prior to frank leukemic transformation in these models is typically 15 to 20 months,11,12 suggesting that additional mutations may be important for disease pathogenesis and progression.13
To gain better insight into the pathogenesis of T-PLL, we performed comprehensive whole-genome sequencing (WGS) in combination with high-resolution copy-number variant (CNV) analysis and whole-exome sequencing (WES) on primary T-PLL samples. Our goal was to provide for the first time a comprehensive portrait of the genomic landscape of T-PLL and to identify actionable somatic mutations that could contribute to disease pathogenesis for the development of more effective targeted therapies.
Materials and methods
Patients and samples
Criteria for diagnosis of T-PLL were based on the 2008 World Health Organization classification.14 Clinical samples were obtained with institutional review board approval from the Department of Hematopathology at the University of Texas MD Anderson Cancer Center, the Department of Pathology at the University of Michigan, and the University of Cologne. For a given patient, samples represented either formalin-fixed, paraffin-embedded tissue or cryopreserved peripheral blood or both. Tumor DNA was extracted from cryopreserved peripheral blood CD4-enriched T cells. For assessment of somatic status of Janus kinase (JAK) signal transducer and activator of transcription (STAT) mutations, we obtained constitutional DNA from immunomagnetically separated nonneoplastic (CD4-negative) peripheral blood leukocytes or available matched nonhematopoietic tissue biopsy material (supplemental Table 1, available on the Blood Web site). Thus, we successfully obtained matched constitutional DNA from 32 of 38 patients (84.2%) with mutations affecting the interleukin-2 receptor gamma (IL2RG)-JAK-STAT axis. A complete description of all methods is provided in supplemental Methods.
WGS, WES, and Sanger resequencing
WGS of index T-PLL cases was performed by Complete Genomics (Mountain View, CA). Analyses of WGS data were performed using custom-designed bioinformatics tools.15 WGS yielded a mean of 351 ± 13 Gb mapped per sample with 97.4% to 97.8% fully called genome fraction and 97.1% to 97.7% fully called exome fraction. The median genomic sequencing depth exceeded 60× in all samples. WES was performed on the Illumina HiSeq following exome capture using Nimblegen EZCap 3 with paired-end 100 bp reads. The average depth of coverage for exome sequencing was 30.4 ± 11.9× with >95% of coding exons sequenced to a depth of 30× or more. After identifying candidate mutations of interest, genomic DNA from tumor and matched constitutional tissue was subjected to bidirectional Sanger resequencing for confirmation or mutation screening. All sequencing reactions were performed using nested sequencing primers. All 77 sequence variants described in this study and listed in Table 1, including those affecting the JAK-STAT pathway, were confirmed by Sanger resequencing. A total of 66 of the 77 sequence variants were present in patients for whom matched constitutional normal tissue was available and were confirmed to be somatic mutations.
Selected recurrently mutated genes in T-PLL by WGS and WES
| Number . | Gene . | Chromosome . | Position . | Reference . | Alternate . | Consequence . | Amino acid residue . | No. of cases . | Confirmed somatic . | Total no. confirmed somatic . | Novel . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL2RG | chrX | 70 328 495 | CATGGAGGC | Frameshift | G268_M270del | 1 | Somatic | 1 | Novel | |
| 2 | 70 327 753 | T | C | Missense | K315E | 1 | Somatic | 1 | Novel | ||
| 3 | JAK1 | chr1 | 65 313 222 | TATCCC | Deletion | R629_D630del | 1 | Somatic | 1 | Novel | |
| 4 | 65 312 347 | C | A | Missense | V658F | 2 | Somatic | 2 | COSMIC | ||
| 5 | 65 311 203 | C | A | Missense | S703I | 1 | Somatic | 1 | COSMIC | ||
| 6 | 65 305 426 | G | C | Missense | T901R | 1 | Somatic | 1 | Novel | ||
| 7 | JAK3 | chr19 | 17 948 006 | G | A | Missense | A573V | 2 | Somatic | 2 | Previous |
| 8 | 17 948 745 | TGCAGTTCT | Deletion | K563_C565del | 1 | Somatic | 1 | Novel | |||
| 9 | 17 945 970 | G | A | Missense | R657W | 1 | N/A | Novel | |||
| 10 | 17 949 121 | T | G | Missense | Q507P | 1 | Somatic | 1 | COSMIC | ||
| 11 | 17 949 108 | C | T | Missense | M511I | 5 | Somatic | 5 | Previous | ||
| 12 | 17 949 108 | C | G | Missense | M511I | 2 | Somatic | 2 | Previous | ||
| 13 | 17 949 108 | C | A | Missense | M511I | 3 | Somatic | 3 | COSMIC | ||
| 14 | STAT5B | chr17 | 40 362 212 | G | C | Missense | T628S | 3 | Somatic | 3 | Novel |
| 15 | 40 359 729 | T | G | Missense | N642H | 11 | Somatic | 11 | Previous | ||
| 16 | 40 359 678 | G | T | Missense | R659C | 1 | Somatic | 1 | Novel | ||
| 17 | 40 359 659 | T | A | Missense | Y665F | 2 | Somatic | 2 | Previous | ||
| 18 | 40 354 787 | T | A | Missense | Q706L | 1 | Somatic | 1 | Novel | ||
| 19 | ATM | chr11 | 108 099 934 | AG | Frameshift | E73fs | 1 | Somatic | 1 | ||
| 20 | 108 115 522 | A | G | Missense | K224E | 1 | Somatic | 1 | |||
| 21 | 108 117 745 | T | G | Missense | L319R | 1 | Somatic | 1 | |||
| 22 | 108 122 676 | AATC | Frameshift | S575fs | 1 | Somatic | 1 | ||||
| 23 | 108 124 604 | A | Frameshift | T655fs | 1 | Somatic | 1 | ||||
| 24 | 108 124 723 | T | G | Missense | L694R | 1 | Somatic | 1 | |||
| 25 | 108 178 665 | C | T | Nonsense | Q1906X | 1 | Somatic | 1 | |||
| 26 | 108 186 590 | A | G | Missense | D2016G | 1 | Somatic | 1 | |||
| 27 | 108 199 938 | T | G | Missense | L2427R | 1 | N/A | ||||
| 28 | 108 199 938 | T | C | Missense | L2427P | 1 | N/A | ||||
| 29 | 108 200 975 | G | A | Missense | D2448N | 1 | Somatic | 1 | |||
| 30 | 108 200 991 | G | C | Missense | R2453P | 1 | Somatic | 1 | COSMIC | ||
| 31 | 108 206 576 | TGA | Deletion | D2720del | 1 | Somatic | 1 | ||||
| 32 | 108 206 581 | G | A | Missense | D2721N | 1 | Somatic | 1 | COSMIC | ||
| 33 | 108 206 594 | A | T | Missense | D2725V | 1 | Somatic | 1 | |||
| 34 | 108 206 595 | T | A | Missense | D2725E | 2 | Somatic | 2 | |||
| 35 | 108 206 600 | T | G | Missense | V2727G | 1 | N/A | ||||
| 36 | 108 216 627 | C | T | Missense | S2859F | 1 | Somatic | 1 | |||
| 37 | 108 218 052 | G | C | Missense | L2877F | 1 | Somatic | 1 | |||
| 38 | 108 224 492 | G | A | Missense | G2890D | 1 | Somatic | 1 | |||
| 39 | 108 218 092 | G | C | Missense | G2891R | 1 | Somatic | 1 | COSMIC | ||
| 40 | 108 224 493 | G | A | Missense | G2891D | 1 | N/A | ||||
| 41 | 108 224 518 | C | G | Missense | I2899M | 1 | N/A | ||||
| 42 | 108 235 809 | G | A | Missense | V2951I | 1 | Somatic | 1 | |||
| 43 | 108 236 080 | G | A | Missense | A3006T | 1 | Somatic | 1 | |||
| 44 | 108 236 086 | C | T | Missense | R3008C | 1 | Somatic | 1 | |||
| 45 | CHEK2 | chr22 | 29 091 856 | G | Frameshift | T146fs | 1 | N/A | Novel | ||
| 46 | 29 083 962 | G | C | Missense | R298G | 1 | Somatic | 1 | COSMIC | ||
| 47 | EZH2 | chr7 | 148 526 858 | A | C | Missense | L110R | 1 | N/A | Novel | |
| 48 | 148 516 705 | G | A | Nonsense | Q284X | 1 | Somatic | 1 | Novel | ||
| 49 | 148 506 474 | C | T | Missense | V624M | 1 | N/A | Novel | |||
| 50 | 148 506 468 | C | T | Missense | A626T | 1 | Somatic | 1 | Novel | ||
| 51 | 148 506 245 | AA | Frameshift | K703fs | 1 | N/A | Novel | ||||
| 52 | FBXW10 | chr17 | 18 653 403 | G | A | Missense | D318N | 1 | Somatic | 1 | Novel |
| 53 | 18 661 631 | C | T | Nonsense | R416* | 1 | N/A | Novel | |||
| 54 | 18 682 350 | AA | Frameshift | K966fs | 1 | Somatic | 1 | Novel | |||
| 77 | 66 |
| Number . | Gene . | Chromosome . | Position . | Reference . | Alternate . | Consequence . | Amino acid residue . | No. of cases . | Confirmed somatic . | Total no. confirmed somatic . | Novel . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL2RG | chrX | 70 328 495 | CATGGAGGC | Frameshift | G268_M270del | 1 | Somatic | 1 | Novel | |
| 2 | 70 327 753 | T | C | Missense | K315E | 1 | Somatic | 1 | Novel | ||
| 3 | JAK1 | chr1 | 65 313 222 | TATCCC | Deletion | R629_D630del | 1 | Somatic | 1 | Novel | |
| 4 | 65 312 347 | C | A | Missense | V658F | 2 | Somatic | 2 | COSMIC | ||
| 5 | 65 311 203 | C | A | Missense | S703I | 1 | Somatic | 1 | COSMIC | ||
| 6 | 65 305 426 | G | C | Missense | T901R | 1 | Somatic | 1 | Novel | ||
| 7 | JAK3 | chr19 | 17 948 006 | G | A | Missense | A573V | 2 | Somatic | 2 | Previous |
| 8 | 17 948 745 | TGCAGTTCT | Deletion | K563_C565del | 1 | Somatic | 1 | Novel | |||
| 9 | 17 945 970 | G | A | Missense | R657W | 1 | N/A | Novel | |||
| 10 | 17 949 121 | T | G | Missense | Q507P | 1 | Somatic | 1 | COSMIC | ||
| 11 | 17 949 108 | C | T | Missense | M511I | 5 | Somatic | 5 | Previous | ||
| 12 | 17 949 108 | C | G | Missense | M511I | 2 | Somatic | 2 | Previous | ||
| 13 | 17 949 108 | C | A | Missense | M511I | 3 | Somatic | 3 | COSMIC | ||
| 14 | STAT5B | chr17 | 40 362 212 | G | C | Missense | T628S | 3 | Somatic | 3 | Novel |
| 15 | 40 359 729 | T | G | Missense | N642H | 11 | Somatic | 11 | Previous | ||
| 16 | 40 359 678 | G | T | Missense | R659C | 1 | Somatic | 1 | Novel | ||
| 17 | 40 359 659 | T | A | Missense | Y665F | 2 | Somatic | 2 | Previous | ||
| 18 | 40 354 787 | T | A | Missense | Q706L | 1 | Somatic | 1 | Novel | ||
| 19 | ATM | chr11 | 108 099 934 | AG | Frameshift | E73fs | 1 | Somatic | 1 | ||
| 20 | 108 115 522 | A | G | Missense | K224E | 1 | Somatic | 1 | |||
| 21 | 108 117 745 | T | G | Missense | L319R | 1 | Somatic | 1 | |||
| 22 | 108 122 676 | AATC | Frameshift | S575fs | 1 | Somatic | 1 | ||||
| 23 | 108 124 604 | A | Frameshift | T655fs | 1 | Somatic | 1 | ||||
| 24 | 108 124 723 | T | G | Missense | L694R | 1 | Somatic | 1 | |||
| 25 | 108 178 665 | C | T | Nonsense | Q1906X | 1 | Somatic | 1 | |||
| 26 | 108 186 590 | A | G | Missense | D2016G | 1 | Somatic | 1 | |||
| 27 | 108 199 938 | T | G | Missense | L2427R | 1 | N/A | ||||
| 28 | 108 199 938 | T | C | Missense | L2427P | 1 | N/A | ||||
| 29 | 108 200 975 | G | A | Missense | D2448N | 1 | Somatic | 1 | |||
| 30 | 108 200 991 | G | C | Missense | R2453P | 1 | Somatic | 1 | COSMIC | ||
| 31 | 108 206 576 | TGA | Deletion | D2720del | 1 | Somatic | 1 | ||||
| 32 | 108 206 581 | G | A | Missense | D2721N | 1 | Somatic | 1 | COSMIC | ||
| 33 | 108 206 594 | A | T | Missense | D2725V | 1 | Somatic | 1 | |||
| 34 | 108 206 595 | T | A | Missense | D2725E | 2 | Somatic | 2 | |||
| 35 | 108 206 600 | T | G | Missense | V2727G | 1 | N/A | ||||
| 36 | 108 216 627 | C | T | Missense | S2859F | 1 | Somatic | 1 | |||
| 37 | 108 218 052 | G | C | Missense | L2877F | 1 | Somatic | 1 | |||
| 38 | 108 224 492 | G | A | Missense | G2890D | 1 | Somatic | 1 | |||
| 39 | 108 218 092 | G | C | Missense | G2891R | 1 | Somatic | 1 | COSMIC | ||
| 40 | 108 224 493 | G | A | Missense | G2891D | 1 | N/A | ||||
| 41 | 108 224 518 | C | G | Missense | I2899M | 1 | N/A | ||||
| 42 | 108 235 809 | G | A | Missense | V2951I | 1 | Somatic | 1 | |||
| 43 | 108 236 080 | G | A | Missense | A3006T | 1 | Somatic | 1 | |||
| 44 | 108 236 086 | C | T | Missense | R3008C | 1 | Somatic | 1 | |||
| 45 | CHEK2 | chr22 | 29 091 856 | G | Frameshift | T146fs | 1 | N/A | Novel | ||
| 46 | 29 083 962 | G | C | Missense | R298G | 1 | Somatic | 1 | COSMIC | ||
| 47 | EZH2 | chr7 | 148 526 858 | A | C | Missense | L110R | 1 | N/A | Novel | |
| 48 | 148 516 705 | G | A | Nonsense | Q284X | 1 | Somatic | 1 | Novel | ||
| 49 | 148 506 474 | C | T | Missense | V624M | 1 | N/A | Novel | |||
| 50 | 148 506 468 | C | T | Missense | A626T | 1 | Somatic | 1 | Novel | ||
| 51 | 148 506 245 | AA | Frameshift | K703fs | 1 | N/A | Novel | ||||
| 52 | FBXW10 | chr17 | 18 653 403 | G | A | Missense | D318N | 1 | Somatic | 1 | Novel |
| 53 | 18 661 631 | C | T | Nonsense | R416* | 1 | N/A | Novel | |||
| 54 | 18 682 350 | AA | Frameshift | K966fs | 1 | Somatic | 1 | Novel | |||
| 77 | 66 |
Ploidy analysis
CNVs were detected using comparative genome hybridization on Nimblegen whole-genome arrays containing 270 000 features (Roche Applied Science) sufficient to detect CNVs of ≥50 kb.
Functional analysis of IL2RG, JAK1, JAK3, and STAT5B mutants
IL2RG, JAK1, JAK3, and STAT5B mutants were generated using standard polymerase chain reaction–based site-directed mutagenesis. For STAT5B reporter assays, HeLa cells were transiently transfected along with a reporter construct to link luciferase activity with signal transducer and activator of transcription 5B (STAT5B) expression. Protein lysates were used to determine phosphorylated STAT5 levels by western blotting. Wild-type STAT5B and its mutants were used for cell proliferation and soft-agar colony-formation assays. For detection of phosphorylated signal transducer and activator of transcription 5 (pSTAT5) by immunofluorescence microscopy in primary cells, the same anti-pSTAT5 antibody described above for western blot analysis was used.
Primary-cell and cell-line culture and pimozide treatment
Cells were thawed and grown overnight in RPMI cell culture medium followed by treatment with pimozide,16 a specific signal transducer and activator of transcription 5 (STAT5) inhibitor, at a final concentration of 10 μM. Cells were harvested at different time points, and protein lysates were subjected to western blot analysis. The human T-lineage lymphoma/leukemia cell lines Jurkat (pSTAT5 negative), HH (pSTAT5 negative), HUT78 (pSTAT5 positive), and SUDHL1 (pSTAT5 positive) were used as controls. Cell proliferation and viability assays were performed as described in supplemental Methods.
Statistical analysis of clinical outcomes
Clinical outcomes data (time to transformation, relapse, or death) were analyzed using standard survival analysis. A complete description of statistical techniques is presented in supplemental Methods.
Results
WGS reveals the genomic complexity of T-PLL and identifies JAK1 mutations
We began by performing WGS of 4 index cases of T-PLL (supplemental Figure 1) that fulfilled established diagnostic criteria14 including characteristic cytologic (supplemental Figure 2A), immunophenotypic, and karyotypic features. In total, WGS yielded a mean of 351 ± 13 Gb mapped reads per sample with 97.4% to 97.8% fully called genome fraction and 97.1% to 97.7% fully called exome fraction. The median genomic sequencing depth exceeded 60× in all samples normalized across the entire genome. As anticipated by clinical karyotyping, confirmatory fluorescence in situ hybridization results (supplemental Figure 2B), and numerous prior studies,5,17-19 3 of 4 index cases harbored the characteristic inv(14) alteration involving the TCL1A/B locus (supplemental Figure 2C-E) and the fourth harbored the less common t(X;14) lesion involving the TCL1 analog MTCP1 (supplemental Figure 2F). Altogether, WGS identified a total of 751 novel structural abnormalities in the 4 index T-PLL genomes. In addition to a focused 9.1-Mb rearrangement in 1 of 4 index genomes consistent with loss of genetic material comprising the ATM locus as anticipated by earlier studies,20 this analysis also highlighted a novel disruption on chromosome 14q24 in 1 of 4 genomes affecting the NUMB gene, which links Notch pathways with T-cell receptor signaling in T lymphocytes,21 and loss of 1.0 to 2.0 Mb of genetic material on chromosome 7q36 in 2 of 4 genomes comprising the histone methyltransferase EZH2 gene and genes in the GIMAP family known to influence T-lymphocyte development and function22 (supplemental Table 2). However, whereas this analysis highlighted the structural genomic complexity of T-PLL and confirmed the presence of translocations involving the TCL1A/B and MTCP1 loci (supplemental Figure 2G), no additional recurrent fusion events were noted. Comprehensive structural alteration data from WGS can be found in supplemental Table 2 and may serve as a framework for future studies of less common large structural alterations contributing to T-PLL pathogenesis.
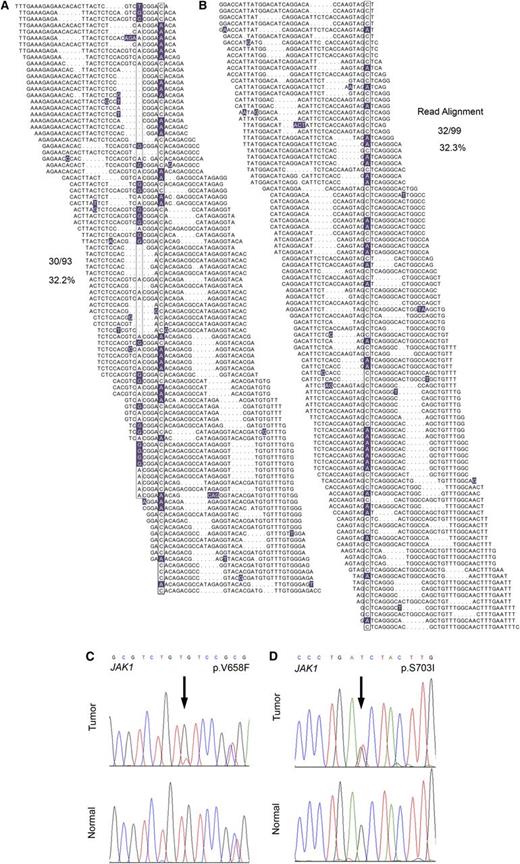
In total, WGS analysis revealed 2358 novel sequence variants and small insertion/deletion events affecting protein-coding regions, most of which have not been previously implicated in the pathogenesis of T-PLL. These included a frameshift mutation in the SET domain of EZH2, a novel missense mutation in CREBBP, a p.V1670I mutation in the NODP (number of developmental processes) domain of NOTCH1, and 3 novel missense mutations in ATM in 3 of 4 index genomes. Comprehensive WGS mutation data are provided in supplemental Table 3. Interestingly, WGS revealed JAK1 mutations in 2 of 4 T-PLL samples (Figure 1A-B; p.V658F23,24 and p.S703I25 ), both of which were confirmed to be somatic by Sanger resequencing (Figure 1C-D) and, similar to recent findings,13,26 represent gain-of-function mutations. The p.V658F identified in JAK1 is analogous to the JAK2 p.V617F mutation characteristic of myeloproliferative neoplasms, and both mutations have been reported in isolated cases of lymphoid neoplasms in the Catalogue of Somatic Mutations in Cancer (COSMIC) database of somatic cancer mutations. Cytokine-receptor/JAK-STAT–mediated cytokine signaling regulates hematopoietic cell ontogeny, and its deregulation has been implicated in the pathogenesis of a number of hematopoietic malignancies.13,26-28
WGS identifies JAK1 mutations in 2 out of 4 index T-PLL cases. (A-B) The total individual reads supporting variant calling of the JAK1 p.V658F (A) and p.S703I (B) mutations in index T-PLL samples are shown. Nucleotides with a deviation from the reference sequence are highlighted. The mutations as well as a synonymous single-nucleotide polymorphism (A) are boxed. Dots in individual reads (A-B) represent unsequenced nucleotides (as opposed to sequence gaps) and are intrinsic to the self-assembling DNA nanoarray next-generation sequencing platform and unchained base-reads analysis approach used for WGS (see supplemental Methods for further details). (C-D) Sanger resequencing confirmation of the somatic acquisition of these mutations is shown.
WGS identifies JAK1 mutations in 2 out of 4 index T-PLL cases. (A-B) The total individual reads supporting variant calling of the JAK1 p.V658F (A) and p.S703I (B) mutations in index T-PLL samples are shown. Nucleotides with a deviation from the reference sequence are highlighted. The mutations as well as a synonymous single-nucleotide polymorphism (A) are boxed. Dots in individual reads (A-B) represent unsequenced nucleotides (as opposed to sequence gaps) and are intrinsic to the self-assembling DNA nanoarray next-generation sequencing platform and unchained base-reads analysis approach used for WGS (see supplemental Methods for further details). (C-D) Sanger resequencing confirmation of the somatic acquisition of these mutations is shown.
CNV analysis confirms abnormalities involving the ATM locus
To more completely define the landscape of large structural alterations in T-PLL, we performed high-resolution CNV analysis using array comparative genome hybridization (aCGH) on an additional 39 T-PLL cases (supplemental Figure 1). Consistent with several previous reports,29-32 aCGH confirmed loss of the 11q23 region in 65.1% of T-PLL samples (28/39; supplemental Figure 3A, arrow) encompassing the ATM locus (supplemental Figure 3B) and isochromosome 8 or large gains of chromosome 8q in 76.9% (30/39) of T-PLL samples (supplemental Figure 3A, arrowhead). Interestingly, a total of 16 of 39 genomes (41.0%) revealed loss of chromosomal material comprising the EZH2 and GIMAP locus on chromosome 7q36 (supplemental Figure 4). Although this locus and genes in the GIMAP family have been shown to be important in the regulation of T-lymphocyte development and survival,33,34 they have not been previously implicated in the pathogenesis of T-PLL. Our CNV analysis suggests that the function of these genes is important in regulation of T-cell proliferation and can be deregulated in malignancy. Additional regions of aneuploidy included frequent alterations of chromosome 22, including loss of 22p and gains of 22q consistent with previous results29,31 as well as a striking new deletion on chromosome 7q36, which encompasses the EZH2 locus. Results of the comprehensive CNV analysis can be found in supplemental Table 4 and represent the list of recurrent copy-number alterations in T-PLL.
WES identifies recurrently altered genes including members of the IL2RG-JAK1-JAK3-STAT5B axis in T-PLL
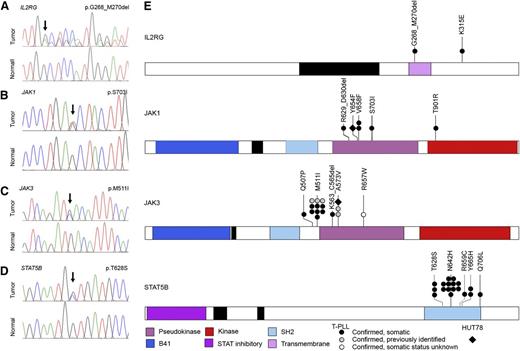
Whereas a significant body of literature describing structural alterations including translocations and CNVs affecting T-PLL exists, the mutational landscape of T-PLL remains poorly characterized. To more completely define the landscape of genetic mutations in T-PLL and to confirm the results of our WGS mutational analysis of the 4 index T-PLL cases, we next performed WES on a total of 36 T-PLL cases (supplemental Figure 1) paired with constitutional DNA for Sanger resequencing confirmation of somatic status for selected sequence variants (66 mutations; Table 1). As anticipated from previous studies, 70.0% (28/40) of T-PLL samples harbored somatic mutations in the tumor suppressor ATM (supplemental Figure 3C-D) predicted to be deleterious to protein function, including frameshift and nonsense mutations as well as missense mutations clustered in the FAT and PI3K domains, similar to earlier reports.8,10 Using this approach, we also identified a number of genetic alterations not previously associated with T-PLL (Table 1), including mutations in CHEK2, a gene encoding a protein kinase activated in response to DNA damage35 (2/40, 5.0%), and novel mutations in EZH2, a member of the Polycomb group family of transcriptional repressors (5/40, 12.5%) frequently mutated in myeloid36 and lymphoid37 malignancy, and FBWX10, a member of the F-box protein family of ubiquitin ligases (3/40, 7.5%). Mutations in these proteins included several deleterious frameshift and nonsense somatic mutations, raising the possibility that ATM, CHEK2, EZH2, and FBW10 might contribute to T-PLL pathogenesis via their roles in DNA repair, epigenetic transcriptional regulation, and proteasomal degradation pathways, respectively. Comprehensive WES data are provided in supplemental Tables 5 and 6. All 77 sequence variants identified by next-generation DNA sequencing and discussed in this paper were confirmed by Sanger resequencing and, in the case of 66 sequence variants where constitutional DNA was available, were confirmed to be somatic (Figures 1C-D and 2A-D, Table 1, and supplemental Tables 1 and 7).
High-frequency IL2RG-JAK1-JAK3-STAT5B mutations in T-PLL. (A-D) Representative IL2RG, JAK1, JAK3, and STAT5B mutations in primary T-PLL cells identified by WGS/WES and confirmed to be somatic by Sanger resequencing of tumor DNA (upper traces) and paired constitutional DNA (lower traces). (E) Schematic representations of mutations in IL2RG, JAK1, JAK3, and STAT5B identified through WGS, WES, or targeted Sanger resequencing of primary T-PLL (circles) and HUT78 cells (diamond). Confirmed somatic mutations are shown as filled symbols; variants where adequate matched constitutional DNA was not available are shown as open symbols. Mutations are clustered in the autoinhibitory pseudokinase domains of JAK1 and JAK3 (purple) or the SH2 domain of STAT5B (light blue) that mediates interactions between JAK and STAT proteins. One additional variant was detected in the kinase domain of JAK1 (red); a single case of T-PLL harbored a somatic 3-amino-acid deletion in the transmembrane domain of IL2RG (purple) as well as a somatic missense mutation in the cytoplasmic domain.
High-frequency IL2RG-JAK1-JAK3-STAT5B mutations in T-PLL. (A-D) Representative IL2RG, JAK1, JAK3, and STAT5B mutations in primary T-PLL cells identified by WGS/WES and confirmed to be somatic by Sanger resequencing of tumor DNA (upper traces) and paired constitutional DNA (lower traces). (E) Schematic representations of mutations in IL2RG, JAK1, JAK3, and STAT5B identified through WGS, WES, or targeted Sanger resequencing of primary T-PLL (circles) and HUT78 cells (diamond). Confirmed somatic mutations are shown as filled symbols; variants where adequate matched constitutional DNA was not available are shown as open symbols. Mutations are clustered in the autoinhibitory pseudokinase domains of JAK1 and JAK3 (purple) or the SH2 domain of STAT5B (light blue) that mediates interactions between JAK and STAT proteins. One additional variant was detected in the kinase domain of JAK1 (red); a single case of T-PLL harbored a somatic 3-amino-acid deletion in the transmembrane domain of IL2RG (purple) as well as a somatic missense mutation in the cytoplasmic domain.
Strikingly, our WES analysis also identified somatic mutations in IL2RG, JAK1, JAK3, and STAT5B, including recurrent lesions not previously reported in T-PLL (Tables 1 and 2 and Figure 2). Altogether, WGS, WES, and targeted Sanger resequencing analysis (supplemental Figure 1) identified mutations in JAK1 (4/50, 8.0%) and JAK3 (15/50, 30.0%; Figure 2B-C,E) that were clustered in the autoinhibitory pseudokinase domain and included some variants detected previously in other malignancies or shown to lead to constitutive activation of JAK-STAT signaling27,38 (including Janus kinase 1 [JAK1] p.V658F23,24 and p.S703I25 and Janus kinase 3 [JAK3] p.M511I,25,39,40 p.A573V,41 and p.R657W40 ). Although recent reports described a similar finding of recurrent mutations in JAK1 and JAK3 in a cohort of T-PLL,13,26 our studies further revealed a novel mutation (p.T901R; 1/50, 2.0%; Figure 2B) in the kinase domain of JAK1. Notably, our analysis also identified for the first time in human cancer a novel mutation in the IL2RG transmembrane domain (p.G268_M270del; 1/50, 2.0%; Figure 2A) in 1 case of T-PLL. A second somatic mutation outside of the transmembrane domain, IL2RG p.K315E, was also identified in the same sample. Importantly, our WES studies identified high-frequency mutations in STAT5B (18/50, 36.0%) in T-PLL. The mutations clustered in the Src-like homology domain (SH2; p.T628S, p.N642H, p.R659C, and p.Y665H; Figure 2D-E). An additional p.Q706L mutation in STAT5B was also identified in a sample with a JAK1 p.V658F mutation that mediates the interaction between JAK and STAT proteins. Significantly, mutations in STAT5B have never been previously reported in T-PLL. In our cohort, STAT5B mutations actually represented the highest frequency of all JAK-STAT family members (36.0%; Tables 1 and 2 and supplemental Table 1). Notably, our WGS and WES studies of 50 cases of T-PLL revealed no evidence of activating STAT3 mutations. In total, 38 out of 50 T-PLL genomes harbored somatic mutations in IL2RG, JAK1, JAK3, or STAT5B (76.0%; Figure 2E). With one exception (JAK1 p.V658F and STAT5B p.Q706L; see Table 2), all cases harbored mutually exclusive mutations in genes comprising the IL2RG-JAK1-JAK3-STAT5B pathway. In the majority of patients with mutations in the JAK-STAT pathway (32/38, 84.2%), matched constitutional normal tissue (eg, CD4− leukocytes from peripheral blood) was available to confirm the somatic status of the mutations (Figure 2 and Table 2). Of the 18 distinct JAK-STAT pathway mutations identified, all but one (JAK3 p.R657W) were confirmed to be somatic (Table 1; 17/18, 94.4%). These data suggest a significant role for mutational activation of the interleukin-2 receptor (IL2R)-JAK1-JAK3-STAT5 axis in T-PLL.
JAK-STAT mutational status associated with patient demographic and clinical diagnostic and prognostic information
| PLL_ID . | Age at Dx . | Sex . | ATM . | ATM . | inv(14)/TCL1+ . | iso(8) . | IL2RG . | JAK1 . | JAK3 . | STAT5B . | JAK-STAT mutation status . | Previous treatment status . | Time to relapse (days) . | Time to death (days) . | Survival . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PLL_index1 | 47 | M | T655fs | X,14 | Yes | S703I | Confirmed somatic | Untreated | 335 | 622 | Dead | ||||
| PLL_index2 | 89 | M | Deletion | inv(14) | Yes | V658F | Q706L | Confirmed somatic | Untreated | 160 | Dead | ||||
| PLL_index3 | 53 | F | H2037R | inv(14) | None: genome | None: genome | None: genome | None: genome | No mutation | Unknown | 1008 | 1077 | Dead | ||
| PLL_index4 | M | Unknown | inv(14) | Yes | None: genome | None: genome | None: genome | None: genome | No mutation | Untreated | 548 | Dead | |||
| PLL_1 | 71 | M | Deletion | E73fs | T628S | Confirmed somatic | Untreated | 1288 | Dead | ||||||
| PLL_2 | 69 | F | None: exome | None: exome | None: exome | None: exome | No mutation | Treated | 2674 | Dead | |||||
| PLL_3 | 81 | F | Deletion | G2891RX | add(14) | M511I | Confirmed somatic | Untreated | 103 | Dead | |||||
| PLL_4 | 68 | M | TCL1+ | T901R | Confirmed somatic | Untreated | Alive | ||||||||
| PLL_5 | 58 | M | L2427R | TCL1+ | N642H | Confirmed somatic | Treated | 791 | 1084 | Dead | |||||
| PLL_6 | 61 | F | Unknown | Unknown | Yes | None: Sanger | None: Sanger | None: Sanger | None: Sanger | No mutation | Untreated | 192 | Dead | ||
| PLL_7 | 71 | M | D2725V | inv(14) | V658F | Confirmed somatic | Untreated | Alive | |||||||
| PLL_8 | 68 | M | Deletion | V2727G | TCL1+ | Y665F | Confirmed somatic | Untreated | 130 | Dead | |||||
| PLL_9 | 68 | M | del(14) | M511I | Confirmed somatic | Treated | 361 | Dead | |||||||
| PLL_10 | 60 | M | Deletion | inv(14) | M511I | Confirmed somatic | Treated | 924 | Alive | ||||||
| PLL_11 | 79 | M | Deletion | D2719del | inv(14) | None: exome | None: exome | None: exome | None: exome | No mutation | Untreated | Alive | |||
| PLL_12 | 80 | M | TCL1+ | Yes | M511I | Confirmed somatic | Untreated | 189 | 190 | Dead | |||||
| PLL_13 | 62 | M | inv(14) | M511I | Confirmed somatic | Treated | 335 | 754 | Dead | ||||||
| PLL_14 | 77 | F | Deletion | I2899M | inv(14) | M511I | Confirmed somatic | Treated | 2522 | 2871 | Dead | ||||
| PLL_15 | 27 | M | N642H | Confirmed somatic | Treated | 2680 | Alive | ||||||||
| PLL_16 | 36 | F | Deletion | D2725E | inv(14) | M511I | Confirmed somatic | Untreated | 561 | Dead | |||||
| PLL_17 | 43 | M | TCL1+ | T628S | Confirmed somatic | Untreated | Alive | ||||||||
| PLL_18 | 71 | M | Deletion | D2721NX | 10,14 | yes | K563_C565del | Confirmed somatic | Untreated | Alive | |||||
| PLL_19 | 75 | F | Deletion | S575fs | A573V | Confirmed somatic | Treated | 973 | 1122 | Dead | |||||
| PLL_20 | Deletion | G2891D | N642H | Confirmed somatic | Unknown | ||||||||||
| PLL_21 | 73 | M | Deletion | A3006T | +8 | N642H | Confirmed somatic | Treated | 212 | 824 | Dead | ||||
| PLL_22 | 74 | F | Unknown | Unknown | inv(14) | +8 | None: Sanger | None: Sanger | None: Sanger | None: Sanger | No mutation | Untreated | Alive | ||
| PLL_23 | 61 | F | Deletion | L2427P | inv(14) | iso(8) | Y665F | Confirmed somatic | Untreated | 486 | Alive | ||||
| PLL_24 | 60 | M | Deletion | H2038D | TCL1+ | Unknown | T628S | Confirmed somatic | Untreated | Alive | |||||
| PLL_25 | 78 | F | Deletion | L694R | inv(14) | G628_S630del and K315E | Confirmed somatic | Untreated | Alive | ||||||
| PLL_26 | 72 | M | TCL1+ | Unknown | None: exome | None: exome | None: exome | None: exome | No mutation | Untreated | 109 | 132 | Dead | ||
| PLL_27 | 45 | M | Deletion | D2448N | TCL1+ | Unknown | N642H | Confirmed somatic | Untreated | 111 | 342 | Dead | |||
| PLL_28 | 62 | F | Unknown | Unknown | TCL1+ | +MYC | None: Sanger | None: Sanger | None: Sanger | None: Sanger | No mutation | Untreated | 120 | Dead | |
| PLL_29 | 63 | M | Deletion | D2016G | t(14q11) | +8q24 | M511I | Confirmed somatic | Untreated | 238 | Dead | ||||
| PLL_30 | 63 | M | Unknown | Unknown | None: Sanger | None: Sanger | None: Sanger | None: Sanger | No mutation | Untreated | Alive | ||||
| PLL_31 | 50 | F | V2951I | TCL1+ | Unknown | Q507P | Confirmed somatic | Untreated | 443 | 560 | Dead | ||||
| PLL_32 | 56 | M | Deletion | R2453P | TCL1+ | +MYC | R657W | Mutation but normal tissue not available | Treated | 823 | 941 | Dead | |||
| PLL_33 | 52 | M | Deletion | L319R | TCL1+ | Unknown | N642H | Confirmed somatic | Untreated | 1643 | 2179 | Dead | |||
| PLL_34 | 58 | M | TCL1+ | N642H | Confirmed somatic | Untreated | 609 | Dead | |||||||
| PLL_35 | 48 | M | Deletion | D2725E and K224E | inv(14) | +MYC | N642H | Confirmed somatic | Untreated | 276 | Alive | ||||
| PLL_36 | 72 | M | L2877F | TCL1+ | Unknown | N642H | Confirmed somatic | Untreated | 129 | Dead | |||||
| PLL_37 | 72 | M | TCL1+ | +MYC | None: exome | None: exome | None: exome | None: exome | No mutation | Untreated | 612 | Dead | |||
| PLL_38 | 68 | M | R3008C | inv(14) | A573V | Confirmed somatic | Untreated | Alive | |||||||
| PLL_39 | 64 | M | Deletion | S2859F | TCL1+ | Unknown | R629_D630del | Confirmed somatic | Untreated | Alive | |||||
| PLL_40 | 71 | M | Q1906X | TCL1+ | Unknown | N642H | Confirmed somatic | Untreated | Alive | ||||||
| PLL_41 | 67 | F | Unknown | Unknown | inv(14) | None: Sanger | None: Sanger | None: Sanger | None: Sanger | No mutation | Untreated | 722 | Dead | ||
| PLL_42 | 67 | M | Unknown | Unknown | Unknown | Unknown | N642H | Confirmed somatic | Untreated | 92 | Dead | ||||
| PLL_43 | 54 | M | inv(14) | Yes | M511I | Confirmed somatic | Unknown | 1461 | 1990 | Dead | |||||
| PLL_44 | 73 | M | M511I | Confirmed somatic | Untreated | 18 | Dead | ||||||||
| PLL_45 | 58 | M | Deletion | inv(14) | Yes | None: Sanger | None: Sanger | None: Sanger | None: Sanger | No mutation | Untreated | 135 | Dead | ||
| PLL_46 | 57 | M | R659C | Confirmed somatic | Untreated | Alive |
| PLL_ID . | Age at Dx . | Sex . | ATM . | ATM . | inv(14)/TCL1+ . | iso(8) . | IL2RG . | JAK1 . | JAK3 . | STAT5B . | JAK-STAT mutation status . | Previous treatment status . | Time to relapse (days) . | Time to death (days) . | Survival . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PLL_index1 | 47 | M | T655fs | X,14 | Yes | S703I | Confirmed somatic | Untreated | 335 | 622 | Dead | ||||
| PLL_index2 | 89 | M | Deletion | inv(14) | Yes | V658F | Q706L | Confirmed somatic | Untreated | 160 | Dead | ||||
| PLL_index3 | 53 | F | H2037R | inv(14) | None: genome | None: genome | None: genome | None: genome | No mutation | Unknown | 1008 | 1077 | Dead | ||
| PLL_index4 | M | Unknown | inv(14) | Yes | None: genome | None: genome | None: genome | None: genome | No mutation | Untreated | 548 | Dead | |||
| PLL_1 | 71 | M | Deletion | E73fs | T628S | Confirmed somatic | Untreated | 1288 | Dead | ||||||
| PLL_2 | 69 | F | None: exome | None: exome | None: exome | None: exome | No mutation | Treated | 2674 | Dead | |||||
| PLL_3 | 81 | F | Deletion | G2891RX | add(14) | M511I | Confirmed somatic | Untreated | 103 | Dead | |||||
| PLL_4 | 68 | M | TCL1+ | T901R | Confirmed somatic | Untreated | Alive | ||||||||
| PLL_5 | 58 | M | L2427R | TCL1+ | N642H | Confirmed somatic | Treated | 791 | 1084 | Dead | |||||
| PLL_6 | 61 | F | Unknown | Unknown | Yes | None: Sanger | None: Sanger | None: Sanger | None: Sanger | No mutation | Untreated | 192 | Dead | ||
| PLL_7 | 71 | M | D2725V | inv(14) | V658F | Confirmed somatic | Untreated | Alive | |||||||
| PLL_8 | 68 | M | Deletion | V2727G | TCL1+ | Y665F | Confirmed somatic | Untreated | 130 | Dead | |||||
| PLL_9 | 68 | M | del(14) | M511I | Confirmed somatic | Treated | 361 | Dead | |||||||
| PLL_10 | 60 | M | Deletion | inv(14) | M511I | Confirmed somatic | Treated | 924 | Alive | ||||||
| PLL_11 | 79 | M | Deletion | D2719del | inv(14) | None: exome | None: exome | None: exome | None: exome | No mutation | Untreated | Alive | |||
| PLL_12 | 80 | M | TCL1+ | Yes | M511I | Confirmed somatic | Untreated | 189 | 190 | Dead | |||||
| PLL_13 | 62 | M | inv(14) | M511I | Confirmed somatic | Treated | 335 | 754 | Dead | ||||||
| PLL_14 | 77 | F | Deletion | I2899M | inv(14) | M511I | Confirmed somatic | Treated | 2522 | 2871 | Dead | ||||
| PLL_15 | 27 | M | N642H | Confirmed somatic | Treated | 2680 | Alive | ||||||||
| PLL_16 | 36 | F | Deletion | D2725E | inv(14) | M511I | Confirmed somatic | Untreated | 561 | Dead | |||||
| PLL_17 | 43 | M | TCL1+ | T628S | Confirmed somatic | Untreated | Alive | ||||||||
| PLL_18 | 71 | M | Deletion | D2721NX | 10,14 | yes | K563_C565del | Confirmed somatic | Untreated | Alive | |||||
| PLL_19 | 75 | F | Deletion | S575fs | A573V | Confirmed somatic | Treated | 973 | 1122 | Dead | |||||
| PLL_20 | Deletion | G2891D | N642H | Confirmed somatic | Unknown | ||||||||||
| PLL_21 | 73 | M | Deletion | A3006T | +8 | N642H | Confirmed somatic | Treated | 212 | 824 | Dead | ||||
| PLL_22 | 74 | F | Unknown | Unknown | inv(14) | +8 | None: Sanger | None: Sanger | None: Sanger | None: Sanger | No mutation | Untreated | Alive | ||
| PLL_23 | 61 | F | Deletion | L2427P | inv(14) | iso(8) | Y665F | Confirmed somatic | Untreated | 486 | Alive | ||||
| PLL_24 | 60 | M | Deletion | H2038D | TCL1+ | Unknown | T628S | Confirmed somatic | Untreated | Alive | |||||
| PLL_25 | 78 | F | Deletion | L694R | inv(14) | G628_S630del and K315E | Confirmed somatic | Untreated | Alive | ||||||
| PLL_26 | 72 | M | TCL1+ | Unknown | None: exome | None: exome | None: exome | None: exome | No mutation | Untreated | 109 | 132 | Dead | ||
| PLL_27 | 45 | M | Deletion | D2448N | TCL1+ | Unknown | N642H | Confirmed somatic | Untreated | 111 | 342 | Dead | |||
| PLL_28 | 62 | F | Unknown | Unknown | TCL1+ | +MYC | None: Sanger | None: Sanger | None: Sanger | None: Sanger | No mutation | Untreated | 120 | Dead | |
| PLL_29 | 63 | M | Deletion | D2016G | t(14q11) | +8q24 | M511I | Confirmed somatic | Untreated | 238 | Dead | ||||
| PLL_30 | 63 | M | Unknown | Unknown | None: Sanger | None: Sanger | None: Sanger | None: Sanger | No mutation | Untreated | Alive | ||||
| PLL_31 | 50 | F | V2951I | TCL1+ | Unknown | Q507P | Confirmed somatic | Untreated | 443 | 560 | Dead | ||||
| PLL_32 | 56 | M | Deletion | R2453P | TCL1+ | +MYC | R657W | Mutation but normal tissue not available | Treated | 823 | 941 | Dead | |||
| PLL_33 | 52 | M | Deletion | L319R | TCL1+ | Unknown | N642H | Confirmed somatic | Untreated | 1643 | 2179 | Dead | |||
| PLL_34 | 58 | M | TCL1+ | N642H | Confirmed somatic | Untreated | 609 | Dead | |||||||
| PLL_35 | 48 | M | Deletion | D2725E and K224E | inv(14) | +MYC | N642H | Confirmed somatic | Untreated | 276 | Alive | ||||
| PLL_36 | 72 | M | L2877F | TCL1+ | Unknown | N642H | Confirmed somatic | Untreated | 129 | Dead | |||||
| PLL_37 | 72 | M | TCL1+ | +MYC | None: exome | None: exome | None: exome | None: exome | No mutation | Untreated | 612 | Dead | |||
| PLL_38 | 68 | M | R3008C | inv(14) | A573V | Confirmed somatic | Untreated | Alive | |||||||
| PLL_39 | 64 | M | Deletion | S2859F | TCL1+ | Unknown | R629_D630del | Confirmed somatic | Untreated | Alive | |||||
| PLL_40 | 71 | M | Q1906X | TCL1+ | Unknown | N642H | Confirmed somatic | Untreated | Alive | ||||||
| PLL_41 | 67 | F | Unknown | Unknown | inv(14) | None: Sanger | None: Sanger | None: Sanger | None: Sanger | No mutation | Untreated | 722 | Dead | ||
| PLL_42 | 67 | M | Unknown | Unknown | Unknown | Unknown | N642H | Confirmed somatic | Untreated | 92 | Dead | ||||
| PLL_43 | 54 | M | inv(14) | Yes | M511I | Confirmed somatic | Unknown | 1461 | 1990 | Dead | |||||
| PLL_44 | 73 | M | M511I | Confirmed somatic | Untreated | 18 | Dead | ||||||||
| PLL_45 | 58 | M | Deletion | inv(14) | Yes | None: Sanger | None: Sanger | None: Sanger | None: Sanger | No mutation | Untreated | 135 | Dead | ||
| PLL_46 | 57 | M | R659C | Confirmed somatic | Untreated | Alive |
Index cases subjected to WGS are listed first. Deletions or point mutations in the 11q23 locus or ATM gene, respectively, are indicated. Karyotypic or immunohistochemical evidence of rearrangements involving the TCL1A/B or MTCP1 loci or TCL1 protein are indicated. Mutations identified in T-PLL cases are shown by gene. Cases for which no mutation was identified are indicated according to which method of mutation detection was employed. The patient’s treatment status at time of specimen collection is also indicated, as are the times from disease diagnosis to relapse or death and the patients’ survival status.
Dx, diagnosis; F, female; M, male.
In silico structural modeling of JAK3 and STAT5B mutations
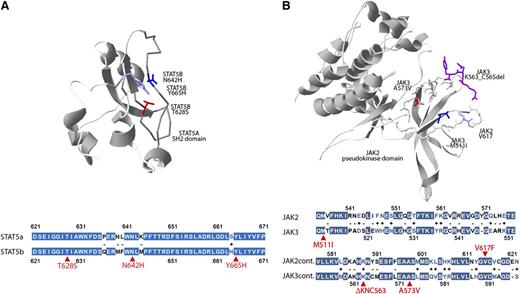
Although the crystal structure of JAK3 has not been solved, the high degree of homology between JAK2 and JAK3 (Figure 3B) permitted us to locate in the JAK2 3-dimensional structure the residues analogous to the recurrent JAK3 p.M511I and p.A573V mutations and the adjacent p.K563_C565del mutation. Plotting these residues onto the 3-dimensional structure of JAK2 revealed the close proximity of these residues and suggest a similar gain-of-function mechanism as seen with JAK2 p.V617F mutation.42 Similarly, the high homology between signal transducer and activator of transcription 5A (STAT5A) and STAT5B (Figure 3A) permitted us to localize the STAT5A residues homologous to the mutated STAT5B residues, which revealed the close 3-dimensional proximity of these residues in the SH2 domain of the predicted phosphotyrosine-binding loop.
Three-dimensional localization of recurrent JAK3- and STAT5B-mutated amino acids. (A) The crystal structure of the SH2 domain of the STAT5A protein (1y1u) highlighting analogous residues of the STAT5B mutations p.N642H (blue), p.Y665H (purple), and p.T628S (red) demonstrating close 3-dimensional proximity of recurrently mutated STAT5B residues. Colored fill indicates an identical amino acid; white, minus indicates disparate residues; white with colored text, + indicates similar residues; and selected mutated residues are indicated in red. (B) The pseudokinase domain of JAK2 (4fvp) highlighting the V617 residue (light blue) recurrently mutated in myeloproliferative neoplasms and analogous residues for JAK3 mutations p.A573V (red), p.M511I (dark blue), and p.K563_C565del (purple). The extent of homology between the STAT5A and STAT5B or JAK2 and JAK3 in the regions of these recurrently mutated residues (red arrows) is highlighted below each respective 3-dimensional structure.
Three-dimensional localization of recurrent JAK3- and STAT5B-mutated amino acids. (A) The crystal structure of the SH2 domain of the STAT5A protein (1y1u) highlighting analogous residues of the STAT5B mutations p.N642H (blue), p.Y665H (purple), and p.T628S (red) demonstrating close 3-dimensional proximity of recurrently mutated STAT5B residues. Colored fill indicates an identical amino acid; white, minus indicates disparate residues; white with colored text, + indicates similar residues; and selected mutated residues are indicated in red. (B) The pseudokinase domain of JAK2 (4fvp) highlighting the V617 residue (light blue) recurrently mutated in myeloproliferative neoplasms and analogous residues for JAK3 mutations p.A573V (red), p.M511I (dark blue), and p.K563_C565del (purple). The extent of homology between the STAT5A and STAT5B or JAK2 and JAK3 in the regions of these recurrently mutated residues (red arrows) is highlighted below each respective 3-dimensional structure.
IL2RG-JAK1-JAK3-STAT5B mutations lead to STAT5 transcriptional hyperactivation
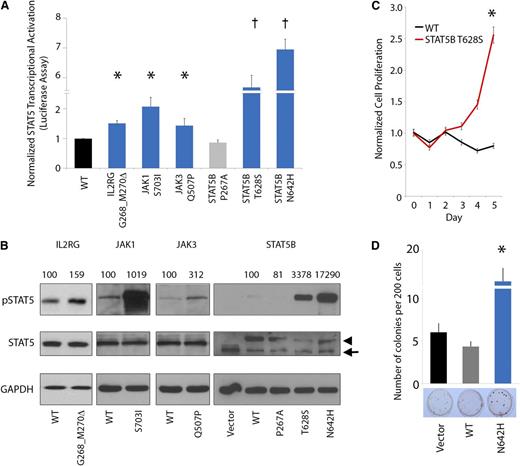
JAK-STAT mutations, including the JAK2 p.V617F mutation, are frequently implicated in the pathogenesis of many cancers and particularly in hematopoietic neoplasia.27 Several of the JAK-STAT mutations or analogous alterations identified in our study have been functionally characterized to demonstrate constitutive activation of JAK-STAT signaling and oncogenic activity.40,41,43-45 Accordingly, we sought to investigate the functional effects of representative novel mutations on JAK-STAT5 activation. In this regard, we assessed transcriptional activation of STAT5B using standard luciferase reporter assays by expressing the mutant IL2RG (p.G268_M270del), JAK1 (p.S703I), JAK3 (p.Q507P), and STAT5B (p.T628S) proteins (Figure 4A) in the HeLa cell line. In each case, expression of mutant IL2RG, JAK1, JAK3, and STAT5B proteins led to elevated STAT5 transcriptional activation (1.4- to 5.7-fold increase in luciferase activity). In particular, expression of the STAT5B p.T628S and p.N642H mutant proteins led to a dramatic increase in STAT5 transcriptional activation.
JAK-STAT mutations lead to increased pSTAT5 signaling, cytokine-independent growth, and enhanced colony formation. (A-B) Mutated IL2RG (p.G628_M630del), JAK1 (p.S703I), JAK3 (p.Q507P), and STAT5B (p.T628S and p.N642H) leads to increased activation of STAT5 transcriptional activity (A, bar graph; n = 3 for each mutant protein in separate experiments; asterisk indicates P < .05; dagger indicates P < .001) and increased phosphorylation of STAT5B (B, western blot; arrowhead indicates exogenous STAT5B, arrow indicates endogenous STAT5B; normalized densitometric pSTAT5/STAT5 ratios are indicated) in HeLa cells. STAT5B p.P267A represents a germline polymorphism. (C) Cytokine-independent cell proliferation in the presence of mutant p.T628S STAT5B protein in the cytokine-dependent Ba/F3 cell line cultured in the absence of growth factors (n = 6; asterisk indicates P < .01). (D) Enhanced colony-forming capacity of STAT5B p.N642H mutant in Jurkat T cells (n = 3; asterisk indicates P < .01).
JAK-STAT mutations lead to increased pSTAT5 signaling, cytokine-independent growth, and enhanced colony formation. (A-B) Mutated IL2RG (p.G628_M630del), JAK1 (p.S703I), JAK3 (p.Q507P), and STAT5B (p.T628S and p.N642H) leads to increased activation of STAT5 transcriptional activity (A, bar graph; n = 3 for each mutant protein in separate experiments; asterisk indicates P < .05; dagger indicates P < .001) and increased phosphorylation of STAT5B (B, western blot; arrowhead indicates exogenous STAT5B, arrow indicates endogenous STAT5B; normalized densitometric pSTAT5/STAT5 ratios are indicated) in HeLa cells. STAT5B p.P267A represents a germline polymorphism. (C) Cytokine-independent cell proliferation in the presence of mutant p.T628S STAT5B protein in the cytokine-dependent Ba/F3 cell line cultured in the absence of growth factors (n = 6; asterisk indicates P < .01). (D) Enhanced colony-forming capacity of STAT5B p.N642H mutant in Jurkat T cells (n = 3; asterisk indicates P < .01).
IL2RG-JAK1-JAK3-STAT5B mutations induce constitutive STAT5 hyperphosphorylation and oncogenic transformation
We further assessed pSTAT5 expression by western blot analysis in cells engineered to express the mutant IL2RG, JAK1, JAK3, and STAT5B proteins (Figure 4B). This analysis confirmed constitutive hyperphosphorylation of STAT5 in each case (1.6- to 173-fold normalized increase in pSTAT5/STAT5 ratio). Moreover, expression of the novel STAT5B p.T628S mutant protein led to increased cell proliferation and interleukin-3–independent growth in Ba/F3 cells (Figure 4C). In addition, STAT5B p.N642H significantly increased colony-forming capacity in the Jurkat T-cell line (Figure 4D). Altogether, these data indicate that the IL2RG, JAK1, JAK3, and STAT5B mutations hyperactivate STAT5 signaling, leading to enhanced, cytokine-independent cell proliferation.
Primary T-PLL cells exhibit constitutive expression of pSTAT5
We next sought to determine the functional consequences of IL2RG-JAK-STAT mutations in primary T-PLL cells. To this end, we first performed immunofluorescence microscopy on primary T-PLL cases harboring IL2RG, JAK3, or STAT5B mutations to assess total cellular levels and nuclear localization of pSTAT5 protein. As shown in Figure 5A, primary leukemic cells from T-PLL 25 and T-PLL 38 demonstrate increased cytoplasmic and nuclear expression of pSTAT5, indicating a common mechanism of pathogenesis in IL2RG-, JAK-, and STAT5B-mutated T-PLL cases. The observation of elevated cytoplasmic levels of pSTAT5 associated with disease development has previously been reported myeloid leukemias.46
Pimozide treatment of primary T-PLL cells to target increased pSTAT5B levels leads to reduced tumor cell growth and apoptosis. (A) Nuclear and cytoplasmic pY699 phosphorylated STAT5 in primary T-PLL samples by immunofluorescence microscopy (representative data are shown for 2 primary T-PLL cases [T-PLL25 and T-PLL38]). (B-C) Effects on HUT78 (pSTAT5-positive) viability following treatment with the JAK3 inhibitor tofacitinib (B; n = 3, P = .1144) and the STAT5 inhibitor pimozide (C; n = 3, asterisk indicates P < .0001). (D-F) Pharmacologic inhibition of pSTAT5 with pimozide leads to decreased viability (D) and diminished pSTAT5 levels (E) in primary T-PLL samples (n = 3 independent replicates for experiments in D; asterisks indicate P < .005; representative data are shown for T-PLL 25 in panel E). HH (pSTAT5 negative) is used as negative control, whereas SUDHL-1 (pSTAT5 positive) is used as a positive control. (F) A pathway diagram illustrates the interaction of IL2RG, JAK1, JAK3, and STAT5B during IL-2 cytokine activation. Cytokine binding to the extracellular portion of membrane-associated IL-2 receptors induces conformational change in the intracellular portion. Associated JAK nonreceptor tyrosine kinases autophosphorylate, leading to STAT recruitment and activation through tyrosine phosphorylation. Activated STAT proteins then dimerize and translocate to the nucleus to regulate transcription of numerous genes involved in differentiation, proliferation, and survival. Mutated components of the IL2R-JAK1-JAK3-STAT5B pathway are highlighted in green. Pimozide treatment inhibits STAT5B phosphorylation, limiting downstream transcriptional activation initiated by mutations in cytokine receptor/JAK-STAT proteins.
Pimozide treatment of primary T-PLL cells to target increased pSTAT5B levels leads to reduced tumor cell growth and apoptosis. (A) Nuclear and cytoplasmic pY699 phosphorylated STAT5 in primary T-PLL samples by immunofluorescence microscopy (representative data are shown for 2 primary T-PLL cases [T-PLL25 and T-PLL38]). (B-C) Effects on HUT78 (pSTAT5-positive) viability following treatment with the JAK3 inhibitor tofacitinib (B; n = 3, P = .1144) and the STAT5 inhibitor pimozide (C; n = 3, asterisk indicates P < .0001). (D-F) Pharmacologic inhibition of pSTAT5 with pimozide leads to decreased viability (D) and diminished pSTAT5 levels (E) in primary T-PLL samples (n = 3 independent replicates for experiments in D; asterisks indicate P < .005; representative data are shown for T-PLL 25 in panel E). HH (pSTAT5 negative) is used as negative control, whereas SUDHL-1 (pSTAT5 positive) is used as a positive control. (F) A pathway diagram illustrates the interaction of IL2RG, JAK1, JAK3, and STAT5B during IL-2 cytokine activation. Cytokine binding to the extracellular portion of membrane-associated IL-2 receptors induces conformational change in the intracellular portion. Associated JAK nonreceptor tyrosine kinases autophosphorylate, leading to STAT recruitment and activation through tyrosine phosphorylation. Activated STAT proteins then dimerize and translocate to the nucleus to regulate transcription of numerous genes involved in differentiation, proliferation, and survival. Mutated components of the IL2R-JAK1-JAK3-STAT5B pathway are highlighted in green. Pimozide treatment inhibits STAT5B phosphorylation, limiting downstream transcriptional activation initiated by mutations in cytokine receptor/JAK-STAT proteins.
Pharmacologic inhibition of STAT5 in primary T-PLL cells results in cell death
To specifically target the JAK-STAT pathway in T-PLL, we selected STAT5 as a rational candidate based on the novelty of STAT5B mutations in T-PLL because STAT5B mutations were the most common JAK-STAT pathway alterations observed in T-PLL (47.4% of all JAK-STAT mutations); because of the convergence of IL2RG, JAK1, and JAK3 signaling on STAT5 activation; because of the critical role of STAT5 in cytokine-induced peripheral T-cell proliferation47 ; and because of the coexistence of JAK1, JAK3, and STAT5B mutations in some T-PLL cases (eg, PLL_index2 in Table 2; 1 of 41 JAK-STAT-mutated T-PLL samples, 2.4%), which would render the tumor cells resistant to treatment with either JAK1 or JAK3 kinase inhibitors. Indeed, treatment of mature T-cell leukemia–derived HUT78 cells, which harbor mutations in both JAK1 and JAK3 (supplemental Figure 5), with the specific JAK3 inhibitor tofacitinib did not result in selective tumor cell killing as compared with JAK-STAT wild-type HH cells (Figure 5B). We therefore investigated the ability of a selective STAT5 inhibitor, pimozide, which was previously shown to be active in myeloid leukemia,48 to mediate specific killing effects on STAT5-activated cells by assessing cell proliferation in pimozide-treated HUT78 cells. In contrast to our observations with the JAK3 inhibitor, treatment with pimozide led to a specific and profound reduction in cell proliferation of HUT78 cells (red lines) as compared with HH cells (Figure 5B, black lines). Next, we sought to determine the effect of pimozide-mediated STAT5 inhibition on primary T-PLL cells. Pimozide treatment of cultured primary T-PLL cells resulted in significant and specific reduction of cell proliferation (Figure 5C) and viability (Figure 5D) with attendant reduction in pSTAT5 levels (Figure 5E). The effects of these mutations and of pharmacologic targeting of the IL2RG-JAK1-JAK3-STAT5B pathway with pimozide are diagrammed in Figure 5F. Taken together, these data indicate that pharmacologic inhibition of activated STAT5 results in tumor cell death in IL2RG-JAK-STAT–mutated primary T-PLL tumor cells.
Chronology of acquisition and impact of JAK-STAT mutations on clinical outcome
To address the possibility that mutations in JAK-STAT family members occurred as a result of chemotherapy, we evaluated the clinical records associated with 49 of the 50 patients. The majority of the patient samples were collected prior to institution of therapy (37/49 [75.5%], compared with 10/49 [20.4%] patients who had received previous therapy; in 2 cases, the chronology of therapy could not be determined unequivocally). No correlation between previous therapy and mutational status was observed (Table 2). These results may indicate that acquisition of JAK-STAT mutations may contribute to the natural evolution of T-PLL.
We assessed the effect of JAK-STAT mutations on clinical outcomes in our cohort of patients. Survival data were available for 49 patients with median follow-up of 20.3 months (range, 0.6 to 195.1 months; Table 2). Although overall survival was not statistically significant, when outcomes in patients with IL2RG-JAK1-JAK3-STAT5B mutations were compared as a group with those without mutations, we observed a trend to shorter time to death in patients specifically harboring JAK3 p.M511I mutations but not other JAK-STAT mutations (supplemental Figure 6). The median overall survival was 27.1 months (95% confidence interval, 20.0 to 42.3) in all patients, whereas the median overall survival in patients harboring the p.M511I mutation was 15.1 months (95% confidence interval, 3.4 to 94.3 months).
Discussion
In the present study, we have provided for the first time a comprehensive portrait of the mutational landscape of T-PLL, an aggressive T-cell malignancy refractory to conventional chemotherapy. Our analysis determined that T-PLL genomes were characterized by significant genomic complexity, including known alterations affecting the TCL1A/B or MTCP1 loci juxtaposed to the TRA locus, the ATM locus, and abnormalities of chromosome 8, including isochromosome 8. WGS and WES revealed mutations or alterations in a number of genes, including those known to be important to T-PLL pathogenesis such as ATM as well as others whose role in T-PLL had not been previously reported such as CHEK2, EZH2, and FBW10, suggesting DNA repair, epigenetic transcriptional regulation, and proteasomal degradation pathways may play an unappreciated role in T-PLL.
Strikingly, our analysis also uncovered a prominent role for mutational activation of JAK-STAT signaling in T-PLL through stereotyped, highly recurrent, largely mutually exclusive gain-of-function mutations in IL2RG (2.0%), JAK1 (8.0%), JAK3 (30.0%), and STAT5B (36.0%). These mutated residues were found to cluster along the linear axis of each gene and to encode amino acids that cluster in 3-dimensional space in domains critical to regulation of signaling activity (such as the pseudokinase domains of JAK1 and JAK3 and the SH2 domain of STAT5B). Moreover, we demonstrate using primary T-PLL cells that the IL2RG, JAK1, JAK3, and STAT5B mutations lead to constitutive STAT5 signaling that can be abrogated via specific STAT5 inhibition with the small-molecule inhibitor pimozide. Taken together, these findings indicate that deregulated JAK-STAT signaling may provide a strong oncogenic stimulus that contributes to the pathogenesis and evolution of T-PLL.
IL2RG encodes the common γ-chain receptor (γc, CD132), a critical regulator of interleukin-2 (IL-2)–induced T-cell signaling and growth but common to many cytokine receptors including IL2R, IL4R, IL7R, IL9R, IL15R, and IL21R.49,50 To our knowledge, our study represents the first report of somatic, gain-of-function mutation in IL2RG in any human malignancy. The significance of this finding is not yet clear, because only 1 patient in this series was identified to have a mutated IL2RG gene. Moreover, whereas low-frequency STAT5B mutations have been reported in <2% of T-large granulocytic leukemias,45 we report high-frequency STAT5B mutations (18/50; 36.0%), which is a novel finding in T-PLL.
In summary, we present the mutational landscape of a large cohort of 50 primary T-PLL cases representing the first such study combining comprehensive genome-wide analysis. In addition to novel alterations affecting epigenetic regulators (EZH2) and DNA repair/checkpoint proteins (CHEK2), we describe a prominent role for activating mutations affecting IL2RG-JAK1-JAK3-STAT5B. Our finding of mutations in genes of the JAK-STAT signaling pathway has therapeutic relevance for patients with T-PLL. US Food and Drug Administration–approved small-molecule inhibitors targeting aberrantly activated JAK-STAT signaling are available for treatment of other hematopoietic malignancies and have been safely and successfully used in clinical settings. Our data suggest that patients with T-PLL could benefit from inhibitors targeting the JAK-STAT pathway, either alone or in combination with current therapies. Our finding of recurrent alteration in JAK-STAT pathway genes represents a promising opportunity to provide more efficacious and less toxic treatment of patients with T-PLL.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Arul Chinnaiyan and Jay Hess for reading the manuscript and providing helpful suggestions; Farah Keyoumarsi for histology and specimen acquisition; Sountharia Rajendran, Nazeena Alvi, Martin Arlt, and Thomas Glover for assistance with aCGH analysis; Ashwini Bhasi and James D. Cavalcoli for assistance with exome bioinformatics analysis; Diane Roulston for providing clinical fluorescence in situ hybridization data; Lloyd M. Stoolman for assistance with clinical sample acquisition; and Constance Eaves, Brendan Tarrier, and Robert Lyons for whole-exome and Sanger resequencing core services.
This work was supported in part by National Institutes of Health, National Cancer Institute grants R01DE119249, R01CA136905 (K.E.J.), R01CA140806 (M.S.L.), and the Department of Pathology at the University of Michigan. M.H. receives support from the German Jose Carreras Leukemia Foundation and the German Research Foundation under HE-3553/4-1 as part of the collaborative research group Consortium for TCR-mediated regulation and oncogenesis in lymphomas of T-cells ‘CONTROL-T’.
Authorship
Contribution: M.J.K., T.V., D.R., A.A.S., M.S.L., and K.S.J.E.-J. designed the study; M.S.L. and K.S.J.E.-J. supervised and oversaw the project; M.J.K. performed WGS and WES bioinformatics analysis with assistance from A.B., B.L.B., J.D.C., B.L., J.Z.L., and A.B.O. and wrote the manuscript with K.S.J.E.-J.; M.J.K. and T.V. prepared all samples for Sanger resequencing, WES, and aCGH and performed and analyzed Sanger resequencing data and aCGH experiments with assistance from A.A.S.; D.R. performed primary cell culture experiments with STAT5 inhibitors; F.C. prepared samples for genomic sequencing and performed fluorescence microscopy for pSTAT5 immunocytochemistry; T.V. and A.A.S. performed functional studies investigating biochemical and in vitro consequences of somatic mutations identified in this study; L.Z. performed statistical analysis of patient outcomes; and A.S., M.H., R.N.M., L.J.M., and N.G.B. provided clinical samples and/or clinical data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kojo S. J. Elenitoba-Johnson, Department of Pathology, University of Michigan Medical School, A. Alfred Taubman Biomedical Science Research Building, 2037 BSRB, 109 Zina Pitcher Pl, Ann Arbor, MI 48109; e-mail: kojoelen@umich.edu; and Megan S. Lim, Department of Pathology, University of Michigan Medical School, A. Alfred Taubman Biomedical Science Research Building, 2037 BSRB, 109 Zina Pitcher Pl, Ann Arbor, MI 48109; e-mail: meganlim@umich.edu.
References
Author notes
M.J.K., T.V., D.R., and A.A.S. contributed equally to this study.

![Figure 5. Pimozide treatment of primary T-PLL cells to target increased pSTAT5B levels leads to reduced tumor cell growth and apoptosis. (A) Nuclear and cytoplasmic pY699 phosphorylated STAT5 in primary T-PLL samples by immunofluorescence microscopy (representative data are shown for 2 primary T-PLL cases [T-PLL25 and T-PLL38]). (B-C) Effects on HUT78 (pSTAT5-positive) viability following treatment with the JAK3 inhibitor tofacitinib (B; n = 3, P = .1144) and the STAT5 inhibitor pimozide (C; n = 3, asterisk indicates P < .0001). (D-F) Pharmacologic inhibition of pSTAT5 with pimozide leads to decreased viability (D) and diminished pSTAT5 levels (E) in primary T-PLL samples (n = 3 independent replicates for experiments in D; asterisks indicate P < .005; representative data are shown for T-PLL 25 in panel E). HH (pSTAT5 negative) is used as negative control, whereas SUDHL-1 (pSTAT5 positive) is used as a positive control. (F) A pathway diagram illustrates the interaction of IL2RG, JAK1, JAK3, and STAT5B during IL-2 cytokine activation. Cytokine binding to the extracellular portion of membrane-associated IL-2 receptors induces conformational change in the intracellular portion. Associated JAK nonreceptor tyrosine kinases autophosphorylate, leading to STAT recruitment and activation through tyrosine phosphorylation. Activated STAT proteins then dimerize and translocate to the nucleus to regulate transcription of numerous genes involved in differentiation, proliferation, and survival. Mutated components of the IL2R-JAK1-JAK3-STAT5B pathway are highlighted in green. Pimozide treatment inhibits STAT5B phosphorylation, limiting downstream transcriptional activation initiated by mutations in cytokine receptor/JAK-STAT proteins.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/9/10.1182_blood-2014-03-559542/4/m_1460f5.jpeg?Expires=1770017020&Signature=bWU5NOOs~bp7USz8S72ug1YwZ9FibS-CIO2bgevyVXRxShWWwCeKWfFl4kfPAGQpt0tlCRrnTRSUOaw-CnQ3Pek4sHf2MZV0PevGyom-BrCdHKXwBaxOLOas2dvpr8WtxRFdQfanO0aPkCKjiT3QT5aRBPJ70dsPG4tCw2oEOjHxh1qUQb94AmrtACUj2Aqpu94Uy5me-GDVGKxCr6dtCcHQ69O3C1ecS6ooX9dFaW2toxQFZfcgOtoU6wBGALGzg5dvgb7DwdU-YbejufBZUluZGsRYt7QHArKOkSUx3uJcIRZC8r9NGvQ656SvqXk1RwkO7kXcbkjXFrcu3ER1sA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)