Key Points
We established hypoxia-resistant cells that can mimic in vivo conditions of hypoxic bone marrow.
Exosomal miR-135b derived from these cell lines enhanced endothelial tube formation under hypoxia via the HIF-FIH signaling pathway.
Abstract
Exosomes are small endosome-derived vesicles containing a wide range of functional proteins, mRNA, and miRNA. Exosomal miRNA from cancer cells helps modulate the microenvironment. In multiple myeloma (MM), the massive proliferation of malignant plasma cells causes hypoxia. To date, the majority of in vitro hypoxia studies of cancer cells have used acute hypoxic exposure (3-24 hours). Thus, we attempted to clarify the role of MM-derived exosomes in hypoxic bone marrow by using MM cells grown continuously in vitro under chronic hypoxia (hypoxia-resistant MM [HR-MM] cells). The HR-MM cells produced more exosomes than the parental cells under normoxia or acute hypoxia conditions, and miR-135b was significantly upregulated in exosomes from HR-MM cells. Exosomal miR-135b directly suppressed its target factor–inhibiting hypoxia-inducible factor 1 (FIH-1) in endothelial cells. Finally, exosomal miR-135b from HR-MM cells enhanced endothelial tube formation under hypoxia via the HIF-FIH signaling pathway. This in vitro HR myeloma cell model will be useful for investigating MM cell–endothelial cell interactions under hypoxic conditions, which may mimic the in vivo bone marrow microenvironment. Although tumor angiogenesis is regulated by various factors, exosomal miR-135b may be a target for controlling MM angiogenesis.
Introduction
Multiple myeloma (MM) is a unique B-cell neoplasm characterized by the accumulation of clonal malignant plasma cells in the bone marrow (BM).1,2 The massive number of plasma cells usually disseminates into multiple bone lesions that are located far from the primary lesion, much like cancer metastasis. The molecular mechanism by which a primary myeloma lesion progresses to multiple lesions has not been fully elucidated. Although autologous stem cell transplantation combined with chemotherapeutic agents such as thalidomide, lenalidomide, and bortezomib can significantly improve response rates and the prognosis of MM patients,3-5 MM remains incurable for the majority of patients because of relapse.6,7
Hypoxia is an important element of the cancer microenvironment and is known to be associated with metastasis. Under hypoxia, cancer cells secrete substances that modulate their hostile microenvironment to promote tumor angiogenesis.8-10 Aberrant angiogenesis has been reported in MM-infiltrated BM,11-13 and increased angiogenic activity is associated with endothelial activation, increased capillary permeability, and hyperperfusion.14-16 Evidence suggests that MM cells promote angiogenic activity via hypoxia-inducible factor (HIF)-1α, a key transcription factor of hypoxia, leading to the overproduction of angiogenic cytokines such as vascular endothelial growth factor (VEGF),17 angiopoietin-1,18 and osteopontin.19
In addition to conventional signaling pathways responding to hypoxia (ie, direct cell-cell contact or VEGF signaling),10 our group and others have shown that exosomes, small endosome-derived vesicles containing a wide range of functional proteins, mRNA, and miRNA, from hypoxic cancer cells help to modulate the microenvironment without contacting the surrounding noncancer cells.20 Previous studies demonstrated that oxygen tension in MM-infiltrated BM was lower than in normal BM, which is already hypoxic in nature.21 The massive proliferation of MM cells produces hypoxic conditions in the tumor, which may lead to more rapid cell proliferation, drug resistance, and angiogenesis.11,22 However, little is known about how hypoxia affects the biological properties of MM cells in vivo. Previous studies using a human tumor syngeneic mouse model (the 5T33M mouse MM model) demonstrated that myelomatous BM is more hypoxic than normal BM.21,23,24 In contrast to those in vivo models, the majority of in vitro hypoxia studies of cancer cells have used acute hypoxic exposure (3-24 hours).
To clarify the role of MM-derived exosomes in hypoxic BM, we established an in vitro chronic hypoxia model using MM cells that show continuous growth in vitro under hypoxic conditions lasting more than 6 months (hypoxia-resistant [HR] cells). Here, we investigated the MM cell–endothelial cell interaction via miR-135b shed from MM cells under hypoxia, which may promote MM disease progression without directly contacting adjacent tissue.
Materials and methods
Cell lines and culture conditions
Human MM cell lines (RPMI8226, KMS-11, U266) and human umbilical vein endothelial cells (HUVECs) were purchased from the Human Science Research Resource Bank (Osaka, Japan) and Lonza Inc. (Allendale, NJ), respectively. See the supplemental Methods on the Blood Web site for details.
Establishment of HR-MM cell lines
Cell lines RPMI8226, KMS-11, and U266 were incubated under hypoxic conditions (1% O2) for 6 to 7 months. The sublines that survived well under long-term hypoxia were designated HR-MM cells RPMI8226-HR, KMS-11-HR, and U266-HR, respectively.
Preparation of exosomal fraction
MM cell lines were seeded at a density of 5 × 105 cells/mL and cultured for 24 hours (unless otherwise indicated) under hypoxic (1% O2) or normoxic (20% O2) conditions in serum-free AIM-V medium (Invitrogen, Carlsbad, CA). The exosomes derived from MM cells were purified by Exoquick Exosome Precipitation Solution (System Biosciences, Mountain View, CA) as described previously.25
Transmission electron microscopy
Exosomes were prepared and fixed as described previously.20 The samples were observed with a transmission electron microscope (JEM-1200EX; JEOL Ltd., Tokyo, Japan) at an acceleration voltage of 80 kV.
Nanoparticle tracking analysis of exosomes
Measurements for nanoparticle tracking analysis were performed using the Nanosight LM10 system (Nanosight, Amesbury, United Kingdom), fixed as described previously.20 The capture settings and analysis settings were performed manually according to the manufacturer’s instructions.
Tube formation assay
The formation of capillarylike structures was assessed as described previously.20 HUVECs (2 × 104 cells/well) were plated on top of Matrigel (280 µL/well) and treated with exosomes (500 µL of exosome fraction/well) derived from MM cells (2.5 × 106 cells) cultured for 24 hours under hypoxic conditions or under normoxic conditions. The total tube area was quantified as mean pixel density obtained from image analysis of 5 random microscopic fields using ImageJ software (http://rsb.info.nih.gov/nih-image/).
miRNA expression profiles
Isolation of cellular and exosomal miRNAs was performed using the miRNeasy kit (Qiagen, Hiden, Germany) as described previously.20 See the supplemental Methods for details. MiRNA profiling in both cells and exosomes was performed using a TaqMan low-density miRNA array (Applied Biosystems, Foster City, CA) according to the manufacturer’s recommendations. The relative expression of each gene was calculated by using the comparative threshold cycle (Ct) method, as described previously.25 RNU6B was used as an invariant control for the cellular miRNA. The synthetic spike control (ath-miR-159) was used as an invariant control for the exosomal miRNA.
Transfection of MM cells with Cy3-labeled pre-mir miRNA precursor and PKH67-labeled exosome transfer
Pre-mir miRNA precursor (has-miR-210 or has-miR-135b; Ambion, Austin, TX) was labeled with Label IT siRNA Tracker Cy3 kit (Mirus, Madison, WI) according to the manufacturer’s instructions. The PKH67-labeled exosomes including Cy3-miR-210 or Cy3-miR-135b were collected as described previously.20
miRNA target reporter luciferase assay
Synthetic oligonucleotides bearing the miR-135b binding sequence (5′-TCACATAGGAATGAAAAGCCATA-3′) or factor-inhibiting hypoxia-inducible factor 1 (FIH-1) 3′-UTR with the miR-135b complementary binding site (5′-TTAGATAGGGTTCCAACTGGGCCTACAAGCTCAAGCCATACATAAAAGGACCTTGGG-3′) were cloned into the firefly luciferase reporter plasmid pMIR-Report (Ambion) according to the manufacturer’s protocol (the seed sequence of miR-135b is shown in bold italics). We also generated the mutated sensor vector in which the seed sequence of miR-135b was changed to CCTAACGC. For measuring luciferase activity, see the supplemental Methods for details.
Knockdown and overexpression experiment of exosomal miR-135b
RPMI8226 cells (1 × 105) or RPMI8226-HR cells (1 × 105) were transfected with anti–miR-135b miRNA inhibitors (Ambion), miR-135b mimic (Ambion), or scrambled control (Negative Control 1; Ambion) or by using HiPerFect (Qiagen). The next day, after a change to fresh media, cells were cultured under hypoxic conditions for 24 hours. The exosomes derived from RPMI8226-HR1%O2/anti–miR-135b cells or RPMI8226-HR1%O2/control were purified as described.
Hypoxia response-element reporter assay
HUVECs were maintained at 50% to 60% confluence and were transduced by either 0.1 µg of pGL4.42(luc2P/HRE/Hygrp) transcription reporter or pGL4.15(luc2P/Hygro) control (Promega, Mannheim, Germany). The luciferase plasmid was cotransfected with 0.01 µg of pGL4.75(hRluc/CMV) control plasmid (Promega) as a control for the transfection efficiency. HUVECs were then treated with RPMI822620%O2-exosome, RPMI82261%O2-exosome, or RPMI8226-HR1%O2-exosome, and the luciferase activity was assessed after 48 hours with the Dual-Glo Luciferase System (Promega) according to the manufacturer’s protocol.
Immunoblotting
The cells were lysed in lysis buffer (Roche, Penzberg, Germany), and equal amounts of protein were separated on sodium dodecyl sulfate–polyacrylamide gels. The exosome pellets isolated from the same amount of culture medium (5 mL) were lysed in 200 µL of lysis buffer (Roche), and the same amounts of lysate (30 µL) were loaded in each lane of the gels. See the supplemental Methods for details of antibodies used for the immunoblots.
In vivo Matrigel plug assay
The exosome derived from RPMI8226/miR135b mimic (RPMI8226-transfected miR-135b mimic) or RPMI8226-HR/anti-miR135b (RPMI8226-HR–transfected anti–miR-135b inhibitor) were mixed with Matrigel (200 µL), and then the mixture was subcutaneously injected into nude mice (female, 8-week-old BALB/c-ν/ν; CLEA Japan, Tokyo, Japan). After 1 week, the Matrigel plugs were harvested and processed for analysis. See the supplemental Methods for details of analysis of capillary density.
Statistical analyses
Data are expressed as mean ± standard deviation (SD). Two treatment groups were compared by Mann-Whitney U test or Student t test. Multiple group comparisons were performed by analysis of variance. GraphPad Prism version 5c for Macintosh (GraphPad Inc., La Jolla, CA) was used for statistical analyses. Results were considered statistically significant when P < .05.
Results
Establishment and characterization of HR-MM cells
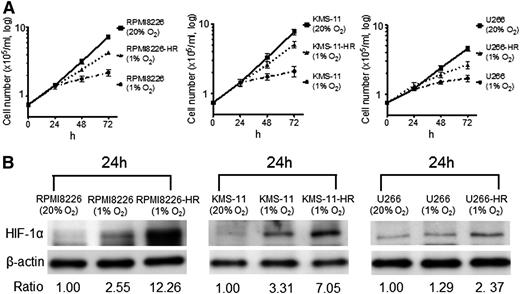
The cell growth of parental cells and cell growth of HR-MM cells were measured at 24, 48, and 72 hours (Figure 1A). Growth of the parental cells was significantly reduced by incubation under hypoxic conditions for 48 to 72 hours, but the HR-MM cells continued to grow, although the proliferation rate was reduced. The expression of HIF-1α was upregulated in parental cells under acute hypoxia for 24 hours, and upregulation of HIF-1α expression was more evident in HR-MM cells under hypoxic conditions (1% O2) compared with parental cells (Figure 1B). Furthermore, we compared the MM surface markers (CD19–, CD138+) between the parental cells and HR-MM cells by flow cytometry (supplemental Figure 1A). We could not find a remarkable change in the CD138 expression pattern, but there was a change in the KMS-11 cells. In RPMI8226 and U266 cells, there was no difference in the expression of 2 genes between parental cells and HR-MM cells (supplemental Figure 1B), and they have maintained the expression of MM cell-surface markers and transcription factors.
The response of HR-MM cells to hypoxia. (A) The growth of parental cells (RPMI8226, KMS-11, U266) and HR-MM cells (RPMI8226-HR, KMS-11-HR, U266-HR) were measured after 24, 48, and 72 hours of normoxic (20% O2) or hypoxic (1% O2) culture conditions. (B) The expression level of HIF-1α protein in parental cells and HR-MM cells cultured under 20% or 1% O2 conditions for 24 hours. Numbers below the panels represent the normalized HIF-1α expression signal by β-actin (ratio).
The response of HR-MM cells to hypoxia. (A) The growth of parental cells (RPMI8226, KMS-11, U266) and HR-MM cells (RPMI8226-HR, KMS-11-HR, U266-HR) were measured after 24, 48, and 72 hours of normoxic (20% O2) or hypoxic (1% O2) culture conditions. (B) The expression level of HIF-1α protein in parental cells and HR-MM cells cultured under 20% or 1% O2 conditions for 24 hours. Numbers below the panels represent the normalized HIF-1α expression signal by β-actin (ratio).
Characterization of exosomes derived from HR-MM cells
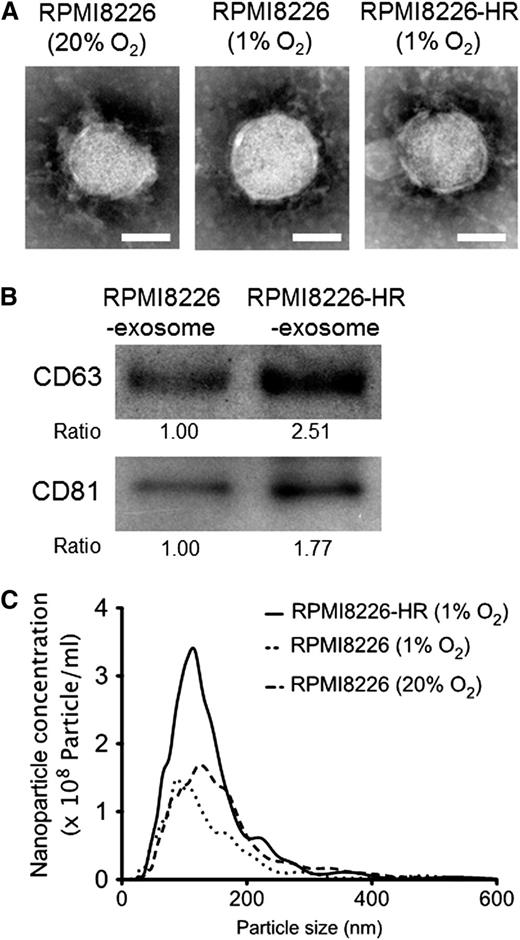
We next compared the size, ultrastructures, and quantity of exosomes between parental and HR-MM cells. Transmission electron microscopy revealed that the size of exosomes was similar between the parental cells (RPMI822620%O2 and RPMI82261%O2) and HR-MM cells (RPMI8226-HR1%O2), and each vesicle showed the classic cup-shaped appearance with the common exosomal markers (CD63 and CD81) (Figure 2A-B). There was no difference between the cell lines in the nanoparticle size distribution of exosomes, but the amount of exosomes secreted from the HR-MM cells was twofold that of the parental cells (Figure 2C). Similar results were observed in the other 2 cell lines, KMS-11-HR and U266-HR (supplemental Figure 2A-C).
Characterization of exosomes derived from HR-MM cells. (A) Transmission electron micrographs of exosomes derived from parental cells (RPMI8226 cells cultured under 20% or 1% O2 conditions) and HR-MM cells (RPMI8226-HR cells cultured under 1% O2 condition). The scale bar represents 50 nm. (B) CD63 and CD81 (common exosomal markers) immunoblots of exosomes derived from RPMI8226 and RPMI8226-HR cells. These exosomes were isolated from 5 mL of culture media of RPMI8226 (5 × 105 cells/mL) or RPMI8226-HR cells (5 × 105 cells/mL) cultured under 20% O2 or 1% O2 conditions for 24 hours, respectively, and the same amounts of exosome lysate (30 µL) were loaded in each lane of the gels. The ratio of CD63 and CD81 expression is indicated below the panels. (C) The nanoparticle concentration and size distribution of the exosomes derived from RPMI8226 cells cultured under 20% or 1% O2 conditions for 24 hours (dotted lines) and RPMI8226-HR cells cultured under 1% O2 conditions for 24 hours (solid lines).
Characterization of exosomes derived from HR-MM cells. (A) Transmission electron micrographs of exosomes derived from parental cells (RPMI8226 cells cultured under 20% or 1% O2 conditions) and HR-MM cells (RPMI8226-HR cells cultured under 1% O2 condition). The scale bar represents 50 nm. (B) CD63 and CD81 (common exosomal markers) immunoblots of exosomes derived from RPMI8226 and RPMI8226-HR cells. These exosomes were isolated from 5 mL of culture media of RPMI8226 (5 × 105 cells/mL) or RPMI8226-HR cells (5 × 105 cells/mL) cultured under 20% O2 or 1% O2 conditions for 24 hours, respectively, and the same amounts of exosome lysate (30 µL) were loaded in each lane of the gels. The ratio of CD63 and CD81 expression is indicated below the panels. (C) The nanoparticle concentration and size distribution of the exosomes derived from RPMI8226 cells cultured under 20% or 1% O2 conditions for 24 hours (dotted lines) and RPMI8226-HR cells cultured under 1% O2 conditions for 24 hours (solid lines).
Remote effect of exosomes: exosomes derived from HR-MM cells induce tube formation in normoxic HUVECs in a dose-dependent manner
To clarify how exosomes derived from HR-MM cells affect endothelial cells that are remote from tumor tissue, we performed an endothelial tube formation assay. HUVECs cultured under normoxic conditions (HUVEC20%O2) were treated with RPMI822620%O2-exosomes, RPMI82261%O2-exosomes, or RPMI8226-HR1%O2-exosomes. RPMI82261%O2-exosomes enhanced the tube formation of HUVECs, whereas RPMI822620%O2-exosomes did not (Figure 3A-B,E-F). Exosomes from HR-MM cells significantly enhanced tube formation of HUVECs compared with the control (Figure 3C,E-F). Because exosome release from HR-MM cells was twofold that of parental cells, we also tested the effect of adding half the amount of RPMI8226-HR1%O2-exosomes; we found that tube formation significantly decreased in an exosome dose–dependent manner (Figure 3C-D,F and supplemental Table 1). These findings indicate that the amount of exosomes affects tube formation in HUVECs under normoxic conditions.
Endothelial tube formation assay in normoxic or hypoxic HUVECs. The formation of tubelike structures was observed under dark field using a cell-permeable dye, Calcein AM (green). Endothelial tube formation of HUVECs cultured under normoxic conditions (20% O2) with (A) RPMI822620%O2-exosomes, (B) RPMI82261%O2-exosomes, (C) RPMI8226-HR1%O2-exosomes, (D) half the amount of RPMI8226-HR1%O2-exosomes, or (E) without exosomes (control; HUVECs20%O2). (F) Quantitative data for the tubelike structures determined by pixel density. RPMI82261%O2-exosomes and RPMI8226-HR1%O2-exosomes significantly enhanced tube formation of HUVECs20%O2 compared with control (HUVECs20%O2) (*P < .01, **P < .001). Because nanoparticle tracking analysis indicated that the HR cells secreted double the amount of exosomes compared with parental cells (Figure 2B), half the amount of RPMI8226-HR1%O2-exosomes was also assessed (RPMI8226-HR1%O2-exosomes vs RPMI8226-HR1%O2-exosome (1/2): #P < .01, Student t test). Endothelial tube formation of HUVECs cultured under hypoxic conditions (1% O2) with (G) RPMI822620%O2-exosomes, (H) RPMI82261%O2-exosomes, (I) RPMI8226-HR1%O2-exosomes, (J) half the amount of RPMI8226-HR1%O2-exosomes, or (K) without exosomes (control; HUVECs1%O2). Because HUVECs were exposed to hypoxic conditions, the induction of tube formation was enhanced in all treatments. (L) Quantitative data for the tubelike structures determined by pixel density. RPMI82261%O2-exosomes and RPMI8226HR1%O2-exosomes enhanced tube formation of HUVECs1%O2 (*P < .01, **P < .001). Even when RPMI8226-HR1%O2-exosomes were reduced to half the dose, induction of tube formation did not decrease (RPMI8226-HR1%O2-exosomes vs RPMI8226-HR1%O2-exosome [1/2]; P = .38, Student t test). Values are mean ± SD. The scale bar indicates 500 µm. exo, exosome.
Endothelial tube formation assay in normoxic or hypoxic HUVECs. The formation of tubelike structures was observed under dark field using a cell-permeable dye, Calcein AM (green). Endothelial tube formation of HUVECs cultured under normoxic conditions (20% O2) with (A) RPMI822620%O2-exosomes, (B) RPMI82261%O2-exosomes, (C) RPMI8226-HR1%O2-exosomes, (D) half the amount of RPMI8226-HR1%O2-exosomes, or (E) without exosomes (control; HUVECs20%O2). (F) Quantitative data for the tubelike structures determined by pixel density. RPMI82261%O2-exosomes and RPMI8226-HR1%O2-exosomes significantly enhanced tube formation of HUVECs20%O2 compared with control (HUVECs20%O2) (*P < .01, **P < .001). Because nanoparticle tracking analysis indicated that the HR cells secreted double the amount of exosomes compared with parental cells (Figure 2B), half the amount of RPMI8226-HR1%O2-exosomes was also assessed (RPMI8226-HR1%O2-exosomes vs RPMI8226-HR1%O2-exosome (1/2): #P < .01, Student t test). Endothelial tube formation of HUVECs cultured under hypoxic conditions (1% O2) with (G) RPMI822620%O2-exosomes, (H) RPMI82261%O2-exosomes, (I) RPMI8226-HR1%O2-exosomes, (J) half the amount of RPMI8226-HR1%O2-exosomes, or (K) without exosomes (control; HUVECs1%O2). Because HUVECs were exposed to hypoxic conditions, the induction of tube formation was enhanced in all treatments. (L) Quantitative data for the tubelike structures determined by pixel density. RPMI82261%O2-exosomes and RPMI8226HR1%O2-exosomes enhanced tube formation of HUVECs1%O2 (*P < .01, **P < .001). Even when RPMI8226-HR1%O2-exosomes were reduced to half the dose, induction of tube formation did not decrease (RPMI8226-HR1%O2-exosomes vs RPMI8226-HR1%O2-exosome [1/2]; P = .38, Student t test). Values are mean ± SD. The scale bar indicates 500 µm. exo, exosome.
Local effect of exosome: increased tube formation by exosomes from HR-MM cells is dose independent in hypoxic HUVECs
In the majority of cases, both MM and endothelial cells might be under hypoxic conditions, especially when endothelial cells are adjacent to MM cells. Therefore, we investigated the local effect of exosomes and whether exosomes derived from HR-MM cells could affect tube formation of HUVECs cultured under hypoxic conditions (HUVECs1%O2). Similar to the results using HUVECs20%O2, RPMI82261%O2-exosomes enhanced the tube formation of HUVECs1%O2 compared with the control (Figure 3H,K-L). Exosomes from HR-MM cells enhanced tube formation of HUVECs1%O2 to an even greater degree (Figure 3I,K-L and supplemental Table 1). It would be logical to assume that tube formation might decrease by half with half the amount of exosomes, but the decrease of tube formation of HUVECs was not so evident at this lower dose. This indicates that the accelerated tube formation does not simply depend on the amount of exosomes.
Cellular and exosomal miRNA profiling of HR-MM cells
We then compared miRNA profiles of cells and exosomes using the 3 parental cells and 3 HR-MM sublines (NCBI, gene expression omnibus; GSE48983). We extracted the cellular and exosomal miRNAs with a fold change of <1.5 and on the basis of Ct value (20-25, 25-30, 30-35). We arbitrarily subdivided miRNA as follows: (1) miRNAs upregulated in acute hypoxia and HR cells, (2) miRNAs upregulated in only acute hypoxia, and (3) miRNAs upregulated in only HR cells (Tables 1-3). The upregulation of miR-210 was found in acute hypoxia and HR cells in all cell lines. In contrast, we could not find any HR-specific miRNA universally expressed in the 3 cell lines. Several miRNAs were HR specific in at least 2 cell lines: miR-425 and miR-135b (RPMI8226-HR and KMS-11-HR cells); miR-335, miR-328, and miR-200c (KMS-11-HR and U266-HR cells); and miR-223 (RPMI8226-HR and U266-HR cells) (Tables 1-3). Among them, the expression level of miR-135b was as high as that of miR-210. Therefore, we investigated miR-135b further.
Cellular and exosomal miRNA profiles of RPMI8226-HR cells
| Ct value . | miRNA . | (1) Acute hypoxia−, RPMI8226-HR↑ . | miRNA . | (2) Acute hypoxia↑, RPMI8226-HR↑ . | ||
|---|---|---|---|---|---|---|
| Fold change (vs parental cell) . | Fold change (vs parental cell) . | |||||
| Exosome . | Cell . | Exosome . | Cell . | |||
| 20-25 | miR-425 | 2.1558 | 2.4911 | miR-210 | 17.486 | 23.9463 |
| miR-27a | 1.7622 | 2.7311 | ||||
| miR-27b | 2.0682 | 4.4697 | ||||
| miR-223 | 2.671 | 6.33 | ||||
| miR-135a | 16.991 | 18.1049 | ||||
| miR-324-3p | 1.5767 | 1.8605 | ||||
| miR-361-5p | 1.71 | 3.4489 | ||||
| miR-383 | 4.1609 | 10.8108 | ||||
| miR-551b | 3.6555 | 2.5412 | ||||
| 25-30 | miR-140-3p | 2.1908 | 2.5292 | |||
| miR-500 | 2.2111 | 4.8255 | ||||
| miR-135b | 7.9551 | 21.8038 | ||||
| miR-483-5p | 2.5079 | 1.6511 | ||||
| miR-362-5p | 1.747 | 2.7095 | ||||
| Ct value . | miRNA . | (1) Acute hypoxia−, RPMI8226-HR↑ . | miRNA . | (2) Acute hypoxia↑, RPMI8226-HR↑ . | ||
|---|---|---|---|---|---|---|
| Fold change (vs parental cell) . | Fold change (vs parental cell) . | |||||
| Exosome . | Cell . | Exosome . | Cell . | |||
| 20-25 | miR-425 | 2.1558 | 2.4911 | miR-210 | 17.486 | 23.9463 |
| miR-27a | 1.7622 | 2.7311 | ||||
| miR-27b | 2.0682 | 4.4697 | ||||
| miR-223 | 2.671 | 6.33 | ||||
| miR-135a | 16.991 | 18.1049 | ||||
| miR-324-3p | 1.5767 | 1.8605 | ||||
| miR-361-5p | 1.71 | 3.4489 | ||||
| miR-383 | 4.1609 | 10.8108 | ||||
| miR-551b | 3.6555 | 2.5412 | ||||
| 25-30 | miR-140-3p | 2.1908 | 2.5292 | |||
| miR-500 | 2.2111 | 4.8255 | ||||
| miR-135b | 7.9551 | 21.8038 | ||||
| miR-483-5p | 2.5079 | 1.6511 | ||||
| miR-362-5p | 1.747 | 2.7095 | ||||
After the cellular and exosomal miRNAs were extracted by a fold change of <1.5 and Ct value, they were subdivided into 2 groups: (1) miRNAs upregulated only in HR-MM cells (Acute hypoxia−, RPMI8226-HR↑), and (2) miRNAs upregulated in both acute hypoxia and HR-MM cells (Acute hypoxia↑, RPMI8226-HR↑).
Cellular and exosomal miRNA profiles of KMS-11-HR cells
| Ct value . | miRNA . | (1) Acute hypoxia−, KMS-11-HR↑ . | miRNA . | (2) Acute hypoxia↑, KMS-11-HR↑ . | ||
|---|---|---|---|---|---|---|
| Fold change (vs parental cell) . | Fold change (vs parental cell) . | |||||
| Exosome . | Cell . | Exosome . | Cell . | |||
| 25-30 | miR-425 | 1.7072 | 2.196 | miR-210 | 13.9761 | 16.5659 |
| miR-335 | 6.5755 | 7.0668 | ||||
| 30-35 | miR-132 | 1.9641 | 5.6163 | |||
| miR-518b | 1.6252 | 3.0374 | ||||
| miR-328 | 1.9408 | 4.8109 | ||||
| miR-200c | 2.3988 | 3.1656 | ||||
| miR-331-5p | 1.7919 | 2.0219 | ||||
| miR-135b | 5.9424 | 2.0893 | ||||
| Ct value . | miRNA . | (1) Acute hypoxia−, KMS-11-HR↑ . | miRNA . | (2) Acute hypoxia↑, KMS-11-HR↑ . | ||
|---|---|---|---|---|---|---|
| Fold change (vs parental cell) . | Fold change (vs parental cell) . | |||||
| Exosome . | Cell . | Exosome . | Cell . | |||
| 25-30 | miR-425 | 1.7072 | 2.196 | miR-210 | 13.9761 | 16.5659 |
| miR-335 | 6.5755 | 7.0668 | ||||
| 30-35 | miR-132 | 1.9641 | 5.6163 | |||
| miR-518b | 1.6252 | 3.0374 | ||||
| miR-328 | 1.9408 | 4.8109 | ||||
| miR-200c | 2.3988 | 3.1656 | ||||
| miR-331-5p | 1.7919 | 2.0219 | ||||
| miR-135b | 5.9424 | 2.0893 | ||||
After the cellular and exosomal miRNAs were extracted by a fold change of <1.5 and Ct value, they were subdivided into 2 groups: (1) miRNAs upregulated only in HR-MM cells (Acute hypoxia−, KMS-11-HR↑), and (2) miRNAs upregulated in both acute hypoxia and HR-MM cells (Acute hypoxia↑, KMS-11-HR↑).
Cellular and exosomal miRNA profiles of U266-HR cells
| Ct value . | miRNA . | (1) Acute hypoxia−, U266-HR↑ . | miRNA . | (2) Acute hypoxia↑, U266-HR↑ . | ||
|---|---|---|---|---|---|---|
| Fold change (vs parental cell) . | Fold change (vs parental cell) . | |||||
| Exosome . | Cell . | Exosome . | Cell . | |||
| 20-25 | miR-193b | 2.0112 | 2.6867 | miR-210 | 17.486 | 33.3568 |
| miR-138 | 2.1346 | 3.1043 | ||||
| 25-30 | miR-328 | 2.1138 | 1.9223 | |||
| let-7e | 1.6542 | 1.8644 | ||||
| miR-204 | 16.0121 | 5.2747 | ||||
| miR-223 | 2.45 | 12.9772 | ||||
| miR-335 | 11.6635 | 21.1083 | ||||
| 30-35 | miR-23a | 2.4124 | 5.4919 | |||
| miR-200c | 3.1872 | 2.8421 | ||||
| miR-185 | 2.4197 | 2.7435 | ||||
| Ct value . | miRNA . | (1) Acute hypoxia−, U266-HR↑ . | miRNA . | (2) Acute hypoxia↑, U266-HR↑ . | ||
|---|---|---|---|---|---|---|
| Fold change (vs parental cell) . | Fold change (vs parental cell) . | |||||
| Exosome . | Cell . | Exosome . | Cell . | |||
| 20-25 | miR-193b | 2.0112 | 2.6867 | miR-210 | 17.486 | 33.3568 |
| miR-138 | 2.1346 | 3.1043 | ||||
| 25-30 | miR-328 | 2.1138 | 1.9223 | |||
| let-7e | 1.6542 | 1.8644 | ||||
| miR-204 | 16.0121 | 5.2747 | ||||
| miR-223 | 2.45 | 12.9772 | ||||
| miR-335 | 11.6635 | 21.1083 | ||||
| 30-35 | miR-23a | 2.4124 | 5.4919 | |||
| miR-200c | 3.1872 | 2.8421 | ||||
| miR-185 | 2.4197 | 2.7435 | ||||
After the cellular and exosomal miRNAs were extracted by a fold change of <1.5 and Ct value, they were subdivided into 2 groups: (1) miRNAs upregulated only in HR-MM cells (Acute hypoxia−, U266-HR↑), and (2) miRNAs upregulated in both acute hypoxia and HR-MM cells (Acute hypoxia↑, U266-HR↑).
Kinetics of miR-210 and miR-135b in hypoxia
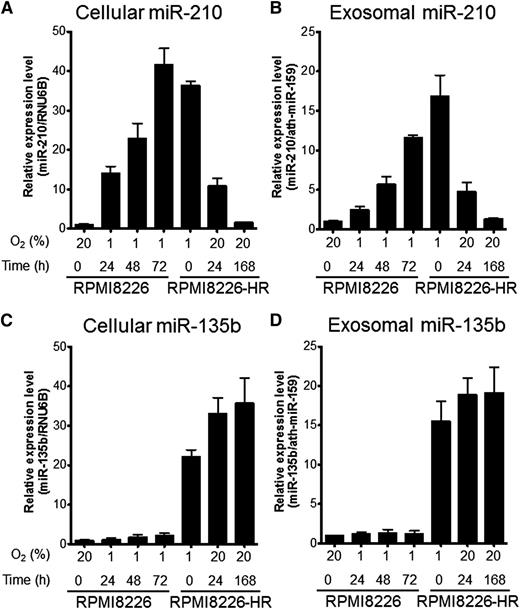
We measured the intracellular or exosomal miR-210 and miR-135b expressions of RPMI8226 and RPMI8226-HR with real-time reverse-transcription polymerase chain reaction. We found different miRNA kinetics of miR-210 and miR-135b during culture under hypoxic conditions. In parental cells, the expression of intracellular and exosomal miR-210 was not found under normoxic conditions, whereas miR-210 expression was upregulated soon after hypoxic exposure (1 hour) and then increased gradually until 72 hours (Figure 4A-B). In HR-MM cells, high miR-210 expression was maintained in hypoxic culture, but the expression gradually disappeared over the course of 1 week in culture under normoxic conditions (Figure 4A-B). These findings indicate that upregulated miR-210 expression is reversible and depends on the oxygen concentration. In contrast, miR-135b was barely detected in cells and exosomes of the parental line. miR-135b upregulation only occurred in HR-MM cells, and the expression level was maintained when cells were cultured under normoxic conditions for 1 week (Figure 4C-D). These results indicate that miR-210 is a universal hypoxia-responsive miRNA with transient expression, whereas miR-135b is an HR-MM cell–specific miRNA with continuous expression.
Time course of miR-210 and miR-135b expression under hypoxic conditions. Parental cells (RPMI8226) and HR-MM cells (RPMI8226-HR) were cultured under normoxic conditions (20% O2) or hypoxic conditions (1% O2) for the indicated times (0, 24, 48, or 72 hours). Cellular and exosomal miRNA expression levels were quantified by quantitative reverse-transcription polymerase chain reaction: (A) cellular miR-210, (B) exosomal miR-210, (C) cellular miR-135b, and (D) exosomal miR-135b. RNU6B and ath-miR-159 were used as the invariant control for cell and exosome, respectively. Values are means ± SD of 3 independent experiments, with each performed on different days.
Time course of miR-210 and miR-135b expression under hypoxic conditions. Parental cells (RPMI8226) and HR-MM cells (RPMI8226-HR) were cultured under normoxic conditions (20% O2) or hypoxic conditions (1% O2) for the indicated times (0, 24, 48, or 72 hours). Cellular and exosomal miRNA expression levels were quantified by quantitative reverse-transcription polymerase chain reaction: (A) cellular miR-210, (B) exosomal miR-210, (C) cellular miR-135b, and (D) exosomal miR-135b. RNU6B and ath-miR-159 were used as the invariant control for cell and exosome, respectively. Values are means ± SD of 3 independent experiments, with each performed on different days.
Exosomal miR-135b derived from HR-MM cells regulates the target gene in HUVECs
We visualized the transport of exosomal miR-135b derived from RPMI8226-HR cells into HUVECs transfected with β-gal control plasmid (HUVECs/β-gal) using the method modified from our previous report.25 After incubation with PKH67-labeled exosomes derived from RPMI8226-HR/Cy3-miR-135b cells, the Cy3-miR-135b signals and PKH67 signals were colocalized in the cytoplasm of HUVECs/β-gal (Figure 5A-F). We performed a luciferase reporter assay to assess whether the exogenous miR-135b via exosomal transport could function like endogenous miRNA in endothelial cells. When HUVECs/β-gal transduced with the reporter plasmid–containing complementary miR-135b binding site (sensor vector, HUVECs/LUC/β-gal) were incubated with RPMI8226-HR1%O2-exosomes, firefly luciferase activity was significantly reduced compared with the control (HUVECs only; **P < .01) (Figure 5G). In contrast, the RPMI8226-HR1%O2-exosomes did not reduce the luciferase activity using a mutated sensor vector of miR-135b (Figure 5G).
Transfer of miR-135b derived from HR-MM cells to HUVECs via exosomes. HUVECs/β-gal (HUVECs transfected with pMIR-reporter β-gal control vector) were cultured with PKH67-labeled exosomes derived from the parental cells (RPMI8226) transfected with Cy3-pre-miR-135b. The Cy3-miR-135b signals were detected in (A) the cytoplasm of HUVECs (red), and (B) green signals indicate PKH67-labeled exosomes. (C) Cy3-miR-135b signals are colocalized with PKH67 in HUVECs (yellow). Parts of areas in (A-C) are enlarged in (D-F), respectively. Nuclear counterstaining was performed using 4′,6-diamidino-2-phenylindole (DAPI) (blue). The scale bar indicates 10 µm. (G) Luciferase reporter vector for assessing miR-135b–specific activity contained complementary miR-135b sequences in its 3′-UTR. The normalized firefly luciferase activity was obtained by firefly luciferase activity/β-gal activity. Sensor vector: luciferase activity of HUVECs/Luc/β-gal (HUVECs cotransfected with luciferase reporter vector and β-gal control vector) treated with RPMI8226-HR1%O2-exosomes was significantly reduced compared with control (HUVECs/Luc/β-gal without exosomes) (**P < .01, n = 3). RPMI8226-HR/anti-miR135b exosomes (miR-135b–deactivated exosomes of RPMI8226-HR cells) could not reduce the luciferase activity in HUVECs (RPMI8226-HR/anti–miR135b-exo vs RPMI8226-HR/con-exo: #P < .01, Student t test, n = 3). In the mutated sensor vector, there was no difference in luciferase activity with or without exosomes. (H-I) Tube formation assay of HUVECs cultured under 1% O2 with RPMI8226-HR1%O2/control exosomes (H) and with RPMI8226-HR1%O2/anti-miR135b exosomes (I). The scale bar indicates 500 µm. (J) The tubelike structures determined by pixel density are reduced by the addition of RPMI8226-HR1%O2/anti-miR135b exosomes compared with control (*P < .05 vs control, Student t test, n = 3). Values are means ± SD of 3 independent experiments, with each performed on a different day. exo, exosome; con, control.
Transfer of miR-135b derived from HR-MM cells to HUVECs via exosomes. HUVECs/β-gal (HUVECs transfected with pMIR-reporter β-gal control vector) were cultured with PKH67-labeled exosomes derived from the parental cells (RPMI8226) transfected with Cy3-pre-miR-135b. The Cy3-miR-135b signals were detected in (A) the cytoplasm of HUVECs (red), and (B) green signals indicate PKH67-labeled exosomes. (C) Cy3-miR-135b signals are colocalized with PKH67 in HUVECs (yellow). Parts of areas in (A-C) are enlarged in (D-F), respectively. Nuclear counterstaining was performed using 4′,6-diamidino-2-phenylindole (DAPI) (blue). The scale bar indicates 10 µm. (G) Luciferase reporter vector for assessing miR-135b–specific activity contained complementary miR-135b sequences in its 3′-UTR. The normalized firefly luciferase activity was obtained by firefly luciferase activity/β-gal activity. Sensor vector: luciferase activity of HUVECs/Luc/β-gal (HUVECs cotransfected with luciferase reporter vector and β-gal control vector) treated with RPMI8226-HR1%O2-exosomes was significantly reduced compared with control (HUVECs/Luc/β-gal without exosomes) (**P < .01, n = 3). RPMI8226-HR/anti-miR135b exosomes (miR-135b–deactivated exosomes of RPMI8226-HR cells) could not reduce the luciferase activity in HUVECs (RPMI8226-HR/anti–miR135b-exo vs RPMI8226-HR/con-exo: #P < .01, Student t test, n = 3). In the mutated sensor vector, there was no difference in luciferase activity with or without exosomes. (H-I) Tube formation assay of HUVECs cultured under 1% O2 with RPMI8226-HR1%O2/control exosomes (H) and with RPMI8226-HR1%O2/anti-miR135b exosomes (I). The scale bar indicates 500 µm. (J) The tubelike structures determined by pixel density are reduced by the addition of RPMI8226-HR1%O2/anti-miR135b exosomes compared with control (*P < .05 vs control, Student t test, n = 3). Values are means ± SD of 3 independent experiments, with each performed on a different day. exo, exosome; con, control.
To investigate whether RPMI8226-HR exosomes mediated angiogenesis is directly dependent on miR-135b, we performed knockdown experiments of exosomal miR-135b. The anti–miR-135b inhibitor disrupted the activity of exosomal miR-135b; RPMI8226-HR/anti–miR135b-exosome and could not reduce the luciferase activity in HUVECs/LUC/β-gal (RPMI8226-HR/anti–miR135b-exo vs RPMI8226-HR/con-exo; #P < .01) (Figure 5G). Furthermore, the knockdown of miR-135b led to a decrease in endothelial tube formation via exosomes (RPMI8226-HR/con-exo vs RPMI8226-HR/anti–miR135b-exo; *P < .05) (Figure 5H-J). These results suggest that the RPMI8226-HR exosome–mediated angiogenesis is dependent on exosomal miR-135b.
Exosomal miR-135b enhances neovascularization in vivo
To analyze angiogenic responses to exosomes (exosomal miR-135b) derived from HR-MM cells, we performed an in vivo Matrigel plug angiogenesis assay to detect the newly-formed blood vessels in the transplanted gel plugs in nude mice (Figure 6A). The density of the neovessels line in Matrigel plugs was quantified by immunohistochemical staining with anti-mouse CD31 (Figure 6B). The plugs containing the exosomes derived from RPMI8226-HR/anti-miR135b significantly reduced the density of CD31+ neovesssels compared with control (the exosomes derived from RPMI8226-HR/con) (Figure 6C). These additional experiments provide evidence of important roles for exosomal miR-135b derived from HR-MM cells on angiogenesis in vivo.
Exosomal miR-135b enhances neovascularization in vivo. (A) Representative light microscopic photographs of Matrigel plugs harvested 1 week after subcutaneous injection into nude mice. The exosomes derived from RPMI8226, RPMI8226-HR/control, RPMI8226-HR/anti-miR135b, or phosphate-buffered saline (as a vehicle control) were mixed with growth factor–reduced Matrigel. (B) The neovasculature induced by the exosome of RPMI8226-HR cells in Matrigel was visualized by immunohistochemical staining of frozen sections with anti-mouse CD31 antibody. Representative photographs reveal abundant vasculature positively stained for CD31 (red). Nuclear counterstaining was performed using DAPI (blue). The scale bar indicates 200 µm. (C) Quantitative data for the neovessels lined in Matrigel plugs determined by pixel density. The exosome derived from RPMI8226-HR cells increased the density of vasculature positive for CD31 in Matrigel plugs compared with vehicle control (*P < .01 vs control, Student t test). The HR exosome–induced increase of neoangiogenesis was canceled by anti–miR-135b disrupting the activity of exosomal miR-135b (RPMI8226-HR/con-exo vs RPMI8226-HR/anti-miR135b-exo; #P < .05, Student t test). Values are mean ± SD. exo, exosome; con, control.
Exosomal miR-135b enhances neovascularization in vivo. (A) Representative light microscopic photographs of Matrigel plugs harvested 1 week after subcutaneous injection into nude mice. The exosomes derived from RPMI8226, RPMI8226-HR/control, RPMI8226-HR/anti-miR135b, or phosphate-buffered saline (as a vehicle control) were mixed with growth factor–reduced Matrigel. (B) The neovasculature induced by the exosome of RPMI8226-HR cells in Matrigel was visualized by immunohistochemical staining of frozen sections with anti-mouse CD31 antibody. Representative photographs reveal abundant vasculature positively stained for CD31 (red). Nuclear counterstaining was performed using DAPI (blue). The scale bar indicates 200 µm. (C) Quantitative data for the neovessels lined in Matrigel plugs determined by pixel density. The exosome derived from RPMI8226-HR cells increased the density of vasculature positive for CD31 in Matrigel plugs compared with vehicle control (*P < .01 vs control, Student t test). The HR exosome–induced increase of neoangiogenesis was canceled by anti–miR-135b disrupting the activity of exosomal miR-135b (RPMI8226-HR/con-exo vs RPMI8226-HR/anti-miR135b-exo; #P < .05, Student t test). Values are mean ± SD. exo, exosome; con, control.
Exosomal miR-135b derived from HR-MM cells regulated the HIF-1 signaling in HUVECs
We performed in silico analysis to determine possible miR-135b targets that may be responsible for its angiogenic function, specifically under hypoxia, using database resources including Targetscan and MiRanda. Seven-hundred eighteen transcripts with conserved miR-135b binding sites were predicted. Among them, we selected 2 candidates with high predicted efficacy calculated by the context scores. One is angiopoietin-2 (ANGPT2), a key regulator of angiogenesis that exerts context-dependent effects on endothelial cells, and the other is FIH-1, which is also known as hypoxic-inducible factor-1α subunit inhibitor. However, we found that only FIH-1 showed direct binding with miR-135b by luciferase reporter assay (supplemental Figure 3). To validate the direct binding between miR-135b and the FIH-1 3′-UTR region, we performed a luciferase reporter assay using the reporter plasmid containing FIH-1 3′-UTR with the miR-135b binding site. Reduction of luciferase activity from the pLuc-FIH1-3′-UTR plasmid was observed in HUVECs cultured with RPMI822620%O2/miR-135b exosome (exosomes derived from RPMI822620%O2 transfected with miR-135b mimics; 70% reduction) or in HUVECs cultured with RPMI8226-HR1%O2-exosome (30% reduction) compared with control (HUVECs only) (Figure 7A, left panel). As expected, deletion of the FIH-1 3′-UTR sequence from the construct abolished the inhibitory effects of miR-135b on luciferase activity (Figure 7A, right panel). To clarify whether FIH-1 expression levels are reduced in RPMI8226-HR cells, we compared FIH-1 between the parental cells (RPMI8226) and HR-MM cells (RPMI8226-HR) using immunoblot analysis. FIH-1 expression was reduced in RPMI8226-HR cells (supplemental Figure 4A). In contrast, FIH-1 protein was not detected in exosomes derived from RPMI8226-HR cells (supplemental Figure 4B). The reduction of FIH-1 expression levels in RPMI8226-HR cells may be induced by endogenous miR-135b, and the FIH-1 protein of RPMI8226-HR cells did not transfer to the other cells via exosomes. These results indicate that exosomal (exogenous) miR-135b interacts with the FIH-1 3′-UTR to exert translational repression in HUVECs.
Exosomal miR-135b targets FIH-1 in hypoxic HUVECs. (A) miR-135b binding sites in the FIH-1 3′-UTR were cloned into pMIR luciferase reporter vector (left, sensor vector). Identical construct mutation was generated (right, mutated vector). RPMI82261%O2/miR-135b exosomes (exosomes derived from RPMI82261%O2 transfected with miR-135b mimics), RPMI82261%O2/control exosomes (derived from RPMI82261%O2 transfected with scramble control miR), or RPMI8226HR1%O2-exosomes were treated with HUVEC/Luc/β-gal (HUVECs cotransfected with pMIR-FIH1-3′-UTR vector and β-gal control vector). Luciferase activity of the sensor vector displayed a significant decrease by treatment with RPMI82261%O2/miR-135b exosome (**P < .01, Student t test, n = 3) and RPMI8226-HR1%O2-exosome (*P < .05, Student t test, n = 3) compared with the control (HUVECs/Luc/β-gal without exosomes). These experiments were performed in triplicate, and the results are shown as mean ± SD. (B) FIH-1 protein expression levels measured by immunoblot after treatment with RPMI822620%O2-exosome, RPMI82261%O2-exosome, or RPMI8226-HR1%O2-exosome of normoxic HUVECs (solid bars) and hypoxic HUVECs (open bars). The intensity of each band was quantified and normalized to FIH-1 expression signals by β-actin (**P < .01 vs control, Student t test, n = 3). (C) Effect of exosomes derived from HR-MM cells (RPMI8226-HR) on HIF-1–dependent transactivation of luciferase activity in normoxic HUVECs (solid bars) and hypoxic HUVECs (open bars) transfected with luciferase reporter genes linked to hypoxia response element (HRE). Luciferase activity was measured using the Dual-Glo Luciferase System (Promega). All assays were performed in triplicate. Means ± SD are shown (**P < .01 vs control; HUVECs1%O2 without exosomes, Student t test, n = 3). exo, exosome.
Exosomal miR-135b targets FIH-1 in hypoxic HUVECs. (A) miR-135b binding sites in the FIH-1 3′-UTR were cloned into pMIR luciferase reporter vector (left, sensor vector). Identical construct mutation was generated (right, mutated vector). RPMI82261%O2/miR-135b exosomes (exosomes derived from RPMI82261%O2 transfected with miR-135b mimics), RPMI82261%O2/control exosomes (derived from RPMI82261%O2 transfected with scramble control miR), or RPMI8226HR1%O2-exosomes were treated with HUVEC/Luc/β-gal (HUVECs cotransfected with pMIR-FIH1-3′-UTR vector and β-gal control vector). Luciferase activity of the sensor vector displayed a significant decrease by treatment with RPMI82261%O2/miR-135b exosome (**P < .01, Student t test, n = 3) and RPMI8226-HR1%O2-exosome (*P < .05, Student t test, n = 3) compared with the control (HUVECs/Luc/β-gal without exosomes). These experiments were performed in triplicate, and the results are shown as mean ± SD. (B) FIH-1 protein expression levels measured by immunoblot after treatment with RPMI822620%O2-exosome, RPMI82261%O2-exosome, or RPMI8226-HR1%O2-exosome of normoxic HUVECs (solid bars) and hypoxic HUVECs (open bars). The intensity of each band was quantified and normalized to FIH-1 expression signals by β-actin (**P < .01 vs control, Student t test, n = 3). (C) Effect of exosomes derived from HR-MM cells (RPMI8226-HR) on HIF-1–dependent transactivation of luciferase activity in normoxic HUVECs (solid bars) and hypoxic HUVECs (open bars) transfected with luciferase reporter genes linked to hypoxia response element (HRE). Luciferase activity was measured using the Dual-Glo Luciferase System (Promega). All assays were performed in triplicate. Means ± SD are shown (**P < .01 vs control; HUVECs1%O2 without exosomes, Student t test, n = 3). exo, exosome.
To determine whether FIH-1 expression is suppressed by adding HR-MM–derived exosomes (exogenous-miR-135b), and whether the suppression depends on the oxygen levels of HUVECs in culture, we compared FIH-1 protein expression levels in normoxic HUVECs (Figure 7B, left panel) and hypoxic HUVECs (Figure 7B, right panel). A reduction of approximately 70% of FIH-1 protein was observed in HUVECs cultured with RPMI8226-HR1%O2-exosomes compared with the control (Figure 7B). There was no significant difference between HUVECs20%O2 and HUVECs1%O2. These findings indicate that the suppression of FIH-1 by HR-MM–derived exosomes was independent of the oxygen levels of HUVECs.
Finally, we investigated the HIF transcriptional activity to clarify the involvement of the HIF-1 and FIH-1 pathways in exosomal miR-135b–mediated tube formation in HUVECs. We transfected HUVECs with a reporter plasmid containing an SV40 promoter-luciferase transcription unit downstream of a 68-bp hypoxia-response element that mediates HIF-1–dependent gene transcription. The reporter-gene expression was markedly increased in HUVECs1%O2 relative to HUVECs20%O2 (Figure 7C, right panel). In HUVECs20%O2, HIF-1 activity was barely detected before and after adding exosomes (Figure 7C, left panel). These findings strongly indicate that exosomal miR-135b accelerated HIF-1 transcriptional activity via inhibition of FIH-1.
Discussion
Hypoxic MM cells release various diffusible factors (eg, VEGF) that promote the angiogenic switch in MM.26,27 Although the hypoxic signaling pathway via VEGF by cell-cell contact has been studied in MM angiogenesis,11,17 the complexity of the hypoxic response in the MM microenvironment needs to be defined more clearly. Here, we provide evidence of exosome-mediated angiogenesis in MM cells under prolonged hypoxia.
One major barrier to investigating the effects of hypoxia on MM cells in vitro was that cells could grow only for several days under hypoxic conditions. Therefore, most studies were performed using MM cells exposed to hypoxia for 24 to 72 hours. In such cases, MM cells might respond to hypoxic stress in a manner similar to that of an ischemic attack rather than long-lasting intratumor hypoxia. Therefore, we established HR-MM cells, which can serve as a surrogate of therapy-resistant MM cells, because the biological effects of hypoxia on tumor cells are known to be resistant to anticancer chemotherapy and are known to increase the risk of tumor metastasis.28-30
In the current study, we investigated the characteristics of exosome release in HR-MM cells. We first noted that the amount of exosomes from HR-MM cells was significantly greater than that of parental cells, and a subset of exosomal miRNA might work in a dose-dependent manner. Exosomal miR-210 induced tube formation in a dose-dependent manner when HUVECs were cultured under normoxic conditions.20 One important finding is that upregulation of miR-210 is not specific to HR-MM cells. When HUVECs were cultured under hypoxic conditions, miR-210 expression in the HUVECs themselves increased. Under such conditions, certain miRNAs, other than miR-210, were likely to play more important roles, especially in local cell-cell communication in MM. We then focused on exosomal miR-135b as an miRNA specific to HR-MM cells. miR-135b was specifically upregulated in both exosomes and HR-MM cells, whereas the expression was barely detected in the parental cells, even when they were cultured under short-term hypoxia. miR-135b is an oncogenic microRNA that has been linked to the progression of several types of cancers, including colon cancer,31,32 osteosarcoma,33 and non–small-cell lung cancer.34 Some targets already reported the direct binding of miR-135b (such as SMAD5).35 To the best of our knowledge, there is no report dealing with an association between those targets and angiogenesis. Zhang et al demonstrated a parallel correlation between miR-135b and HIF-1α; miR-135b is more involved with mechanisms of hypoxic response.36 We noted that exosomal miR-135b derived from chronic HR cells functioned as a signaling factor to transmit a hypoxic response. miR-135b may also contribute to angiogenesis in anaplastic large-cell lymphoma, implying that it plays a central role in regulating several signaling pathways.36-38 In HR-MM cells, we identified the direct target of miR-135b, FIH-1, which is an asparaginyl hydroxylase enzyme binding to HIF-1α that inhibits its transactivation function.39 Exosomal miR-135b may have the ability to improve the surrounding BM microenvironment by HR-MM cells via the HIF-FIH signaling pathway.
This study has several limitations. Tumor angiogenesis is known to be regulated by various kinds of factors, including cytokines, chemokines, and growth factors derived from the BM microenvironment such that a complex mechanism may support MM disease progression and resistance to chemotherapy.40,41 Exosomes carry various molecules, including proteins, lipids, mRNAs, and miRNA. Thus, further studies are required to understand how soluble factors and/or exosomal contents affect miR-135b in the BM microenvironment in MM. There is another issue that needs to be addressed. A recent study by King et al has clearly demonstrated that hypoxia promotes the release of exosomes in breast cancer cells: the amount of exosomes increased when cells were incubated under hypoxia.42 Therefore, it is important to take into consideration the amount of exosomes released in hypoxia. In the current study, we demonstrated both the quality and quantity of exosomes from HR-MM cells; however, it is still unknown whether known members of the HIF pathway affect exosome pathways or processing. Most importantly, the clinical relevance of miR-135b expression in MM patients is still uncertain. We and others have reported miRNA expression profiles in MM, but miR-135b expression was barely detected in MM cells and plasma,43 which is consistent with our results using parental MM cell lines. In addition, we have determined the exosomal miR-135b expression level in plasma of 15 MM patients and 5 healthy volunteers. However, the expression levels of exosomal miR-135b in MM patients were significantly lower compared with those of healthy volunteers (supplemental Figure 5). The most plausible explanation for this phenomenon is that exosomal miR-135b might play a major role in local area rather than circulating plasma. For this reason, plasma miR-135b expression is not a suitable prognostic factor at present, but exosomal miR-135b might be a potent molecular target related to local tumor angiogenesis. We also investigated exosomes from the primary myeloma cells of 2 MM patients (supplemental Figure 6), and the findings indicate that at least a subset of MM patients exhibits high miR-135b expression, whereas MM patients are a highly heterogeneous group. Even when miR-135b was elevated in exosomes from MM cells of patient 2 (supplemental Figure 5), plasma miR-135b expression level was low. Furthermore, the Matrigel plug assay may provide evidence of an important role for exosomal miR-135b derived from HR-MM cells on angiogenesis in vivo (Figure 6).
In conclusion, we constructed an in vitro HR-MM cell model that reflects prolonged intratumor hypoxia. The majority of previous studies refer to acute hypoxia, which may differ somewhat from in vivo conditions. To our knowledge, this is the first report to deal with cell-cell communication via exosomes under chronic hypoxia. Using this model, we provide new evidence that hypoxia-driven accelerated tube formation is attributable to exosomal miR-135b shed from HR-MM cells by targeting the HIF-1/FIH signaling pathway.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank C. Kobayashi and Y. Yamamoto for their technical assistance.
This study was supported in part by the Private University Strategic Research-Based Support Project (S1311016) from the Ministry of Education, Culture, Sports, Science, and Technology (Tokyo, Japan); and the Kobayashi Foundation for Cancer research (Tokyo, Japan).
Authorship
Contribution: T.U., K.O., and J.H.O. designed the research and wrote the paper; T.U., H.T., and K.A. performed research and analyzed data; and S.Y. provided patient samples.
Conflict-of-interest disclosure: K.O. received research support from Celgene KK, Chugai Pharmaceutical KK, and Janssen Pharma KK.
Correspondence: Tomohiro Umezu, Tokyo Medical University, 6-7-1 Nishi-shinjuku, Shinjuku, Tokyo 160-0023, Japan; e-mail: t_umezu@tokyo-med.ac.jp.

![Figure 3. Endothelial tube formation assay in normoxic or hypoxic HUVECs. The formation of tubelike structures was observed under dark field using a cell-permeable dye, Calcein AM (green). Endothelial tube formation of HUVECs cultured under normoxic conditions (20% O2) with (A) RPMI822620%O2-exosomes, (B) RPMI82261%O2-exosomes, (C) RPMI8226-HR1%O2-exosomes, (D) half the amount of RPMI8226-HR1%O2-exosomes, or (E) without exosomes (control; HUVECs20%O2). (F) Quantitative data for the tubelike structures determined by pixel density. RPMI82261%O2-exosomes and RPMI8226-HR1%O2-exosomes significantly enhanced tube formation of HUVECs20%O2 compared with control (HUVECs20%O2) (*P < .01, **P < .001). Because nanoparticle tracking analysis indicated that the HR cells secreted double the amount of exosomes compared with parental cells (Figure 2B), half the amount of RPMI8226-HR1%O2-exosomes was also assessed (RPMI8226-HR1%O2-exosomes vs RPMI8226-HR1%O2-exosome (1/2): #P < .01, Student t test). Endothelial tube formation of HUVECs cultured under hypoxic conditions (1% O2) with (G) RPMI822620%O2-exosomes, (H) RPMI82261%O2-exosomes, (I) RPMI8226-HR1%O2-exosomes, (J) half the amount of RPMI8226-HR1%O2-exosomes, or (K) without exosomes (control; HUVECs1%O2). Because HUVECs were exposed to hypoxic conditions, the induction of tube formation was enhanced in all treatments. (L) Quantitative data for the tubelike structures determined by pixel density. RPMI82261%O2-exosomes and RPMI8226HR1%O2-exosomes enhanced tube formation of HUVECs1%O2 (*P < .01, **P < .001). Even when RPMI8226-HR1%O2-exosomes were reduced to half the dose, induction of tube formation did not decrease (RPMI8226-HR1%O2-exosomes vs RPMI8226-HR1%O2-exosome [1/2]; P = .38, Student t test). Values are mean ± SD. The scale bar indicates 500 µm. exo, exosome.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/25/10.1182_blood-2014-05-576116/4/m_3748f3.jpeg?Expires=1763474540&Signature=uDohQnpVHqDql7O5vIs28pikHgi0vxWBhra09~9n3XReHar1v689NhdYeV2Ag6UPyGa19s~5cyoz~In-3jN12CY~-45RjwWhbdlvJPv~Ge2-ZPJKUDaniEdshmj3oKng9McB82RJbIcnMTTP6Wxk98~ZzSZkBTFG86CtlE4G3u2bDfksc47t1BjmLLtBrFKudOWqFbhj7EayE3mlimGVvt1wZ2zHpBveLxw0u9lPqgdoZuxCvNZGiB4YfjtR1iBcJjsekCo4SNIkFy0n5klsn2jJ5~oWFg5y7sQiGPwSTBnFx98BEYJrjWdbW6um8PyQQ92qOfKvSMf2to9xZn-Oyg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal