Key Points
This study has identified a novel mechanism by which TF expression is posttranscriptionally regulated in macrophages.
The mechanism involves the control of mRNA stability by a cooperation between PARP-14 and TTP.
Abstract
Tissue factor (TF) (CD142) is a 47 kDa transmembrane cell surface glycoprotein that triggers the extrinsic coagulation cascade and links thrombosis with inflammation. Although macrophage TF expression is known to be regulated at the RNA level, very little is known about the mechanisms involved. Poly(adenosine 5′-diphosphate [ADP]-ribose)-polymerase (PARP)-14 belongs to a family of intracellular proteins that generate ADP-ribose posttranslational adducts. Functional screening of PARP-14–deficient macrophages mice revealed that PARP-14 deficiency leads to increased TF expression and functional activity in macrophages after challenge with bacterial lipopolysaccharide. This was related to an increase in TF messenger RNA (mRNA) stability. Ribonucleoprotein complex immunoprecipitation and biotinylated RNA pull-down assays demonstrated that PARP-14 forms a complex with the mRNA-destabilizing protein tristetraprolin (TTP) and a conserved adenylate-uridylate-rich element in the TF mRNA 3′ untranslated region. TF mRNA regulation by PARP-14 was selective, as tumor necrosis factor (TNF)α mRNA, which is also regulated by TTP, was not altered in PARP-14 deficient macrophages. Consistent with the in vitro data, TF expression and TF activity, but not TNFα expression, were increased in Parp14−/− mice in vivo. Our study provides a novel mechanism for the posttranscriptional regulation of TF expression, indicating that this is selectively regulated by PARP-14.
Introduction
Tissue factor (TF) (CD142) is a 47kDa transmembrane cell surface glycoprotein that triggers the extrinsic coagulation cascade.1 Moreover, activation of protease-activated receptors by coagulation factors links TF to inflammation.2 TF, therefore, plays a central role in diverse pathologic processes including atherosclerosis, thrombosis, sepsis, and tumor growth.3-7
Monocytes and macrophages are the predominant source of TF in myeloid cells.8-10 TF expression in these cells is low to undetectable basally, but is induced transcriptionally by inflammatory mediators, such as bacterial lipopolysaccharide (LPS).11 TF messenger RNA (mRNA) transcripts are stable over 2-hours after LPS treatment in THP-1 monocytic cells12 and in endothelial cells,13 but then decay, which leads to a time window for TF mRNA translation into protein. TF mRNA stability is regulated by a sequence at the distal end of the 3′-untranslated region (UTR) and is likely to involve 1 or more adenylate-uridylate (AU)-rich elements (AREs).14 However, the basic molecular mechanisms involved have not been described.
Tristetrapolin (TTP) is a CCCH tandem zinc finger protein that binds AREs in the 3′ UTRs of target mRNAs and recruits mRNA-degrading enzymes.15-17 Phosphorylation of TTP by MK2, a kinase activated by p38 mitogen-activated protein kinase (MAPK), leads to its inactivation and thereby stabilization of mRNA targets, whereas dephosphorylation via serine–threonine phosphatase PP2A restores its mRNA destabilizing activity.16,18,19 TTP contributes to the degradation of many mRNAs relevant to inflammation, including tumor necrosis factor (TNF)α, but little is known about whether its activity on separate mRNA targets is differentially regulated.20,21
There are at least 17 intracellular proteins containing a poly (adenosine 5′-diphosphate [ADP]-ribose) polymerase (PARP) domain.22 PARP-1, the canonical PARP protein, has been extensively studied and it is of central importance to DNA repair and transcriptional regulation.22 In contrast, the functional roles of many of the other PARP proteins are less well understood. PARP-14 (also known as ADP-ribosyltransferase diphteria toxin-like 8) is a protein (∼205 kDa) in which enzymatic function is likely to be restricted to ADP-ribosyl monotransferase activity.23 It is known to be a nuclear coactivator of signal transducer and activator of transcription-6–mediated gene transcription in B cells.24-26 Although studies to date on PARP proteins have mainly focused on their nuclear activities, PARP-14 is also expressed, along with several other PARP proteins, in the cytoplasm and may have roles in RNA regulation.24,27 Herein, we report that PARP-14 regulates TF expression at the posttranscriptional level by interacting selectively with TTP.
Materials and methods
A detailed description of all reagents and experimental procedures is provided in the supplemental Methods on the Blood Web site. Isolation and culture of mouse bone-marrow–derived macrophage (BMDM) and human peripheral blood-derived macrophages (PBM), RNA extraction, quantitative reverse-transcriptase polymerase chain reaction (RT-PCR), small interfering RNA (siRNA) knockdown, measurement of mRNA decay, cloning and mutation of TF mRNA 3′UTR, in vitro RNA transcription, protein coimmunoprecipitation, western blotting, luciferase reporter assay, and TNFα enzyme-linked immunosorbent assay were performed using standard techniques. Research was conducted in accordance with the Declaration of Helsinki.
Mice
Parp14−/− and Ttp−/− mice were generated as described and maintained as heterozygous breeding pairs.25,28 Ttp−/− mice were of mixed 129 and C57BL/6 background and Parp14−/− mice had been backcrossed onto a C57BL/6 background for 12 generations. All experiments with Ttp−/− and Parp14−/− mice were conducted using respective age- and sex-matched litter-mate wild-type (WT) progeny as controls. All in vivo procedures were covered with the United Kingdom’s Home Office approval.
TF activity assays
TF activity was measured using a validated one-step plasma recalcification clotting assay for human TF,29 with a minor adaptation for measuring mouse TF.
RIP
Ribonucleoprotein complex immunoprecipitation (RIP) assays were performed as previously described.30 Macrophage lysates were incubated with protein-G agarose beads precoated with either rabbit anti-TTP, rabbit anti–PARP-14 or normal rabbit IgG. The beads were then washed and incubated in ribonuclease-free DNase I to remove genomic DNA contamination. The beads were washed again and incubated in NT2 buffer containing 0.1% sodium dodecyl sulfate and 0.5 mg/mL proteinase K for 15 minutes at 55°C to digest the protein bound to the beads and to release protein-bound RNA. Then RNA was extracted using TRIzol reagent (Sigma-Aldrich, Gillingham, United Kingdom [UK]) and analyzed by RT-PCR. The polymerase chain reaction products were separated in a 2% agarose gel containing ethidium bromide and visualized under UV light.
RNA-biotin pull-down assays
Macrophage lysates were incubated with either the sense (test) or anti-sense (control) biotinylated transcript (2 μg) for 1 hour. Magnetic streptavidin-coated beads (Dynabeads; Invitrogen, Paisley, UK) were then added and incubated at 4°C for 1 hour. The beads were separated using magnetic separation and washed 5 times in ice cold lysis buffer. Bound proteins were then eluted by heating to 4 minutes. Pulled-down proteins were detected using sodium dodecyl sulfate-polyacrylamide gel electrophoresis, western blotting, and immunodetection according to standard methods. The anti-sense strands used in the study do not contain any AREs and therefore served as an internal control for their respective reactions. The experiment was further controlled by detecting for tubulin, which served as a marker for nonspecific protein binding.
Statistical analyses
All continuous variables were expressed as either mean ± standard error of the mean (SEM) or medians, depending on normality. Where data are expressed as mean ± SEM, the unpaired Student t test (2-tailed) was used for comparison of groups. Where the data are expressed as medians, the Mann-Whitney U test (2-tailed) was used. Statistical significance was set at P = .05.
Results
Increased TF expression in PARP-14–deficient cells
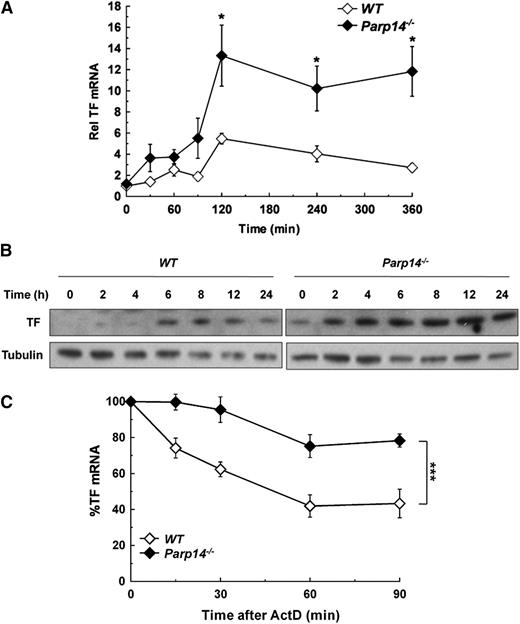
We examined the effect of PARP-14 deficiency on macrophage TF expression while functionally screening Parp14−/− mice for abnormalities relevant to inflammation and thrombosis. Figure 1A shows that TF mRNA levels were significantly increased in Parp14−/− compared with WT macrophages after stimulation with LPS (1 µg/mL). PARP-14 deficiency led to TF protein being detectable by western blot analysis in unstimulated cells and present at increased levels at each of the time-points after LPS stimulation (Figure 1B). Furthermore, absence of PARP-14 also led to significant increases in unstimulated and LPS-stimulated TF functional activity in a turbimetric clotting assay (supplemental Figure 1).
Overexpression of TF mRNA and protein in PARP-14–deficient macrophages with stabilization of TF mRNA. WT and Parp14−/− BMDM were treated with LPS (1 µg/mL) and then analyzed for TF expression at the times shown by (A) RT-PCR, with mRNA levels normalized to unstimulated WT cells (n = 5 experiments). (B) Western blot analysis, with WT and Parp14−/− lysates were run on the same gels and processed simultaneously, but separated for clarity. The figure shows representative blots of five independent experiments. (C) TF mRNA decay in WT and Parp14−/− macrophages after addition of actinomycin D (5 µg/mL) to cultures that had been treated with LPS for 2 hours (n = 6 experiments). Data in (A) and (C) are expressed as mean ± SEM. *P < .05; ***P < .001 analyzed using a 2-tailed Student t test. ActD, actinomycin D; Rel, relative.
Overexpression of TF mRNA and protein in PARP-14–deficient macrophages with stabilization of TF mRNA. WT and Parp14−/− BMDM were treated with LPS (1 µg/mL) and then analyzed for TF expression at the times shown by (A) RT-PCR, with mRNA levels normalized to unstimulated WT cells (n = 5 experiments). (B) Western blot analysis, with WT and Parp14−/− lysates were run on the same gels and processed simultaneously, but separated for clarity. The figure shows representative blots of five independent experiments. (C) TF mRNA decay in WT and Parp14−/− macrophages after addition of actinomycin D (5 µg/mL) to cultures that had been treated with LPS for 2 hours (n = 6 experiments). Data in (A) and (C) are expressed as mean ± SEM. *P < .05; ***P < .001 analyzed using a 2-tailed Student t test. ActD, actinomycin D; Rel, relative.
TF mRNA is stabilized in PARP-14–deficient cells
Previous reports using the human monocytic cell line THP-1 and endothelial cells have indicated that TF mRNA is more stable at the start of the transcriptional response to LPS than later on.12,13 We verified that this was also the case in human PBM and in mouse BMDM by adding actinomycin D (5 µg/mL) to block transcription at different times after LPS stimulation and then measuring TF mRNA decay. In human PBM, TF mRNA half-life (t1/2) dropped after 2 hours of LPS stimulation, correlating with the time (t1/2 estimated from decay over 90 minutes = 141 ± 2 minutes, 88 ± 16 minutes, and 36 ± 18 minutes at 2, 4, and 6 hours, respectively; r2 = 0.998) (supplemental Figure 2A). A similar pattern was observed with mouse BMDM (t1/2 = 52 ± 8 minutes, 26 ± 7 minutes and 15 ± 6 minutes at 2, 4, and 6 hours, respectively; r2 = 0.948) (supplemental Figure 2B). Therefore, we tested whether increased TF expression in Parp14−/− cells was related to altered mRNA stability, focusing on the 2 hour time-point after LPS stimulation. As shown in Figure 1C, TF mRNA decay was significantly delayed in Parp14−/− macrophages (t1/2 = 181 ± 11 minutes compared with 60 ± 5 minutes in WT cells (mean ± SEM; n = 6; P < .001) (Figure 1C).
TF mRNA is stabilized in TTP-deficient cells
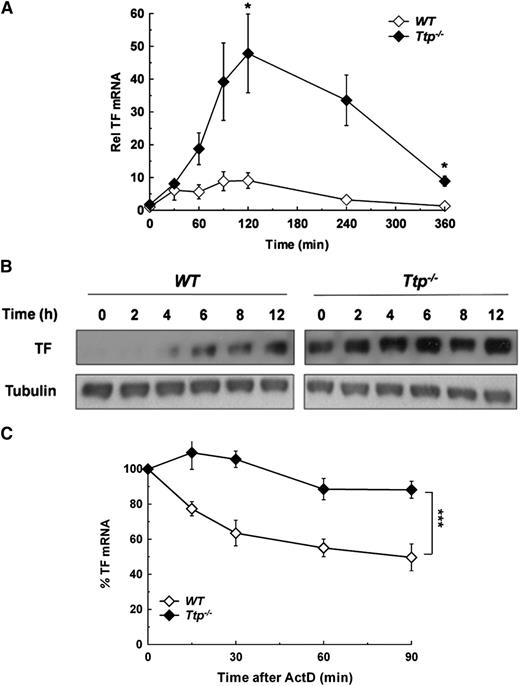
Both human and mouse TF mRNA 3′UTR have several AREs, including a 17-nucleotide sequence (AUAAUUUAUUUAAUAUA) containing a 15-nucleotide palindrome (positions 1-15) harboring 2 overlapping UUAUUUAAU nonamers (one 3′ to 5′, and the other 5′ to 3′). This is highly conserved between species and is a candidate binding site for TTP (supplemental Figure 3). To explore a parallel role for TTP in TF regulation, steady state TF mRNA and protein levels were determined in WT and TTP deficient (Ttp−/−) BMDM. As in Parp14−/− cells, TF mRNA levels were significantly increased in Ttp−/− compared with WT cells after LPS stimulation (Figure 2A), and TF protein was detectable by western blot analysis in unstimulated Ttp−/− cells and present at increased levels at each of the time-points after LPS stimulation (Figure 2B). Furthermore, TTP deficiency also significantly increased the TF activity of LPS-stimulated BMDM (supplemental Figure 4). As shown in Figure 2C, the increase in steady-state TF mRNA in Ttp−/− BMDM was associated with a significant increase in TF mRNA stability at 2 hours after LPS stimulation (t1/2 = 347 ± 62 minutes in Ttp−/− cells and 82 ± 14 minutes in WT; mean ± SEM of 3 experiments; P < .001). In line with the experiments using knockout cells, siRNA knockdown of either PARP-14 or TTP led to a significant increase in LPS-induced TF mRNA expression in human PBM (supplemental Figure 5).
Deficiency in TTP results in overexpression of TF mRNA due to mRNA stabilization. (A) WT and Ttp−/− murine BMDM were stimulated with LPS (1 µg/mL) for the durations shown. TF mRNA levels were then determined at each time point and normalized to levels in unstimulated WT macrophages (n = 3 experiments). (B) WT and Ttp−/− BMDM were stimulated with LPS (1 µg/mL) for the durations shown, after which western blotting was used for cell lysates. The blots shown are representative of 5 experiments and are all from the same gel and are processed simultaneously, with WT and Ttp−/− lanes separated for clarity. (C) WT and Ttp−/− macrophages were stimulated with LPS (1 µg/mL) for 2 hours, after which actinomycin D (5 µg/mL) was added and TF mRNA decay was assessed. All data are expressed as mean ± SEM and analyzed using a 2-tailed Student t test. NS, not significant. *P < .05; ***P < .001. ActD, actinomycin D; Rel, relative.
Deficiency in TTP results in overexpression of TF mRNA due to mRNA stabilization. (A) WT and Ttp−/− murine BMDM were stimulated with LPS (1 µg/mL) for the durations shown. TF mRNA levels were then determined at each time point and normalized to levels in unstimulated WT macrophages (n = 3 experiments). (B) WT and Ttp−/− BMDM were stimulated with LPS (1 µg/mL) for the durations shown, after which western blotting was used for cell lysates. The blots shown are representative of 5 experiments and are all from the same gel and are processed simultaneously, with WT and Ttp−/− lanes separated for clarity. (C) WT and Ttp−/− macrophages were stimulated with LPS (1 µg/mL) for 2 hours, after which actinomycin D (5 µg/mL) was added and TF mRNA decay was assessed. All data are expressed as mean ± SEM and analyzed using a 2-tailed Student t test. NS, not significant. *P < .05; ***P < .001. ActD, actinomycin D; Rel, relative.
Both anti-TTP and PARP-14 antibodies immunoprecipitate TF mRNA
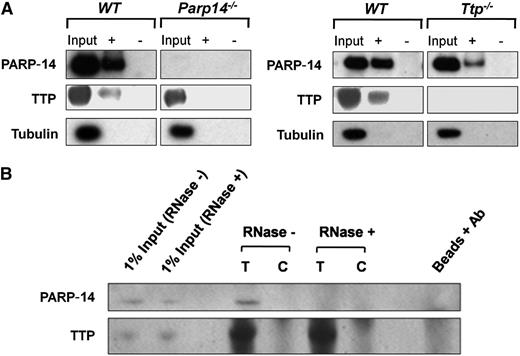
To address whether PARP-14 and/or TTP interact with the TF mRNA 3′UTR, lysates were prepared from LPS-stimulated WT, Ttp−/−, and Parp14−/− macrophages. RIP assays were then performed using anti-TTP or anti–PARP-14 antibodies or control IgG, and the precipitated mRNA amplified by RT-PCR. Figure 3A demonstrates that TF transcripts, but not hypoxanthine guanine phosphoribosyl transferase (HPRT) (or glyceraldehyde-3-phosphate dehydrogenase not shown) transcripts were pulled down from WT lysates by either anti-PARP-14 or anti-TTP but not by IgG control, indicating that both TTP and PARP-14 associate with TF mRNA. Specificity of the antibodies was demonstrated by failure of anti-TTP and anti-PARP-14 to pull down TF mRNA from Ttp−/− or Parp14−/− cell extracts respectively. Absence of bands when the assays were conducted on material pulled down from WT lysates, but without reverse transcription-excluded involvement of contaminating genomic DNA (not shown).
TTP and PARP-14 interact with a conserved ARE in TF mRNA 3′UTR. (A) ribonucleprotein complexes were immunoprecipitated by anti-TTP (T), anti–PARP-14 (P) or IgG negative control (C) from lysates of LPS-treated (1 µg/mL for 2 hours) BMDM and shown by RT-PCR to contain TF mRNA when derived from WT cells, but not when derived from Parp14−/− or Ttp−/− cells. HPRT provides a negative control mRNA for nonspecific pull-down. This figure shows representative gels from 3 independent experiments. (B) Western blot analysis of proteins isolated with streptavidin-coated magnetic beads from lysates of LPS-treated macrophages (1 µg/mL for 2 hours) incubated with biotinylated sense (+) or anti-sense (-) murine TF 3′UTR truncations. TTP and PARP-14 only associated with the sense full-length construct containing the ARE4 segment. In this and other figures, tubulin served as a negative marker for nonspecific protein binding to beads. (C) Similar western blots of proteins pulled down using WT TF 3′ UTR or mutant TF 3′ UTR with guanine substitutions in the conserved 17-nucleototide AREs in the critical ARE4 segment, containing a 15-nucleotide palindrome harboring 2 overlapping UUAUUUAAU nonamers (one 3′ to 5′ and the other 5′ to 3′: shown with dashed lines on AREWT). Intra-ARE mutation affecting both nonamers (AREMut-4) abolished TTP and PARP-14 binding. (D) Nonbiotinylated TF 3′UTR transcript (AREWT) inhibited TTP and PARP-14 binding to biotinylated TF mRNA 3′UTR, whereas the nonbiotinylated mutant TF 3′UTR transcript (AREMut-4) did not.
TTP and PARP-14 interact with a conserved ARE in TF mRNA 3′UTR. (A) ribonucleprotein complexes were immunoprecipitated by anti-TTP (T), anti–PARP-14 (P) or IgG negative control (C) from lysates of LPS-treated (1 µg/mL for 2 hours) BMDM and shown by RT-PCR to contain TF mRNA when derived from WT cells, but not when derived from Parp14−/− or Ttp−/− cells. HPRT provides a negative control mRNA for nonspecific pull-down. This figure shows representative gels from 3 independent experiments. (B) Western blot analysis of proteins isolated with streptavidin-coated magnetic beads from lysates of LPS-treated macrophages (1 µg/mL for 2 hours) incubated with biotinylated sense (+) or anti-sense (-) murine TF 3′UTR truncations. TTP and PARP-14 only associated with the sense full-length construct containing the ARE4 segment. In this and other figures, tubulin served as a negative marker for nonspecific protein binding to beads. (C) Similar western blots of proteins pulled down using WT TF 3′ UTR or mutant TF 3′ UTR with guanine substitutions in the conserved 17-nucleototide AREs in the critical ARE4 segment, containing a 15-nucleotide palindrome harboring 2 overlapping UUAUUUAAU nonamers (one 3′ to 5′ and the other 5′ to 3′: shown with dashed lines on AREWT). Intra-ARE mutation affecting both nonamers (AREMut-4) abolished TTP and PARP-14 binding. (D) Nonbiotinylated TF 3′UTR transcript (AREWT) inhibited TTP and PARP-14 binding to biotinylated TF mRNA 3′UTR, whereas the nonbiotinylated mutant TF 3′UTR transcript (AREMut-4) did not.
TTP and PARP-14 associate with TF mRNA 3′UTR via a conserved ARE
Pull-down assays were performed from lysates of LPS-stimulated BMDM using in vitro transcribed and biotinylated sense (ie, test) and anti-sense (ie, control) RNA constructs representing full-length and truncated mouse TF mRNA 3′UTR (supplemental Figure 6). Figure 3B shows that TTP and PARP-14 were each pulled down by full-length TF 3′UTR, but not by truncations lacking the conserved 17-nucleotide AREs (located in the ARE4 segment). As shown in supplemental Figure 7, TTP and PARP-14 were also pulled down by biotinylated human TF 3′UTR. Constructs of mouse TF mRNA 3′UTR containing guanine substitution mutant sequences of the conserved 17-nucleotide AREs were generated to examine the interactions with TTP and PARP-14 further (Figure 3C). Neither TTP nor PARP-14 binding was affected by AREMut-1. AREMut-2, and AREMut-3, which each had mutations in a single nonamer, reduced but did not abolish TTP and PARP-14 binding. In contrast, AREMut-4, with mutations shared by both nonamers, completely abolished binding of both proteins. The effect of the mutation in AREMut-4 was confirmed with a competition assay (Figure 3D). This showed that nonbotinylated unmutated transcripts, but not nonbiotinylated AREMut-4 transcripts, significantly reduced TTP and PARP-14 binding to the biotinylated TF mRNA 3′UTR captured by the beads. Taken together, these data substantiate the functional importance of the conserved sequence for interaction with both TTP and PARP-14.
Regulation of TF mRNA 3′ UTR with the conserved ARE in a luciferase reporter assay
As shown in supplemental Figure 3, TF mRNA 3′ UTR also has binding sites for microRNA (miR) 19a/19b (miR-A site) and miR 20a/20b/106b (miR-B site), which have been previously shown to regulate TF mRNA.31-33 To examine the relative impact of the conserved 17-nucleotide AREs compared with the miR-A and miR-B sites, we established a luciferase mRNA stability reporter assay in RAW 264.7 cells. These are of mouse macrophage origin and are known to express hypophosphorylated TTP basally without stimulation34 and to express PARP-14 (supplemental Figure 8). We tested the effect of full-length unmutated TF 3′ UTR in comparison with a panel of 3′UTR constructs with 7-nucleotide substitutions in the miR-A, miR-B, and/or ARE sites (supplemental Figure 9). As shown in Figure 4A, these substitutions in the miR-A, miR-B, or ARE sites each modestly increased luciferase activity compared with unmutated full-length TF 3′ UTR. Furthermore, combining miR-A or miR-B site substitutions, singly or together, with the ARE mutation had clear additive effects. Similar data were obtained when the guanine substititions in the ARE were restricted to the 3 in AREMut-4 (not shown). Finally, we ligated the ARE4 segment in isolation into the luciferase reporter vector to address whether it was independently able to regulate the reporter assay. As shown in Figure 4B, the ARE4 segment significantly reduced luciferase reporter activity and this was partially rescued by changing the conserved ARE sequence to AUAAgggggggAAUAUA.
The TF 3′UTR distal ARE regulates luciferase mRNA. Luciferase reporter constructs containing 3′ UTR inserts derived from the TF mRNA sequence were transfected into RAW 264.7 cells, and luciferase activity was then assayed after 16 hours. (A) Shows the effects on luciferase activity of full-length TF 3′ UTR compared with a panel of constructs with 7 guanine substitutions in the conserved ARE in the ARE4 segment (= ARE*), in the miR 19a/19b binding site (= miR-A*) or the miR 20a/20b/106b binding site (= miR-B*), alone or in combination. ARE mutation significantly increased luciferase activity, and this was augmented by the substitutions in miR-A and miR-B. P values refer to comparisons with unmutated 3′ UTR. (B) Shows the destabilizing effect of inserting the isolated ARE4 segment of the TF 3′UTR into the luciferase reporter construct, relative to the luciferase coding region with no 3′ UTR insert. Also shown is the significant rescue of luciferase activity by the 7 guanine substitution (as in ARE* in [A]). Data are mean + SEM of 4 experiments, analyzed by one-way analysis of variance. *P < .05; ***P < .001.
The TF 3′UTR distal ARE regulates luciferase mRNA. Luciferase reporter constructs containing 3′ UTR inserts derived from the TF mRNA sequence were transfected into RAW 264.7 cells, and luciferase activity was then assayed after 16 hours. (A) Shows the effects on luciferase activity of full-length TF 3′ UTR compared with a panel of constructs with 7 guanine substitutions in the conserved ARE in the ARE4 segment (= ARE*), in the miR 19a/19b binding site (= miR-A*) or the miR 20a/20b/106b binding site (= miR-B*), alone or in combination. ARE mutation significantly increased luciferase activity, and this was augmented by the substitutions in miR-A and miR-B. P values refer to comparisons with unmutated 3′ UTR. (B) Shows the destabilizing effect of inserting the isolated ARE4 segment of the TF 3′UTR into the luciferase reporter construct, relative to the luciferase coding region with no 3′ UTR insert. Also shown is the significant rescue of luciferase activity by the 7 guanine substitution (as in ARE* in [A]). Data are mean + SEM of 4 experiments, analyzed by one-way analysis of variance. *P < .05; ***P < .001.
TTP and PARP-14 combine in associating with the TF 3′UTR
Assays were performed next to determine the effect of PARP-14–deficiency on TTP interaction with TF 3′UTR and vice versa. Biotinylated TF 3′UTR transcripts were added to LPS-stimulated lysates from WT, Ttp−/−, and Parp14−/− mouse BMDM, following which RNA-protein complexes were pulled down by streptavidin-coated beads and proteins identified by western blot analysis. Figure 5A shows that pull-down of TTP by the TF 3′UTR was not detectable in the absence of PARP-14. Conversely, pull-down of PARP-14 was reduced in the absence of TTP. The interaction between TTP and PARP-14 appeared to be RNA-dependent, as PARP-14 could be coimmunoprecipitated with TTP using anti-TTP antibody, but not after treatment of the lysates with ribonuclease (RNase) (Figure 5B).
Interaction of TTP and PARP-14 with TF mRNA is interdependent. (A) Western blots of proteins isolated with streptavidin beads from lysates of LPS-treated WT, Ttp−/−, and Parp14−/− macrophages (1 µg/mL, 2 hours) incubated with sense (+) or anti-sense (-) biotinylated murine TF 3′UTR. TF 3′UTR did not pull down TTP in the absence of PARP-14 or PARP-14 in the absence of TTP. (B) Western blots of proteins immunoprecipitated from LPS-stimulated WT lysates with anti-TTP antibody (T) or goat IgG control (C). Coimmunoprecipitation of PARP-14 was abolished by prior treatment of the lysate with ribonuclease-postitive (RNase+), showing that the association of PARP-14 with TTP was dependent on RNA.
Interaction of TTP and PARP-14 with TF mRNA is interdependent. (A) Western blots of proteins isolated with streptavidin beads from lysates of LPS-treated WT, Ttp−/−, and Parp14−/− macrophages (1 µg/mL, 2 hours) incubated with sense (+) or anti-sense (-) biotinylated murine TF 3′UTR. TF 3′UTR did not pull down TTP in the absence of PARP-14 or PARP-14 in the absence of TTP. (B) Western blots of proteins immunoprecipitated from LPS-stimulated WT lysates with anti-TTP antibody (T) or goat IgG control (C). Coimmunoprecipitation of PARP-14 was abolished by prior treatment of the lysate with ribonuclease-postitive (RNase+), showing that the association of PARP-14 with TTP was dependent on RNA.
PARP-14 does not regulate TNFα mRNA in vitro
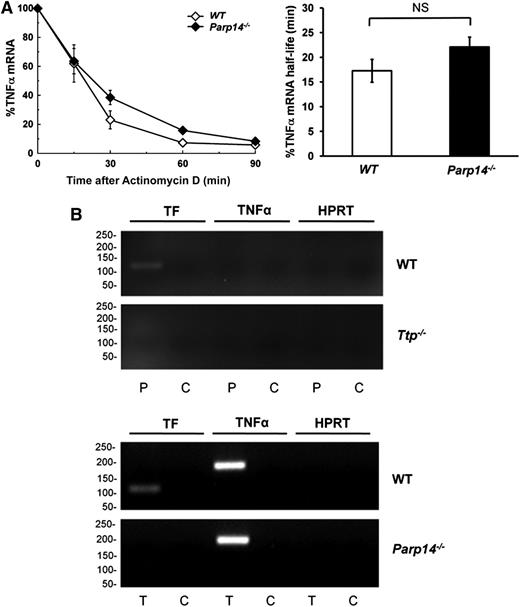
Next, we questioned whether PARP-14 coregulates TNFα mRNA, a well-established TTP target. We found that PARP-14–deficiency made no significant difference to TNFα mRNA or protein expression in BMDM, either when unstimulated or stimulated with LPS for varying durations (supplemental Figure 10). Moreover, there was no significant difference in TNFα mRNA decay curves between WT and Parp14−/− macrophages at 2 hours after LPS stimulation (WT t1/2 = 18 ± 3 minutes, Parp14−/− t1/2 = 23 ± 3 minutes; P = .281) (Figure 6A). At a molecular level, TNFα mRNA independence from PARP-14 was shown using a RIP assay, which showed that TNFα mRNA was pulled down from lysates of LPS-stimulated WT macrophage lysates by anti-TTP, but not by anti-PARP-14 antibodies (Figure 6B). Furthermore, TNFα, but not TF mRNA was immunoprecipitated by anti-TTP from Parp14−/− macrophage lysates. As expected, TF mRNA was pulled down by anti-PARP-14 from lysates of LPS-treated WT, but not Ttp−/− macrophages, and by anti-TTP from lysates of LPS-treated WT, but not Parp14−/− macrophages.
Regulation of TNFα mRNA is independent of PARP-14. (A) TNFα mRNA decay in WT and Parp14−/− BMDM after LPS-stimulation (1 μg/mL) for 2 hours (n = 6 experiments). TF mRNA half-lives are expressed as mean ± SEM, and analyzed using a 2-tailed Student t test. (B) RT-PCR of TF and TNFα mRNA immunoprecipitated by anti-PARP-14 (P) or anti-TTP (T) antibodies compared with rabbit IgG control (C) from lysates of LPS-stimulated (1 μg/mL for 2 hours) WT, Ttp−/− (left), or Parp14−/− (right) BMDM. HPRT acts as a negative control for nonspecific mRNA pull-down. The experiment shows that TNFα mRNA was immunoprecipitated from lysates of LPS-stimulated WT macrophage lysates by anti-TTP, but not by anti-PARP-14 antibodies. Also shown is that TNFα, but not TF mRNA was immunoprecipitated by anti-TTP from Parp14−/− macrophage lysates. TF mRNA was pulled down by anti-PARP-14 from lysates of LPS-treated WT, but not Ttp−/− macrophages, and by anti-TTP from lysates of LPS-treated WT, but not Parp14−/− macrophages. NS, nonsignificant.
Regulation of TNFα mRNA is independent of PARP-14. (A) TNFα mRNA decay in WT and Parp14−/− BMDM after LPS-stimulation (1 μg/mL) for 2 hours (n = 6 experiments). TF mRNA half-lives are expressed as mean ± SEM, and analyzed using a 2-tailed Student t test. (B) RT-PCR of TF and TNFα mRNA immunoprecipitated by anti-PARP-14 (P) or anti-TTP (T) antibodies compared with rabbit IgG control (C) from lysates of LPS-stimulated (1 μg/mL for 2 hours) WT, Ttp−/− (left), or Parp14−/− (right) BMDM. HPRT acts as a negative control for nonspecific mRNA pull-down. The experiment shows that TNFα mRNA was immunoprecipitated from lysates of LPS-stimulated WT macrophage lysates by anti-TTP, but not by anti-PARP-14 antibodies. Also shown is that TNFα, but not TF mRNA was immunoprecipitated by anti-TTP from Parp14−/− macrophage lysates. TF mRNA was pulled down by anti-PARP-14 from lysates of LPS-treated WT, but not Ttp−/− macrophages, and by anti-TTP from lysates of LPS-treated WT, but not Parp14−/− macrophages. NS, nonsignificant.
PARP-14 is required for TF mRNA destabilization by p38 MAPK
Phosphorylation of TTP via p38 MAPK signaling leads to its inactivation and hence stabilization of TTP target mRNA. Thus, inhibition of p38 MAPK accelerates the decay of TTP target mRNA. As expected, we found that the p38 MAPK inhibitors SB203580 and SB202190 destabilized TF mRNA in LPS-stimulated WT mouse macrophages (supplemental Figure 11A) and in human PBM (supplemental Figure 12). In contrast, the prolonged mRNA stability in Ttp-/- macrophages was resistant to inhibition of p38 MAPK, indicating the requirement for TTP (supplemental Figure 11B). Similarly, the increased TF mRNA stability seen in Parp14−/− macrophages was also resistant to p38 MAPK inhibition (supplemental Figure 13), an effect not due to the reduction in p38 MAPK phosphorylation (supplemental Figure 14). Taken together, these data are consistent with the regulation of TF mRNA stability by p38 MAPK requiring both TTP and PARP-14.
PARP-14 regulates TF, but not TNFα mRNA in vivo
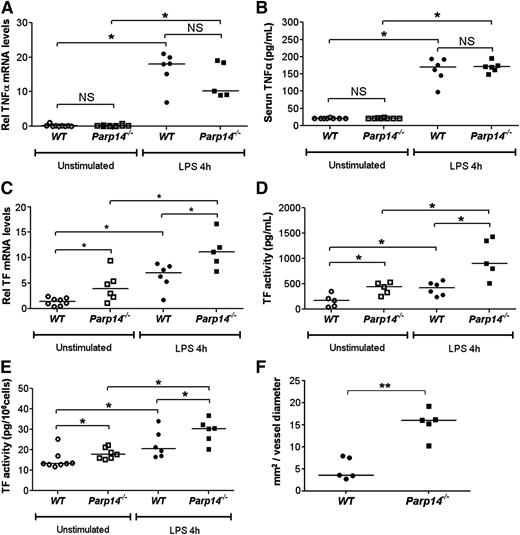
Investigation of the role of TTP on mRNA in Ttp−/− mice in vivo is confounded by inflammation attributable to delayed TNFα mRNA degradation and spontaneous release of TNFα 28. In contrast, Parp14−/− mice appear healthy, and there was no evidence of increased TNFα mRNA in lung extracts (Figure 7A), or of increased TNFα in serum (Figure 7B), either in unstimulated or LPS-stimulated Parp14−/− mice. Importantly, and as predicted by the in vitro experiments, TF mRNA was significantly increased in lung tissue (Figure 7C) and TF activity was significantly increased in the lung (Figure 7D) and circulating leukocytes (Figure 7E) of Parp14−/− mice under the same conditions. Finally, we established using intravital microscopy that thrombus formation after laser injury is significantly enhanced in cremaster muscle arterioles in Parp14−/− compared with WT mice (P < .01) (Figure 7F; supplemental Figure 15).
TF and thrombogenicity are upregulated in vivo in PARP-14–deficient mice, but TNFα expression is not affected. (A-E) WT and Parp14−/− mice were studied unstimulated or 4 hours after intraperitoneal injection of LPS (5 μg) (n = 6-8 mice per group). (A) TNFα and (C) TF mRNA in lung tissue were measured by quantitative RT-PCR. (B) Serum TNFα was measured by enzyme-linked immunosorbent assay. (D-E) TF activity was measured in lung tissue and peripheral blood leukocytes using a turbimetric clotting assay (n = 6-8 mice per group). (F) Comparison by intravital microscopy of thrombus formation in cremaster arterioles. Each point represents average maximal thrombus volume normalized to vessel diameter for each individual mouse (n = 5 mice per group). All bars are medians. *P < .05; **P < .01. h, hours; NS, not significant using a 2-tailed Mann-Whitney U test.
TF and thrombogenicity are upregulated in vivo in PARP-14–deficient mice, but TNFα expression is not affected. (A-E) WT and Parp14−/− mice were studied unstimulated or 4 hours after intraperitoneal injection of LPS (5 μg) (n = 6-8 mice per group). (A) TNFα and (C) TF mRNA in lung tissue were measured by quantitative RT-PCR. (B) Serum TNFα was measured by enzyme-linked immunosorbent assay. (D-E) TF activity was measured in lung tissue and peripheral blood leukocytes using a turbimetric clotting assay (n = 6-8 mice per group). (F) Comparison by intravital microscopy of thrombus formation in cremaster arterioles. Each point represents average maximal thrombus volume normalized to vessel diameter for each individual mouse (n = 5 mice per group). All bars are medians. *P < .05; **P < .01. h, hours; NS, not significant using a 2-tailed Mann-Whitney U test.
Discussion
In this paper, we describe a mechanism by which expression of TF, a key mediator of thrombosis and inflammation, is selectively regulated at a posttranscriptional level, and we provide evidence for the first time that PARP-14 is directly involved in the control of mRNA stability. The finding that PARP-14 is needed by TTP for the control of TF but not TNFα mRNA stability provides a new insight into how different TTP target mRNAs relevant to thrombosis and inflammation are differentially regulated.
We found that TF mRNA levels, TF protein expression detected by western blot analysis and TF activity were each enhanced in BMDM derived from Parp14−/− or Ttp−/−. It is important to stress that the different dynamics of TF mRNA and protein turnover, together with a variable fraction of TF protein being functionally competent, means that levels of the 3 variables are unlikely to correlate precisely, either basally or after LPS treatment. In support of our observations on mouse BMDM, we also found that TF mRNA was increased in LPS-stimulated human PBM after either PARP-14 or TTP siRNA knockdown, and TF activity was significantly increased ex vivo in blood leukocytes obtained from unstimulated Parp14−/− mice.
To our knowledge, TF has not been previously identified as a TTP target gene. We found that TF mRNA was more stable in Ttp−/− macrophages than in WT cells and was resistant to destabilization by p38 MAPK inhibitors. These observations strongly suggest that TTP is a critical RNA-binding protein regulating TF mRNA stability and is controlled in so doing by p38 MAPK. TTP can be phosphorylated on several sites, including Ser-52 and Ser-178, and this is thought to prevent binding of the CCR4-CAF1 deadenylase complex, and thereby stabilizes mRNA.35,36 Conversely, the phosphatase PP2A dephosphorylates TTP and renders it active.19 Consistent with our data, a recent study conducted in one of our laboratories (A.R.C.) has found that TF mRNA is underexpressed in macrophages from a constitutively active TTP knock-in mouse in which the 2 sites of inhibitory phosphorylation by MK2 are mutated to alanines (Tim Smallie et al, manuscript in preparation).
As with Ttp-/- macrophages, increased TF mRNA stability in LPS-stimulated Parp14−/− was not reversed by p38 MAPK inhibition. This is most readily explained by reduced TTP engagement with TF 3′UTR in the absence of PARP-14. Thus, by using RIP and in vitro transcribed RNA pull-down assays, we obtained data that suggest that PARP-14 and TTP require each other for binding optimally to a highly conserved ARE in the TF mRNA 3′UTR. It is possible that TTP-PARP-14 and RNA form a ternary complex, but further experiments using isolated PARP-14, TTP, and TF mRNA are needed to resolve this conclusively. A tentative model of the interaction of PARP-14-TTP and TF 3′ UTR is shown in a diagram in supplemental Figure 16. TTP is known to bind to AREs via its 2 central zinc zingers that coordinate Zn in a disc-like structure.37,38 Furthermore, PARP-14 may bind RNA via an RNA recognition motif at the amino terminus. Although our mutational analysis indicated that pull down of the 2 proteins was abolished by a 3-nucleotide substitution affecting both of the overlapping UUAUUUAAU nonamers in the conserved palindromic ARE, the precise binding sites of the individual proteins and their means of interacting with each other now requires further molecular analyses.
A previous study has suggested that PARP proteins, including PARP-14, localize in stress granules and may be involved in interactions with miR.27 The stability of TF mRNA is known to be regulated by miR 19a/19b and by miR201/20b/106b, acting at distinct binding sites proximal to the conserved ARE engaged by the interaction of PARP-14 and TTP.31-33 Using a luciferase mRNA stability reporter assay, we found that the 2 miR sites and the conserved ARE each contribute to reducing reporter activity in the assay, and have additive effects in combination. Clearly more work is now warranted to understand the molecular interactions between ARE-mediated and miR-mediated regulation of mRNA stability in this model, both under basal conditions and dynamically after LPS or cytokine activation.
TNFα mRNA is a canonical TTP target, and it was surprising to find that LPS-induced TNFα mRNA and protein expression were normal in Parp14−/− macrophages. The finding that TNFα transcripts were pulled down in a RIP assay from Parp14−/− cells by anti-TTP argues that PARP-14 is not required for TTP interactions with TNFα 3′UTR mRNA. It is possible that the direct interaction of TTP with TNFα mRNA is of higher affinity than that with TF mRNA, and thus sufficient to bind without an accessory protein. Alternatively, a different accessory protein(s) may be required for optimal TTP binding to TNFα. Further work will be directed at identifying whether there is a subset of TTP target genes coregulated by PARP-14, and if so, then whether these are functionally related.39
Mice deficient in TTP spontaneously develop a chronic inflammatory state due to dysregulated expression of TNFα.28 As Parp14−/− mice appear healthy, we examined their expression of TNFα and TF in vivo. The finding that Parp14−/− mice have normal TNFα mRNA expression in the lungs and normal TNFα levels in serum, either when unstimulated or after LPS treatment, provides an in vivo validation that TNFα mRNA is not a PARP-14 target. On the other hand, increased TF mRNA in the lungs, increased TF activity in the lungs and circulating leukocytes of Parp14−/− mice, and increased thrombus formation in vivo are all consistent with our in vitro data showing increased TF in the absence of PARP-14. Although we have not directly shown that the increased thrombus formation is due to increased TF expression, the laser-injury model is known to be TF-dependent.40 As the ex vivo and in vivo measurements were made at a tissue level, further work is needed to establish the differential roles of PARP-14 in TF expression in cells other than monocyte-macrophages, and indeed to determine whether there are differences between monocyte and macrophage subsets.
The question remains whether PARP-14 mono-ADP-ribose transferase activity is required for regulating TF mRNA activity. There are no specific chemical inhibitors of PARP-14 catalytic activity available, and results using nonselective PARP inhibitors are hard to interpret. Indeed, we have found that nonselective inhibitors (eg, PJ34 and 3-aminobenzamide) destabilize TF mRNA (but not TNFα mRNA) in a manner similar to that shown above with p38 inhibitors (data not shown). Further work is needed to establish which proteins in the system became ADP-ribosylated and whether PARP-14 or other PARP proteins are responsible. Resolution of these questions will await the development of specific PARP-14 inhibitors or genetically engineered knock-in mice with enzymatically inactive PARP-14.
Posttranscriptional controls on mRNA safeguard against inappropriate transcriptional leak, couple steady-state mRNA levels to transcription, and provide the means for accelerated mRNA decay to terminate gene expression.41 In the case of TF, the existence of posttranscriptional regulation had been indicated in vitro by a reduction in TF mRNA stability over time after cellular activation.12,13 Our study now provides a mechanism regulating TF posttranscriptionally in macrophages and reinforces the relevance of mRNA stability control for preventing inappropriate TF expression and excessive procoagulant activity, both in vitro and in vivo. Posttranscriptional control of TF should now be considered alongside transcriptional42-44 and posttranslational45-47 regulation as one of the critical levels at which expression of this crucial protein is regulated. Further studies are warranted to determine whether polymorphisms in the regulatory or coding sequences of PARP-14 and/or TTP are associated with variability in TF expression and disease. We are not aware of any functional polymorphisms, mutations, or alternatively-spliced isoforms of PARP-14 in either mouse or human, but polymorphisms in the promoter and coding regions of TTP in humans have been linked to TTP expression levels and may influence risk or severity of cancer and inflammatory disorders such as rheumatoid arthritis.48-50
In conclusion, our study establishes posttranscriptional regulation of TF mRNA as an important level of control of TF expression and raises the question as to what degree altered expression of TF in disease states relates to dysfunctional control of mRNA stability. We have discovered interactions between PARP-14, a new protein in posttranscriptional regulation, and TTP, an established protein regulator of mRNA stability, and presented evidence that PARP-14 allows the actions of TTP to be selectively regulated. Thus, although we have focused on the regulation of TF mRNA turnover, our findings have significantly wider implications for posttranscriptional regulatory biology in general and TTP-mediated mRNA decay in vascular inflammation in particular.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors gratefully acknowledge Deborah J. Stumpo (National Institute of Environmental Health Sciences) for help with the Ttp−/− mice. The work was funded by grants from the British Heart Foundation.
Authorship
Contribution: M.B.I., M.J., and D.O.H. designed research; M.B.I., M.J., J.C., Y.L., S.-C.Y., G.H., F.N.G., and K.J.W. performed research; M.B.I., M.J., J.C., Y.L., S.-C.Y., G.H., F.N.G., K.J.W., and D.O.H. analyzed data; F.P.M., S.H.C., and P.J.B. contributed critical mice; M.A.L., A.R.C., N.M., M.B., and J.L.D. advised on the design of experiments and interpretation of data; and M.B.I., M.J., D.O.H., M.A.L., A.R.C., N.M., M.B., and J.L.D. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dorian O. Haskard, Vascular Sciences Section, National Heart and Lung Institute, Imperial College, Du Cane Rd, Hammersmith Hospital, London, W12 ONN, UK; e-mail d.haskard@imperial.ac.uk.
References
Author notes
M.B.I. and M.J. contributed equally to this study.
J.L.D. and M.B. contributed equally to this study.


![Figure 4. The TF 3′UTR distal ARE regulates luciferase mRNA. Luciferase reporter constructs containing 3′ UTR inserts derived from the TF mRNA sequence were transfected into RAW 264.7 cells, and luciferase activity was then assayed after 16 hours. (A) Shows the effects on luciferase activity of full-length TF 3′ UTR compared with a panel of constructs with 7 guanine substitutions in the conserved ARE in the ARE4 segment (= ARE*), in the miR 19a/19b binding site (= miR-A*) or the miR 20a/20b/106b binding site (= miR-B*), alone or in combination. ARE mutation significantly increased luciferase activity, and this was augmented by the substitutions in miR-A and miR-B. P values refer to comparisons with unmutated 3′ UTR. (B) Shows the destabilizing effect of inserting the isolated ARE4 segment of the TF 3′UTR into the luciferase reporter construct, relative to the luciferase coding region with no 3′ UTR insert. Also shown is the significant rescue of luciferase activity by the 7 guanine substitution (as in ARE* in [A]). Data are mean + SEM of 4 experiments, analyzed by one-way analysis of variance. *P < .05; ***P < .001.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/24/10.1182_blood-2014-07-588046/4/m_3646f4.jpeg?Expires=1770242287&Signature=qtOsXYg5vi0xx1lBkDltZioPJcu4ANCbGGKi6yXn7HnRiwFhLX-NM4weEt2KpAc0-tJhwSWAj2uaUwY2dLdm4gIxWXDUk4ydWxM~xn-zKzawVTpR4Z7BUH5d5u4nL9J094QajlaXf34M5-t2Iwt92uwKGgKEiqWY9jRC7XmiTN1qpPsDmgOD3GnG4uZbgGfh1EOhkrNlq5v78l7UwfnZde1khyksHy9as3PdIquEEy-L4fCQkOjbcmcoBvHBcKV4u7tRghPQJX0pbt7pDBUDWn65WhoC9Yxe-aAKApBVVT679s~xfHDXFTduR2Z~YZqvTInjVEYztuVnr2nmhRws9w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal